Introduction
Neonatal candidemia is a significant cause of morbidity and mortality [
1]. Different species of
Candida cause early and late-onset neonatal sepsis. In the present era, non-
albicans Candida species, including
C. tropicalis,
C. glabrata,
C. parapsilosis,
C. krusei, result in two-thirds of the candidemia cases [
2,
3]. However, rare and uncommon
Candida species causing neonatal candidemia have been reported in the literature [
3]. These rare species can be identified readily and accurately by automated systems such as VITEK2, MALDI-TOF.
Candidemia is common in neonates with particular risk factors such as very low birth weight, low gestational age, maternal vaginal colonization, use of antibiotics and instrumentation during parturition [
4]. In addition, poor immune system and immunological memory make the neonates highly vulnerable to infections [
5]. Neonatal candidemia cases are usually managed by intravenous administration of fluconazole. However, the emergence of fluconazole-resistant
Candida species causing neonatal sepsis results in treatment failure [
6]. Therefore, we did a prospective study to determine the predisposing risk factors for neonatal candidemia with its causative etiology, time to positivity of blood culture, and antifungal minimum inhibitory concentration (MIC) against the isolated fungal pathogens at a tertiary care center in North India.
Methods
This prospective study was approved by the Institutional Ethical Committee, (Dean/2021/EC/2462 dated 15 Feb 2021). We obtained written and informed consent from the parents of the admitted neonates who were suspected cases of sepsis. The neonates had lethargy, poor feeding, irritability, fever, hypothermia, and respiratory distress at the time of admission in a tertiary care center in North India which covers the population of eastern Uttar Pradesh and western Bihar. We did the prospective study for a duration of two years from January 2021 to December 2022. We categorized the suspected neonatal sepsis event into early and late-onset sepsis. The events occurring within the 72 hours of life were considered as early onset neonatal sepsis (EoNS), whereas late-onset neonatal sepsis events (LoNS) occurred after 72 hours of life.
Here, we initially collected blood at the time of admission in all neonates. Moreover, we also did a blood culture later on as per days of suspected sepsis. For blood culture, we directly inoculated the aseptically collected blood into BacT/ALERT blood culture bottles and incubated it in the automatic blood culture monitoring system. Once there was a sign of culture positivity by detecting the fluorescence, the flagged positive blood culture broth was immediately processed for Gram stain and subculture on blood, chocolate and Sabouraud’s dextrose agar. The Gram’s stain report of the flagged positive blood culture was immediately conveyed to the neonatologist telephonically. In addition, we categorized the fungal pathogens according to their time to positivity of flagged positive blood culture. We isolated the strains and stocked them in glycerol broth at -20°C. One stock was sent to the Mycology Reference Centre, PGIMER Chandigarh, India for MALDI-TOF based identification.
Initially, we identified the causative fungal pathogens by standard phenotypic methods, using germ tube test, growth at 42°C, sugar fermentation and assimilation test. Subsequently, we performed MALDI-TOF-based identification of all
Candida strains by the on-plate formic acid extraction method. Here, we emulsified a loopful of fresh-grown yeast isolate in 1 mL of sterile double-distilled water and vortexed vigorously. Subsequently, we centrifuged the suspension at 13,000 rpm for 2 min and collected the pellet. We repeated the step twice and re-suspended the pellet in 50 mL sterile double distilled water. Then, we spotted one microliter of the suspension on the MALDI target plate and airdried. We overlaid the spot with 0.5 L of 98% formic acid and left it for air drying. Finally, we added 0.8 L of matrix solution (50 L acetonitrile, 2.5 L trifluoroacetic acid, 1 mg cyano-4-hydroxycinnamic acid and 47.5 L sterile double distilled water) on each spot and air dried. We performed MALDI-TOF MS analysis on MALDI Microflex LT mass spectrometer (Bruker Daltonik GmbH, Germany) and analyzed the results based on the score values recommended by the manufacturer. Here, values ≥2 log score for species identification, 1.7–1.99 for genus identification, and <1.7 as unreliable were considered [
7].
