Abstract
Introduction: Integrons are genetic systems that may confer antibiotic resistance to Pseudomonas aeruginosa. Biofilm formation can facilitate gene exchange and can accelerate the development of antibiotic resistance. The aim of this work was to assess the distribution of resistance integrons including class 1, 2 and 3 among biofilmand non-biofilm producing clinical strains of P. aeruginosa. We also aimed to investigate the relationship between the existence of these integrons and the isolates’ resistance patterns. Methods: Specimens were obtained from patients showing evidence of infection. P. aeruginosa isolates were identified using conventional techniques, while disk diffusion test was used to detect their antimicrobial susceptibilities. Biofilm formation was detected by the tissue culture plate technique, while classes of integrons were detected by polymerase chain reaction. Results: Out of 106 P. aeruginosa isolates, 55.7% were class 1 integron-positive while 19.8% were class 2 integron-positive. However, class 3 integrons were not detected. Significant associations were found between class 1 integrons and resistance toward amikacin, gentamicin, cefepime, ceftazidime and ciprofloxacin. Class 2 integrons were associated with amikacin, ceftazidime and cefepime resistance. Of pseudomonal isolates, 61.3% were biofilm producing. Biofilm production was associated significantly with the existence of class 1 integrons (p<0.001) and class 2 integrons (p=0.039). Conclusions: About two thirds of isolated strains harbored resistance integrons, which emphasized their significance in our locality. The frequencies of class 1 and 2 integrons were significantly higher among biofilm forming isolates. Ongoing surveillance and infection control strategies are necessary to limit spread of integrons.
Introduction
Pseudomonas aeruginosa is a common opportunistic nosocomial pathogen [1,2]. This organism develops resistance to antibiotics through alterations of antibiotic permeability, production of bacterial enzymes, formation of biofilms and usa of efflux pumps [3]. The extensive use of antimicrobials is the most probable primary cause of escalation in antibiotic resistance among P. aeruginosa strains, which is a growing problem globally [1]. In addition, the spread of resistance is caused by the transfer of genetic components such as integrons harboring resistance genes [1,3].
Integrons are bacterial genetic systems capable of acquiring gene cassettes, stockpiling and expressing them using an internal promoter [4,5]. An integron consists of a dsDNA segment that is characterized by the presence of 3 essential elements, Pc promoter, intI gene and attI recombination site [6]. The intI gene encodes for the integrase enzyme that mediates recombination between the integron attachment site (attI) and the gene cassette recombination site (attC) [5]. The Pc promoter is needed for the expression of gene cassettes [6].
Gene cassettes are small genetic components which present in a circular form when they are free. Each gene cassette contains a single promoter-less gene besides a recombination site (attC) that enables its capture by integrons [7]. Through site-specific recombination, the integrons can obtain numerous gene cassettes, which may harbor determinants responsible for antibiotic resistance [6].
Mobile and chromosomal integrons are the two main types of integrons [6]. Chromosomal integrons are located in the chromosomes of marine bacteria such as Vibrio species and are usually not related to resistance. On the other hand, mobile integrons are present on mobile elements such as plasmids and transposons. Mobile integrons carry mostly gene cassettes responsible for antibiotic resistance and therefore, they are alternatively known as resistance integrons [6]. Based on sequence variations in intI gene, the resistance integrons are categorized into different classes [1]. Class 1 integrons are the most common among clinical strains and they can harbor several gene cassettes causing resistance. Class 2 integrons were linked to mobile transposon Tn7; however, they are not as common as class 1 integrons [1]. In 1995, class 3 integrons were initially identified in a Japanese Serratia marcescens isolate and they were reported to carry the metallo-β-lactamase blaIMP gene [8]. This was one of the few reports of class 3 integrons so far [1].
Bacteria can either live freely or as part of surface-adherent communities known as biofilms. A biofilm is a special structure that consists of organized groups of bacterial cells enclosed within an extracellular matrix [9]. Biofilm aids bacterial adherence and enables bacterial cells to withstand harsh external conditions and to evade the host defenses. In addition, biofilms shield the bacteria from antibiotics and facilitate the exchange of genetic resistance determinants. Therefore, biofilms perform a crucial role in the development of resistance within different bacteria, including P. aeruginosa [10,11].
The aim of this work was to assess the distribution of resistance integrons including class 1, 2 and 3 among biofilmand non-biofilm producing clinical strains of P. aeruginosa. We also aimed to investigate the relationship between the existence of these integrons and the isolates’ resistance patterns.
Methods
Selection of patients and collection of samples
A cross-sectional study was conducted during the period from December 2022 to September 2023. We included inpatients showing a clinical picture suggestive of infection from different clinical departments of Mansoura University Hospitals, Mansoura, Egypt. Different samples, according to the infection site, were aseptically collected from the study candidates, including; burn wounds, endotracheal aspirates, urine, blood and surgical wounds. All specimens were transferred immediately to the Medical Microbiology and Immunology Department for further processing.
Identification of Pseudomonas aeruginosa isolates
Standard bacteriological techniques were used to process the collected samples. Inoculated plates of MacConkey, blood and cetrimide agar media (Oxoid, UK) were incubated for 24 hours at a temperature of 37°C. Strains of P. aeruginosa were identified by conventional techniques including Gram-stained films, colonial morphology, growth at 42°C, production of pigment, different biochemical reactions such as oxidase test, citrate test, catalase test and triple sugar iron test.
Antimicrobial susceptibility testing
Antimicrobial susceptibilities of P. aeruginosa strains were identified through disk diffusion test according to the Clinical and Laboratory Standard Institute (CLSI) guidelines [12]. In order to carry out the disk diffusion test, MuellerHinton agar (MHA) (Oxoid, UK) and multiple antibiotic disks (Oxoid, UK) were used; cefepime (30 μg), gentamicin (10 μg), ceftazidime (30 μg), imipenem (10 μg), piperacillin (100 μg), aztreonam (30 μg), amikacin (30 μg), ciprofloxacin (5 μg), tobramycin (10 μg) and ticarcillin (75 μg). The inhibition zone diameter for each mentioned disk was measured and then interpreted, following the CLSI standards [12]. According to CLSI recommendations, P. aeruginosa ATCC 27853 was used for quality control [12]. Multidrug resistance (MDR) was defined when pseudomonal isolates exhibited resistance to at least one agent in three or more classes of antibiotics [1,13,14]. All pseudomonal strains were kept in tryptic soy broth (TSB), after supplementation with glycerol 15%, then stored at a temperature of -20°C for future investigations [15].
Detection of biofilm formation
The tissue culture plate technique was executed to detect biofilm production among P. aeruginosa isolates [11,16,17]. A freshly cultured loopful of P. aeruginosa isolate was inoculated in TSB (10 mL) that contained 1% glucose and the broth was then incubated at a temperature of 37°C for 24 hours. Then, every bacterial suspension was adjusted to match the turbidity of 0.5 McFarland prior to further 1:100 dilution by fresh medium. By using sterile tissue culture plates, 200 μL of the diluted bacterial suspensions were placed in each well. Also, both Staphylococcus epidermidis ATCC 31484 (positive control) and ATCC 12228 (negative control) were tested. A sterile broth was included to guarantee sterility as well as to detect non-specific binding.
The plate was incubated for 24 hours at a temperature of 37 °C, lightly tapped, and then washed with two hundred microliters of phosphate-buffered saline. Such washing process was carried out four times to guarantee that all free bacteria were removed. On the other hand, the biofilms formed by the adherent bacteria were fixed by adding 2% sodium acetate to the wells for 30 minutes. After the biofilms were fixed, crystal violet (0.1%) was applied for 30 minutes to stain these biofilms. Afterward, deionized water was added to wash away any remaining stains. Following air drying of the plate, 150 μL of 95% ethanol were added into every well in order to resolubilize the fixed color. The plate was then covered to reduce evaporation and left at room temperature for 30 minutes.
Finally, the measurement of optical densities (ODs) of colored biofilms was carried out by a micro-ELISA reader (570 nm wavelength). These OD values reflect the isolates’ adhesion to the wells and, hence, biofilm production. This test was carried out in triplicate and the average of the OD readings was determined. As previously described, isolates with OD values <0.120 were identified as non-biofilm producers while isolates with OD values ≥0.120 were considered as biofilm producers [11,16,17].
Detection of resistance integrons class 1, 2 and 3
The genomic DNA was extracted from the isolates using the boiling method [18]. Following overnight culture, three to five colonies of each P. aeruginosa strain were suspended into 1 mL of sterile water. The resulting suspensions were kept at a temperature of 100°C for 10 minutes in water bath. Then, centrifugation was performed for 10 minutes at 13,000 rpm. The resulting supernatants, containing genomic DNA, were placed into sterile Eppendorf tubes and kept frozen at -20°C in order to carry out the following polymerase chain reaction (PCR). Primers targeting intI1, intI2 and intI3 genes were used to detect class 1, 2 and 3 resistance integrons, respectively [1]. These primer sets included: F5′GGTGTGGCGGGCTTCGTG3′; R-5′GCATCCTCGGTTTTCTGG3′ for intI1 gene (480 bp), F-5′ CTAGAATAGGCTGTATAGGCAGA3′; R5′GAGTGACGAAATGTATGACAAG3′ for intI2 gene (850 bp) and F5′CAGTCTTTCCTCAAACAAGTG3′; R-5′ TACATCCTACAGACCGAGAAA3′ for intI3 gene (702 bp) [1].