We also determined fluconazole, itraconazole and amphotericin B MIC against the isolated yeast by broth microdilution method using CLSI M27-A3 guidelines [
8]. Briefly, the yeast was subcultured over potato dextrose agar (PDA) and kept at 35°C for 3-5 days. Grown yeast colonies over PDA were dissolved in normal saline to make 0.5 McFarland having 0.5×10
6 cells/mL, which were subsequently diluted 1:100 times followed by 1:20 dilution in RPMI 1640 to achieve 5×10
2 2.5×10
3 cells/mL [
8].
The antifungal powders were procured from Sigma Pharmaceutics, India. The powders were dissolved in DMSO to have a concentration of 1024 µg/mL, which were subsequently diluted in RPMI 1640 to have 128 µg/mL for fluconazole and 32 µg/mL for itraconazole and amphotericin B. Flat bottom microtitre plates were used to determine the MIC where 100 µL of the prepared yeast suspension were added in the well 100 µL of drug in two-fold dilution. Subsequently, the plates were incubated at 35°C overnight to determine the MIC. For amphotericin B, the drug concentration resulting in no visible growth was taken as MIC, whereas for fluconazole and itraconazole, it was the concentration that resulted in prominent growth reduction [
8]. We calculated the range of MIC with MIC
50 and MIC
90 against each isolated yeast-like fungal pathogen.
All data were checked for normal distribution using Kolmogorov–Smirnov test for normality and Bartlett’s test for homogeneity of variances, followed by Chi-square (χ2) analysis. All statistical analyses were performed using the statistical software MINITAB 18 (Minitab Inc, USA).
Results
During the study period from July 2020 to March 2022, 333 culture-positive neonatal sepsis events occurred. Among these, 32.1% (107/333) of the events were caused by yeast-like pathogens. Among 107 candidemia events, 48 (44.9%) were EoNS, whereas 59 (55.1%) were LoNS events, which did not differ significantly (χ
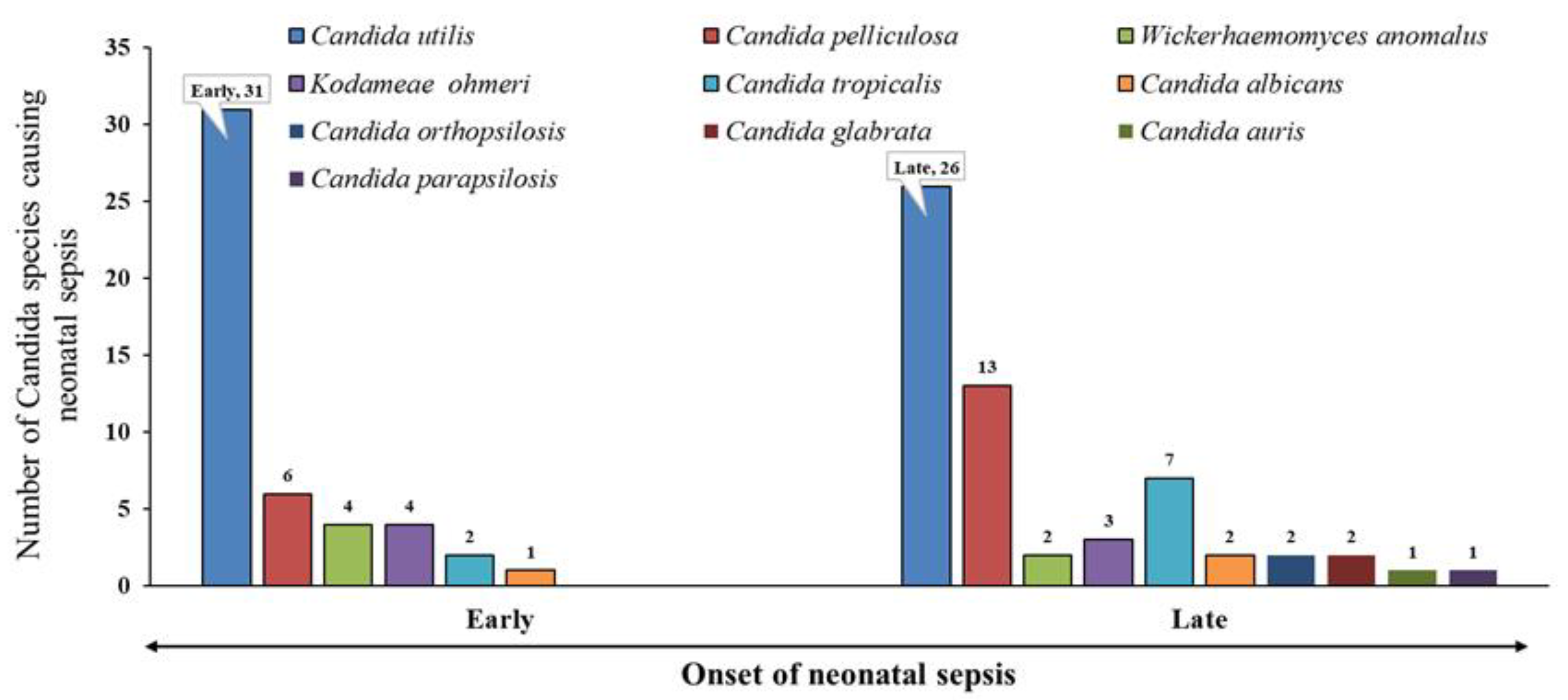
2=1.00; p=0.317; df=1). A range of yeast-like fungal pathogens caused both events; however,