A final volume of 25 μL, containing the template DNA, Taq PCR Master Mix (QIAGEN, UK) and the gene primers, was used for PCR. The PCR assay for intI1 gene included an initial denaturation cycle at 94°C for 5 minutes, 35 cycles of denaturation (1 min at 94°C), annealing (1 min at 50°C) and extension (1 min at 72°C), with a final extension cycle for 5 min at 72°C. For amplification of intI2 and intI3 genes, similar conditions were used apart from the annealing temperature, which was 47°C for intI2 gene and 52°C for intI3 gene [1]. Distilled sterile water was used as a negative control in all PCR reactions, while identified intI1 gene and intI2 gene positive strains from a previous study were used as positive controls [19]. The resulting amplification products were visualized by utilizing agarose gel electrophoresis and 1 kb DNA ladder marker (Enzynomics, South Korea) which consist of DNA fragments with known sizes. Therefore, the size of each amplification product was determined by comparing it against the marker’s fragments (Figure 1). In addition, PCR assays were repeated twice in order to confirm the results.
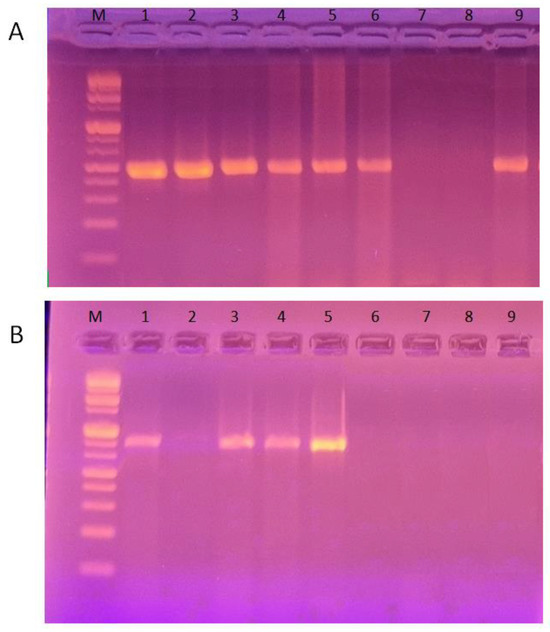
Figure 1.
Detection of class 1 and 2 resistance integrons among P. aeruginosa isolates by PCR. (A) PCR amplification of intI1 gene yielding DNA fragments of 480 bp. (B) PCR amplification of intI2 gene to produce DNA fragments of 850 bp. Lane M shows the DNA molecular marker (1 kb DNA ladder).
Statistical analysis
The SPSS program (v 22.0, IBM Corp, USA) was used for data processing. Descriptive data are displayed in the form of frequency (numberpercent) or mean and standard deviation. The Chi-squared test was executed to compare results when categorical variables were evaluated. When the p value was <0.05, statistical significance was reported.
Ethics statement
The institutional research board of the Faculty of Medicine, Mansoura University approved the research protocol (R.22.11.1967). Following a thorough explanation, an informed written consent was gathered from every participant in the study. Written informed consents were acquired from the legal guardians of unconscious patients.
Results
A total of 106 P. aeruginosa isolates were identified in our study. These strains were recovered from various clinical samples including; burn wounds (35, 33.0%), endotracheal aspirates (27, 25.5%), urine (23, 21.7%), blood (13, 12.3%) and surgical wounds (8, 7.5%). Of the study subjects, 55 (51.9%) were females and 51 (48.1%) were males with a mean age of 42±19 years.
The identified P. aeruginosa isolates showed the highest resistance against ceftazidime (68, 64.2%) and gentamicin (66, 62.3%) followed by cefepime (64, 60.4%), imipenem (60, 56.6%), ticarcillin (56, 52.8%), tobramycin (55, 51.9%), amikacin (54, 50.9%), piperacillin (54, 50.9%), aztreonam (53, 50.0%) and ciprofloxacin, which had the lowest resistance rate among pseudomonal isolates (51, 48.1%). Seventy-three P. aeruginosa strains exhibited MDR patterns with a percentage of 68.9%.
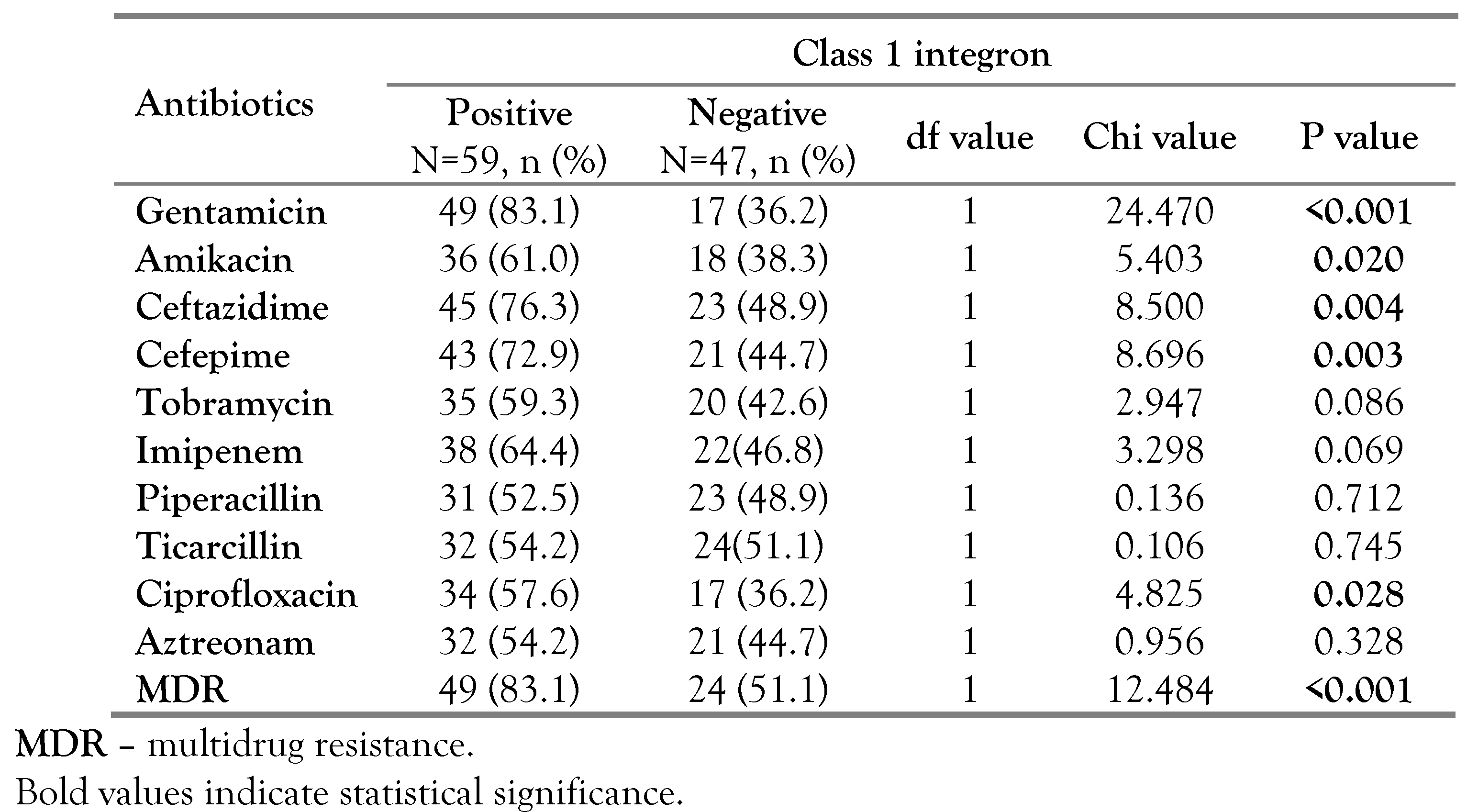
Molecular examination has revealed that out of 106 P. aeruginosa isolates, 50 had only intI1 gene, 12 had only intI2 gene and 9 harbored both genes. Based on these molecular findings, 59 P. aeruginosa isolates (55.7%) were class 1 integronpositive while 21 (19.8%) were class 2 positive. On the other hand, class 3 integrons could not be detected in any of the P. aeruginosa strains.
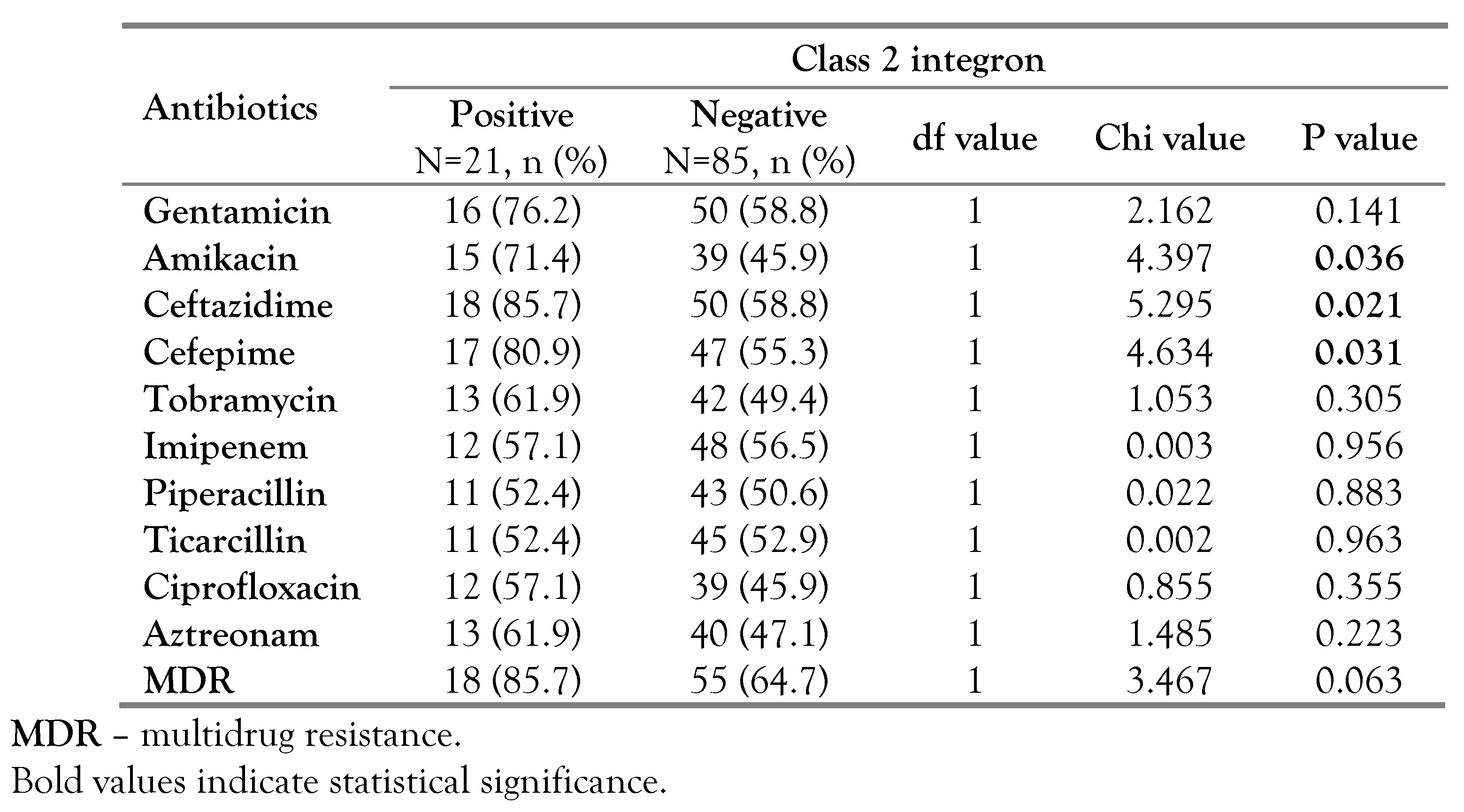
In our study, we found significant associations between the existence of class 1 integron and resistance toward gentamicin (p<0.001, OR=8.647, 95%CI: 3.503-21.346), ceftazidime (p=0.004, OR=3.354, 95%CI: 1.4647.682), amikacin (p=0.020, OR=2.522, 95%CI: 1.148-5.541), ciprofloxacin (p=0.028, OR=2.400, 95%CI: 1.092-5.277) and cefepime (p=0.003, OR=3.327, 95%CI: 1.477-7.498). Furthermore, out of the 59 class 1 integron-positive strains, 49 (83.1%) had MDR profiles, which was also significant (p<0.001, OR=4.696, 95%CI: 1.931-11.419) as shown in Table 1. Moreover, significant associations were