Candida utilis was the predominant one (
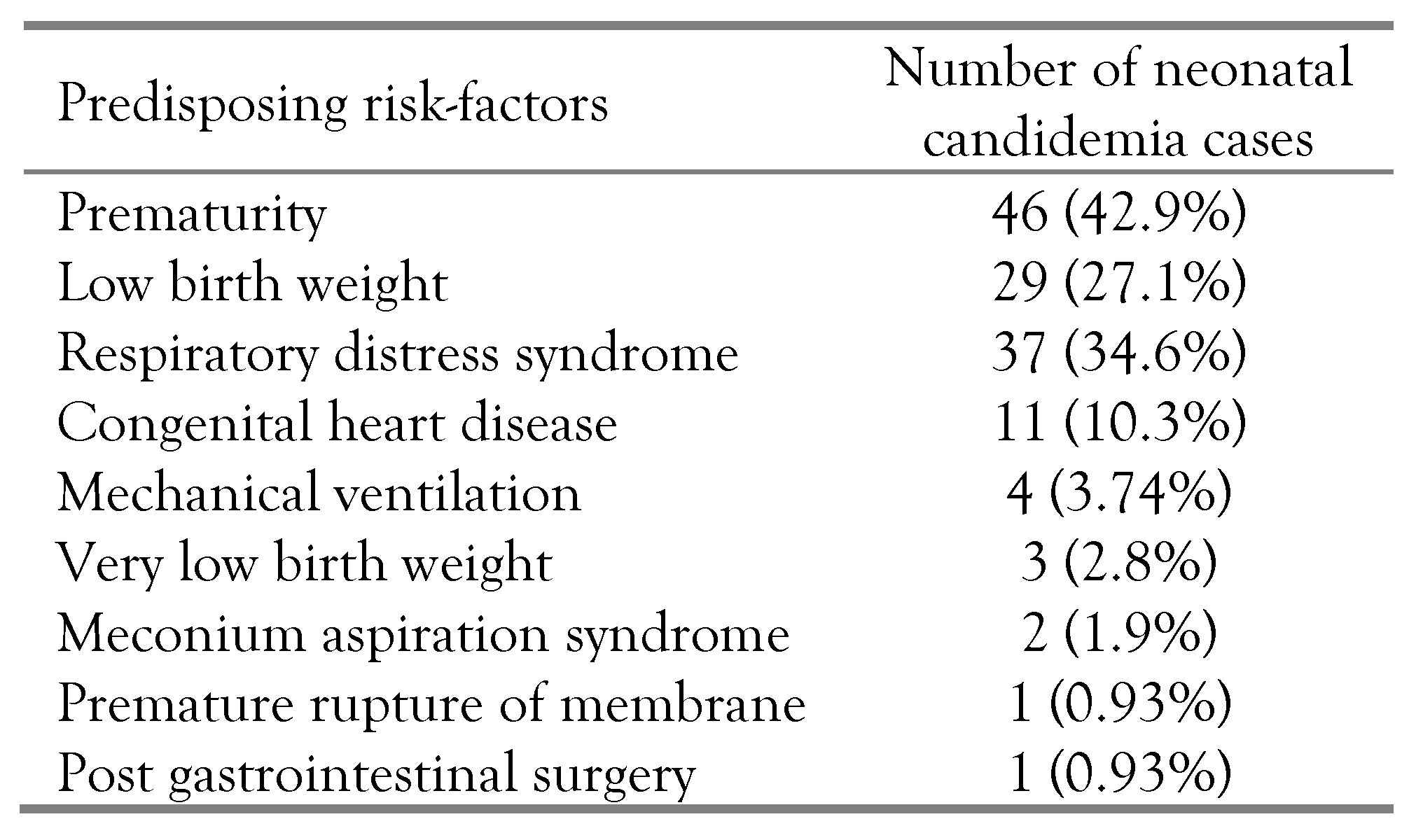
Figure 1). We retrospectively analyzed the predisposing risk factors for neonatal candidemia and summarized them in
Table 1. Prematurity was the most common predisposing risk factor in 43% (n=46) of the enrolled neonates having candidemia, followed by low birth weight (27%, n=29). Only three very low birth weight babies (<1500 grams) had candidemia. However, premature rupture of membranes and post-gastrointestinal surgery were the least common predisposing risk factors, being present in 0.93% (n=1) of the enrolled neonates having candidemia. The variations amongst the predisposing risk factors were statistically significant (χ
2=196.54; p<0.0001; df=10).
In the present study, 97.2% (n=104) of the yeast-like isolates were non-albicans Candida. These strains were characterized at the Mycology Reference Centre, PGIMER Chandigarh, India and identified Candida utilis (n=57, 53.3%) as the predominant pathogen, followed by Candida pelliculosa (n=19, 17.7%), Candida tropicalis (n=9, 8.4%), Kodamaea ohmeri (n=7, 6.5%), Wickerhamomyces anomalus (n=6, 5.6%). Moreover, there were only three isolates of Candida albicans (n=3, 2.8%). The variation in the abundance of the pathogens was statistically significant (χ2=110.84; p<0.0001; df=5).
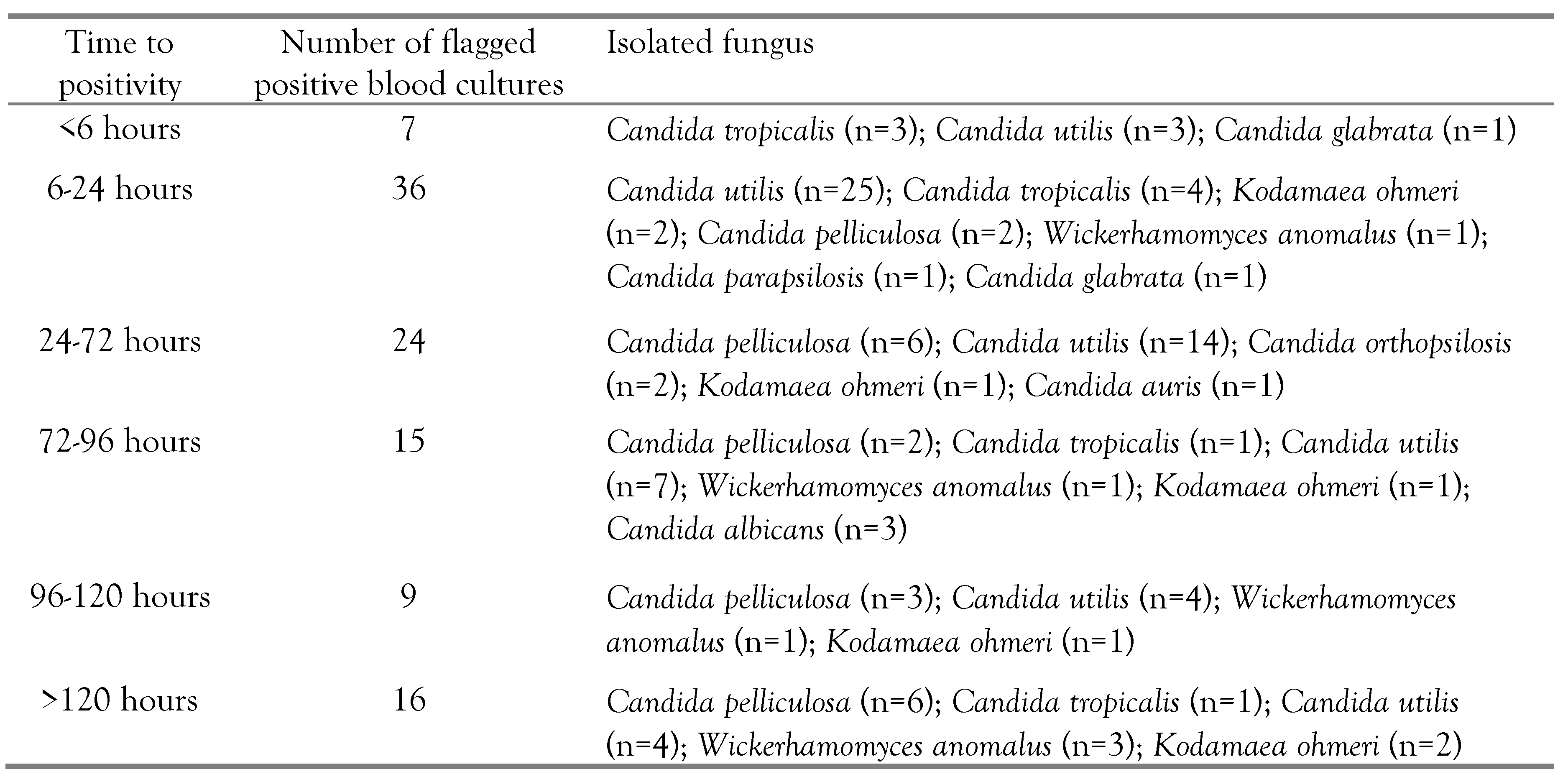
Neonatal candidemia is diagnosed by blood culture, where blood culture bottles are kept in an automatic blood culture monitoring system. We have tabulated the time to positivity for different
Candida species, isolated from the blood of the neonates having sepsis (
Table 2). We noted 62.6% (n=67) of the blood culture positivity within 72 hours of incubation. Among these 67 positive blood cultures, 43 flagged positive within 6-24 hours of incubation. BacT/ALERT blood culture bottles are usually incubated for five days. However, we incubated the bottles for seven days and got sixteen more isolates of yeast-like pathogens, which might have been missed if the bottles had been discarded early.