found between the existence of class 2 integron and resistance toward amikacin (p=0.036, OR=2.949, 95%CI: 1.044-8.330), ceftazidime (p=0.021, OR=4.200, 95%CI: 1.149-15.355) and cefepime (p=0.031, OR=3.436, 95%CI: 1.066-11.073). Unlike class 1 integron, no significant association was found between class 2 integron and MDR, Table 2.

Table 1.
The relation between class 1 integron and antibiotic resistance among P. aeruginosa isolates.

Table 2.
The relation between class 2 integron and antibiotic resistance among P. aeruginosa isolates.
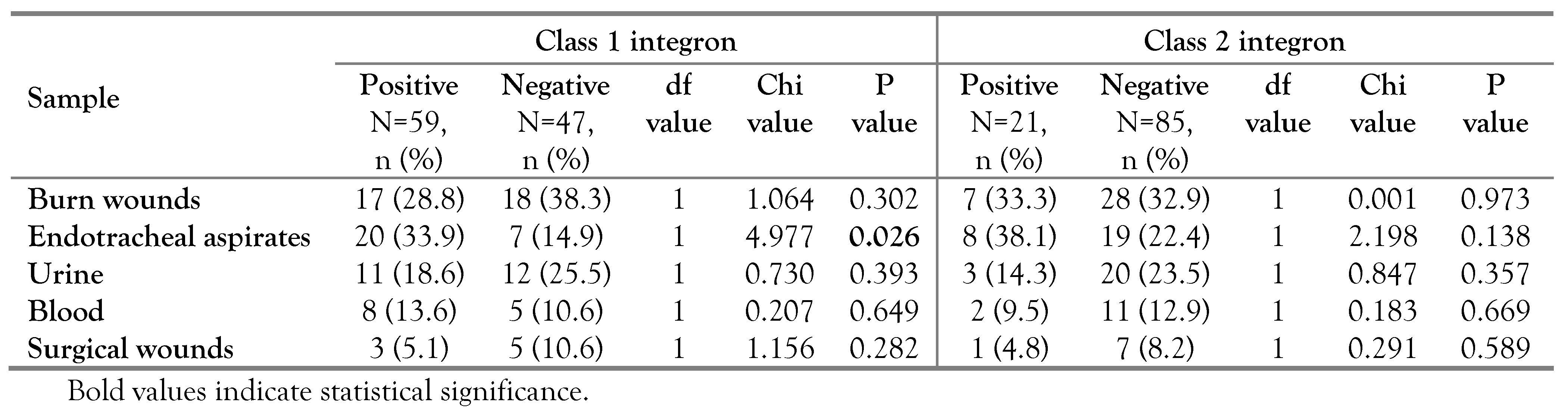
Class 1 and 2 integron-positive strains were isolated from various clinical samples including: endotracheal aspirates, burn wounds, urine, blood and surgical wounds. Notably, a significant association was found between the existence of integrons class 1 and endotracheal aspirates (p=0.026, OR=2.930, 95%CI: 1.114-7.708). On the other hand, none of the collected samples showed significant association with the existence of class 2 integron among pseudomonal isolates, Table 3.

Table 3.
Distribution of class 1 and 2 integrons among P. aeruginosa isolates recovered from various clinical samples.
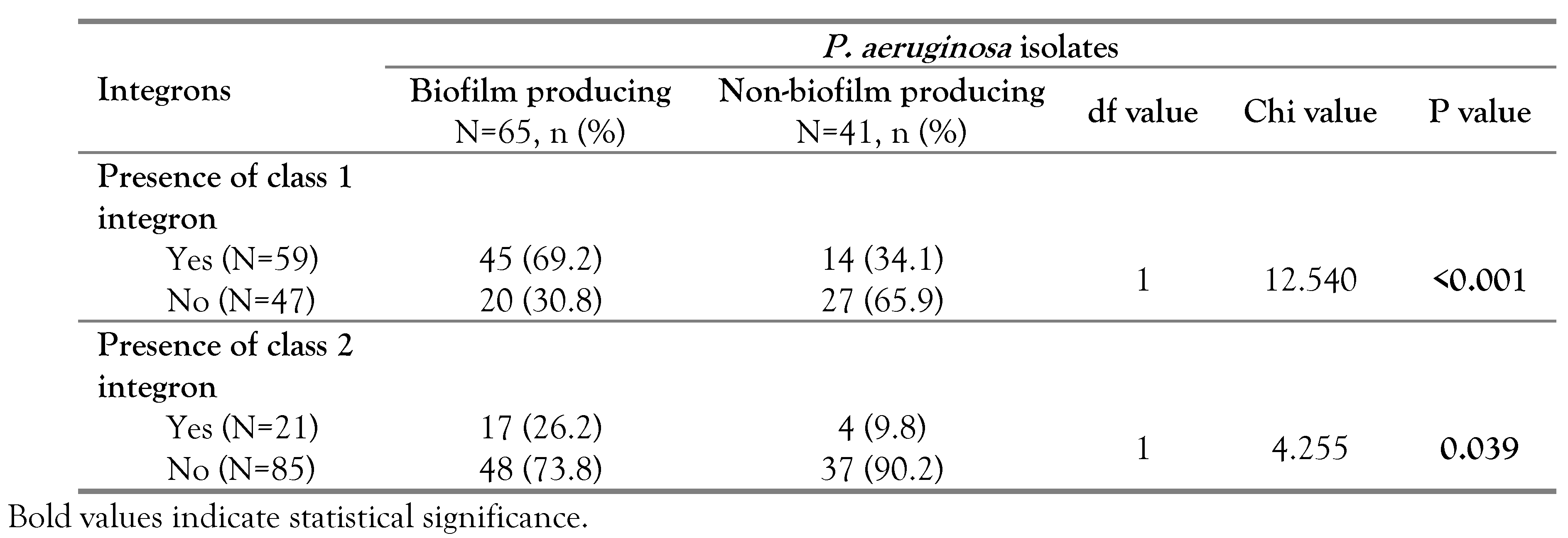
Based on the results of the tissue culture plate method, 61.3% (65/106) of P. aeruginosa isolates were biofilm producing and 38.7% (41/106) were non-biofilm producing. Integrons class 1 and 2 were detected among both biofilm and non-biofilm producing pseudomonal isolates. Notably, significant associations were found between biofilm formation and the existence of class 1 integron (p<0.001, OR=4.339, 95%CI: 1.886-9.983) and class 2 integron (p=0.039, OR=3.276, 95%CI: 1.016-10.561), Table 4.

Table 4.
Distribution of class 1 and 2 integrons among biofilm and non-biofilm producing isolates of P. aeruginosa.