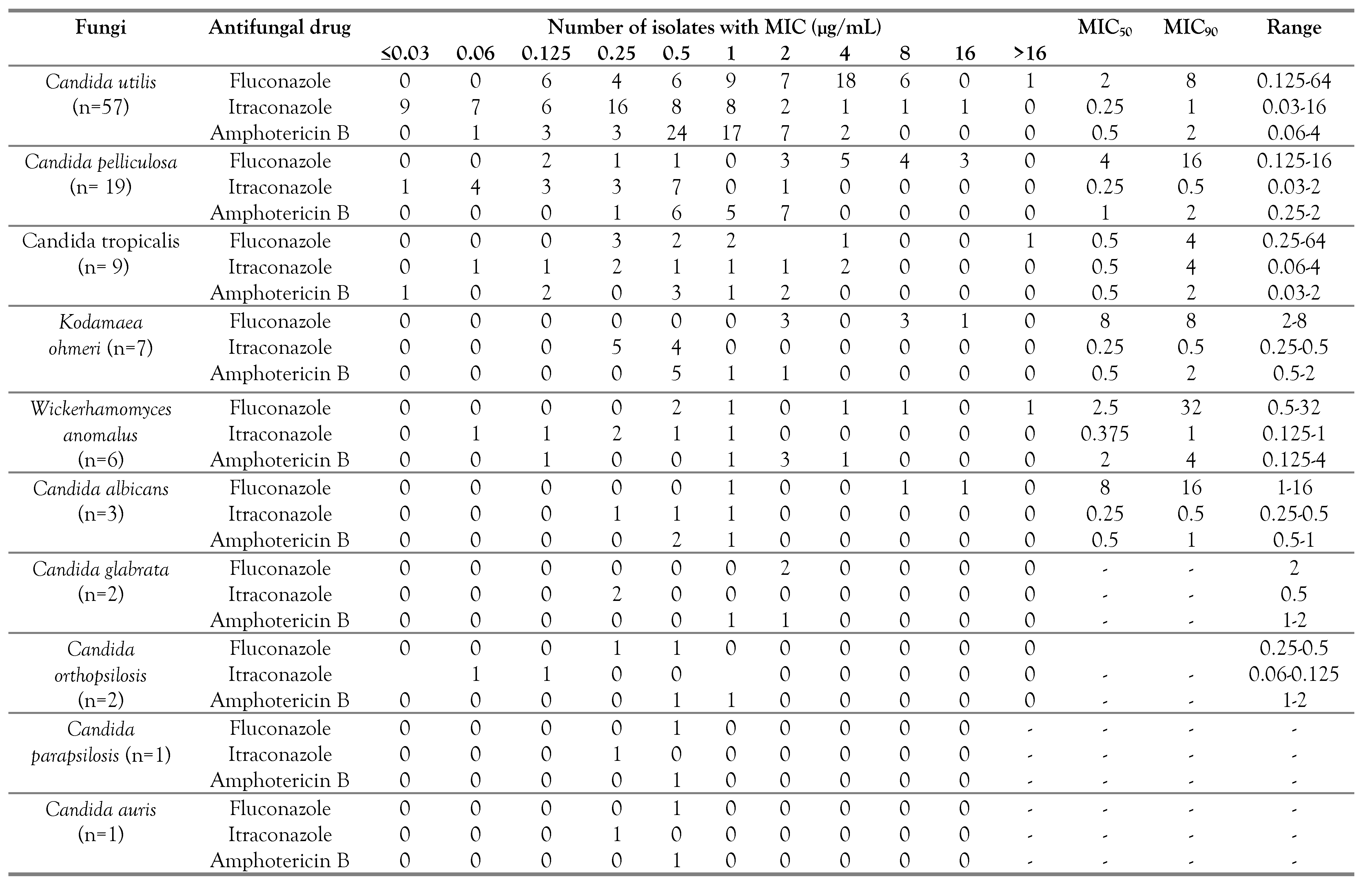
The antifungal MIC ranges, with MIC
50 and MIC
90 obtained upon antifungal susceptibility testing against the isolated
Candida species, have been summarized in
Table 3. Among the tested antifungals, fluconazole had higher MIC than itraconazole and amphotericin B. Overall, 90% of the isolated strains of
Candida species had fluconazole MIC of ≤8 µg/mL. However, each strain of
C. utilis,
C. tropicalis, and
W. anomalus had ≥16 µg/mL fluconazole MIC, which is difficult to achieve in serum at normal doses. In addition, five strains of
C. utilis had >1 µg/mL itraconazole MIC. We have also categorized the isolated
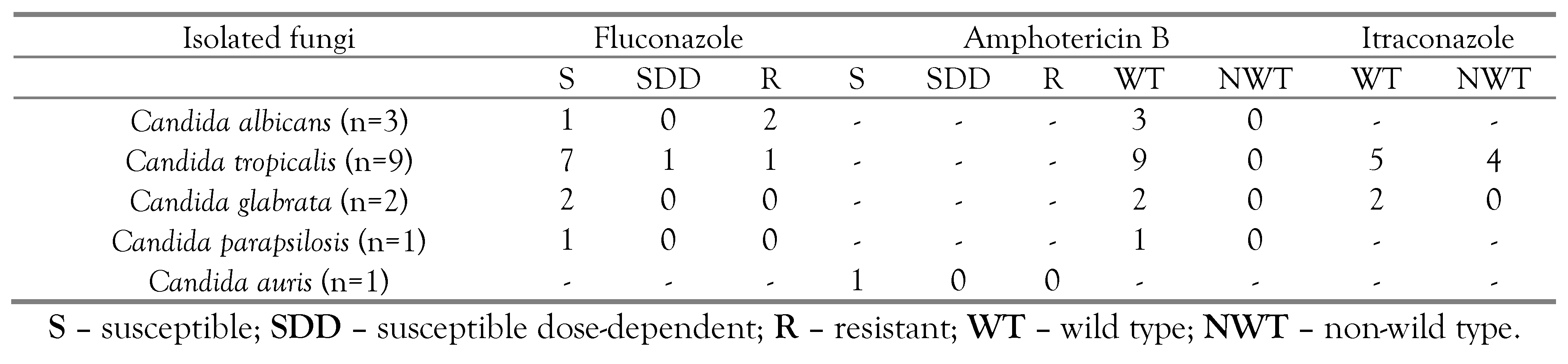
Candida species into susceptible, resistant, wild type and non-wild type strains in
Table 4.
Amphotericin B, a fungicidal drug, is also used in the management of fluconazole-resistant neonatal candidiasis cases. Amphotericin B had varied MIC against the isolated yeast-like pathogens. Most of the isolated strains had <1 µg/mL amphotericin B MIC. However, 9 strains of C. utilis, 7 strains of C. pelliculosa, 4 strains of W. anomalus and 2 strains of C. tropicalis had >1 µL amphotericin B MIC. Each isolated strain of C. parapsilosis and C. auris had lower fluconazole, itraconazole and amphotericin B MIC (<0.5 µg/mL).
Discussion
Candidemia is the fourth most common cause of sepsis, with high morbidity and mortality [
9]. Very low birth weight and preterm babies are highly prone to candidiasis. Moreover, premature rupture of membranes, prolonged labor, instrumentation during parturition, antibiotic use, maternal vaginal colonization by
Candida, and prolonged catheter in situ are the other predisposing risk factors for neonatal candidiasis. The clinical manifestations of neonatal candidemia are usually non-specific and undifferential, however, failure to feed and hypo/hyperthermia are the usual clinical manifestations.