Discussion
Integrons have recently attracted lots of attention due to their crucial role in the spread and expression of antibiotic resistance genes particularly among Gram-negative bacteria [5]. Moreover, while investigating genetic determinants responsible for MDR among P. aeruginosa, the emerging role of integrons has been appreciated [1]. In our study, we have found that 68.9% of P. aeruginosa isolates exhibited MDR profiles, which was higher than 60.7%, reported by a former study in Egypt [19]. Such finding highlighted the growing problem of MDR among these isolates in our locality. Other earlier studies have reported frequencies of MDR among P. aeruginosa at 75% in Iran and 90.1% in China, which were higher than our findings [1,20].
Molecular work conducted in our study has revealed that 55.7% of pseudomonal isolates carried class 1 integron. A higher frequency of class 1 integron positive P. aeruginosa clinical isolates has been reported by Zarei-Yazdeli and colleagues in Iran with a percentage of 82.6% [1]. Other studies conducted on various clinical samples in China and Northern Iran have revealed that 40.8% and 60% of isolated P. aeruginosa strains were positive for class 1 integron, respectively [3,20]. Such differences could be explained by multiple factors such as geographical variations, selective pressure imposed by indiscriminate usage of antibiotics and the variable implementation of infection control precautions.
In our study, class 2 integron was detected among 19.8% of P. aeruginosa isolates, which was similar to what had been reported earlier in China (19.5%) and slightly higher than in Iran (15.3%) [1,21]. However, in Thailand, the reported frequency was zero [22]. On the other hand, we could not detect class 3 integron in any of the P. aeruginosa isolates in our work, which was consistent with previous reports [1,22].
In our effort to evaluate the relations between the presence of different integron classes among pseudomonal isolates and their antibiotic resistance profiles, the existence of integron class 1 was significantly associated with resistance to the following antimicrobials: gentamicin, amikacin, ceftazidime, ciprofloxacin and cefepime. Similarly, class 2 integron was found to be in significant association with resistance to amikacin, ceftazidime and cefepime. These findings can be explained, as resistance integrons frequently carry gene cassettes conveying antimicrobial resistance towards a broad range of antibiotics [7]. in line with our findings, ZareiYazdeli and colleagues reported a significant association between the presence of class 1 integron and gentamicin resistance [1]. They have also reported an association between class 2 integron and resistance to amikacin, ceftazidime and tobramycin [1]. A similar significant association between intI gene presence and resistance to various antibiotics was reported by EbrahimiShahabi et al [3]. Moreover, we have reported a significant association between harboring class 1 integron and MDR pattern, which was consistent with other studies [1,20]. On the other hand, antimicrobial resistance exhibited by integronnegative pseudomonal isolates can be attributed to resistance determinants encoded by chromosomes or carried by mobile elements other than integrons.
In the current study, we reported a significant association between the existence of class 1 integron and endotracheal aspirates. Such finding could be attributed to the location of these ventilated patients in the intensive care units (ICUs) where they usually undergo several invasive procedures and receive multiple antimicrobial treatments resulting in a high selective pressure. Accordingly, improper implementation of infection control precautions in the ICUs could facilitate the circulation of mobile integrons, which can account for our findings.
In our study, biofilm production was detected among 61.3% of P. aeruginosa isolates. In line with our findings, Zahedani et al. reported that 59.3% of P. aeruginosa isolates were biofilm producers [23]. However, biofilm production among P. aeruginosa isolates was reported at a much higher percentage (97.5%) by Bakht et al [15].
In the current study, class 1 and 2 integrons had higher frequencies among biofilm than nonbiofilm forming isolates. According to these results, biofilm production was significantly associated with the existence of integrons class 1 and class 2. In agreement with our findings, previous studies have reported similar associations between biofilm production and antibiotic resistance genetic elements [17,24]. Such findings can be explained by the enhanced genetic exchange among different bacteria residing in the biofilm communities [10]. Within the biofilm, bacterial members are close together and crowded, which can facilitate the transfer of genetic determinants, including mobile integrons, which can also justify our findings [9,10]. Bacterial evolution inside biofilm communities can also result in substantial genetic diversity [25]. Furthermore, biofilms are linked to persistent infections that could accelerate the circulation of integrons and other resistance elements [9].
Different strategies have been developed to combat P. aeruginosa-related infections; however, they have had only modest success because of the variety and multiplicity of resistance mechanisms that enable this bacterium to escape the effect of various antibiotics [2]. It should be mentioned that 9 P. aeruginosa strains in our study were positive for class 1 and 2 integrons as well as for biofilm production. Out of these 9 isolates, 8 (88.9%) were MDR, which highlighted the possible collaboration between these resistance factors that can pose additional challenge on clinicians in treating pseudomonal infections. Therefore, further studies are needed to unravel the interactions between different resistance mechanisms and to design novel antibacterial drugs that can be used in our fight against this bacterium. In order to minimize the spread of resistance elements among pseudomonal strains, well-established infection control guidelines, antimicrobial stewardship programs and continuous monitoring for resistant strains are essential. Our study was subject to certain limitations as the interpretation of our results could be more solid with a larger number of P. aeruginosa isolates. Also, our research did not include outpatients; therefore, studies with different populations are recommended.