The fungal pathogens causing neonatal sepsis are diagnosed by blood culture where fungal growth in the blood culture system is indicated by the fluorescence indicator present at the base of BacT/ALERT blood culture bottles [
10]. Here, the fungal load in the blood culture bottle and the growth rate determine the time to positivity in the blood culture system. We noted that twothirds of the blood cultures flagged positive within 72 hours of incubation. Routinely, BacT/ALERT automatic blood culture bottles are incubated for a duration of 5 days. However, on prolonged incubation for 7 days, we isolated 16 more yeast-like pathogens. Fungi are slow-growing microbes that produce CO
2 in little amounts. Therefore, prolonged continuous incubation should be done in cases of suspected candidemia.
The spectrum of fungi causing neonatal candidemia varies from region to region. Previously,
Candida albicans was the most common cause of neonatal candidemia. However, in the recent era, the emergence of non-
albicans Candida has been reported around the globe. Among these,
C. tropicalis, C
. glabrata,
C. parapsilosis and
C. krusei account for nearly two-third of candidemia events. However, rare
Candida species like
C. auris,
Wickerhamomyces anomalus,
Kodamaea ohmeri,
C. pelliculosa and
C. orthopsilosis have been reported to cause neonatal sepsis [
9,
11]. This altered spectrum and upsurgence of newer/rare
Candida species has been attributed to the availability of sophisticated diagnostic facilities like VITEK-2 and MALDI-TOF. In the present study, we noted the emergence of
Candida utilis as the most common fungus causing neonatal sepsis. Moreover, other rarely reported fungi like
C. pelliculosa, W. anomalus,
Kodamaea ohmeri were also isolated. In the literature,
Candida utilis causing sepsis has been reported around the globe [
12,
13].
C. utilis is an edible yeast, whose telemorphic stage,
Cyberlindnera jadinii, is being used as yeast additive in the food industry [
14]. Therefore, in each case of suspected neonatal candidemia, a blood culture should be done.
Neonatal candidemia cases are treated with azoles and polyenes groups of antifungal agents. Azoles primarily inhibit the 14α-lanosterol demethylase enzyme, resulting in the accumulation of 14α-methylated sterols in the membranes, leading to fungal cell death [
15]. Polyenes antifungals form aqueous pores in the fungal cell wall, resulting in the leakage of intracellular components, ultimately resulting in cell lysis. Among azoles, fluconazole is widely used in the management of neonatal candidemia due to its
fAUC
0–24/MIC of at least 100, easy availability, low cost, and prolonged postantifungal effect [
16]. We noted that most of the isolated
Candida species had <4 µg/mL fluconazole MIC except 7 strains each for
C. utilis and
C. pelliculosa, 4 strains of
K. ohmeri, 2 strains each of
W. anomalus and
C. albicans and 1 strain of
C. tropicalis. In the literature, fluconazole higher MIC/resistant strains have been reported. Fluconazole is mostly ineffective against
Candida glabrata,
C. krusei, and
C. auris strains. Thus
, amphotericin B can be used in such cases. Most of the isolated yeast-like pathogens had <1 µg/mL amphotericin B MIC except for 9 strains of
C. utilis, 7 strains of
C. pelliculosa, 4 strains of
W. anomalus, 2 strains of
C. tropicalis, and 1 strain of
K. ohmeri. In the literature,
W. anomalus,
K. ohmeri strains with >8 µg/mL fluconazole MIC and >1 µg/mL amphotericin B MIC have been reported [
17,
18,
19].
Antifungal MIC results against the isolated
Candida species observed in the present study are in accordance with other published studies. Borman et al. have reported >10% fluconazole resistant
C. jadinii and 30% of strains of
C. jadinii having higher fluconazole MIC (4 µg/mL) [
20]. In addition, the emergence of
C. tropicalis strains with higher fluconazole MIC has been reported in the literature, attributed to the over-expression of the efflux pump and mutation in the
erg11 gene [
21].
In the published literature,
Candida tropicalis,
C. glabrata,
C. krusei and
C. albicans have been reported as common causes of neonatal candidemia. However, we identified only 3 strains of
C. albicans and 2 strains of
C. glabrata. Although most of the
C. albicans strains in literature are sensitive to commonly used antifungals. However, we identified 2 strains of
C. albicans with ≥8 µg/mL fluconazole MIC. This variation in MIC may be explained by the frequent use of fluconazole. Another 2 isolated
C. glabrata strains had 2 µg/mL fluconazole MIC. However,
C. glabrata strains are considered to be intrinsically fluconazole-resistant [
22,
23]. We also found 1 strain of
C. auris, which is deemed a superbug as the species is usually resistant to available antifungals [
24,
25]. However, the isolated
C. auris strain had 0.5, 0.25 and 0.5 µg/mL fluconazole, itraconazole and amphotericin B MIC, respectively.
We noted the emergence of rare fungi species causing neonatal candidemia. Moreover, a varied antifungal MIC against the isolated Candida strains was observed. Therefore, in each suspected case of neonatal candidemia, blood culture and causative pathogen isolation and antifungal MIC determination should be done. However, we didn’t search the source of Candida utilis causing neonatal candidemia. Despite this, the study will enrich the available data regarding the epidemiology of rare yeast-like pathogens causing neonatal candidemia with their antifungal MIC.