Conclusions
In our study, about two thirds of P. aeruginosa isolates were positive for resistance integrons, which underlined the importance of these resistance elements in our locality. The presence of integrons was significantly associated with resistance towards several antibiotics. Notably, biofilm formation was significantly associated with the presence of class 1 as well as class 2 integrons among P. aeruginosa isolates. Such findings emphasized the possible synergistic effect between these resistance mechanisms. Our study provided important data on the distribution of key resistance determinants among P. aeruginosa strains and their possible impact on antibiotic resistance patterns, which can help physicians to cure these resistant infections. Furthermore, these data can be used in surveillance and to improve infection control policies in healthcare settings.
Author Contributions
AMS developed the study design, processed the samples, drafted the manuscript and analyzed the data. NMM shared in the laboratory study, the draft preparation and data analysis of the study. All authors read and approved the final version of the manuscript.
Funding
None to declare.
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author, upon reasonable request.
Acknowledgments
The authors would like to show their gratitude to all members of the laboratory and medical teams for their continuous assistance.
Conflicts of interest
All authors—none to declare.
References
- Zarei-Yazdeli, M.; Eslami, G.; Zandi, H.; et al. Prevalence of class 1, 2 and 3 integrons among multidrug-resistant Pseudomonas aeruginosa in Yazd, Iran. Iran J Microbiol. 2018, 10, 300–6. [Google Scholar]
- Sambrano, H.; Castillo, J.C.; Ramos, C.W.; et al. Prevalence of antibiotic resistance and virulent factors in nosocomial clinical isolates of Pseudomonas aeruginosa from Panamá. Braz J Infect Dis. 2021, 25, 101038. [Google Scholar] [CrossRef]
- Ebrahimi-Shahabi, R.; Ferdosi-Shahandashti, E.; Sabbagh, P. Genetic cassettes profiling of class I integron and antimicrobial susceptibility profiles among Pseudomonas aeruginosa isolates collected from patients in north of Iran. Jundishapur J Microbiol. 2022, 15, e120079. [Google Scholar] [CrossRef]
- Richard, E.; Darracq, B.; Loot, C.; Mazel, D. Unbridled integrons: a matter of host factors. Cells. 2022, 11, 925. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Gillings, M.R.; Penesyan, A.; Qi, Q.; Rajabal, V.; Tetu, S.G. The natural history of integrons. Microorganisms. 2021, 9, 2212. [Google Scholar] [CrossRef]
- Akrami, F.; Rajabnia, M.; Pournajaf, A. Resistance integrons: a mini review. Caspian J Intern Med. 2019, 10, 370–6. [Google Scholar] [CrossRef]
- Partridge, S.R.; Tsafnat, G.; Coiera, E.; Iredell, J.R. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009, 33, 757–784. [Google Scholar] [CrossRef]
- Arakawa, Y.; Murakami, M.; Suzuki, K.; et al. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995, 39, 1612–1615. [Google Scholar] [CrossRef] [PubMed]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: formation, architecture, antibiotic resistance, and control strategies. Braz J Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Aparna, M.S.; Yadav, S. Biofilms: microbes and disease. Braz J Infect Dis. 2008, 12, 526–30. [Google Scholar] [CrossRef] [PubMed]
- Sultan, A.M.; Nabiel, Y. Tube method and Congo red agar versus tissue culture plate method for detection of biofilm production by uropathogens isolated from midstream urine: which one could be better? African J Clin Exp Microbiol. 2019, 20, 60–6. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. In 31st ed.; CLSI supplement M100; CLSI: Malvern, PA, USA, 2021. [Google Scholar]
- Seliem, W.A.; Sultan, A.M. Etiology of early onset neonatal sepsis in neonatal intensive care unit Mansoura, Egypt. J Neonatal Perinatal Med. 2018, 11, 323–330. [Google Scholar] [CrossRef]
- Sultan, A.M.; Ahmed, M.A. Distribution of chlorhexidine resistance genes among Staphylococcus aureus clinical isolates: the challenge of antiseptic resistance. Germs. 2022, 12, 461–71. [Google Scholar] [CrossRef]
- Bakht, M.; Alizadeh, S.A.; Rahimi, S.; et al. Phenotype and genetic determination of resistance to common disinfectants among biofilm-producing and nonproducing Pseudomonas aeruginosa strains from clinical specimens in Iran. BMC Microbiol. 2022, 22, 124. [Google Scholar] [CrossRef]
- Bakir, S.H.; Ali, F.A. Comparison of different methods for detection of biofilm production in multi-drug resistance bacteria causing pharyngotonsillitis. Int J Res Pharm Biosci. 2016, 3, 13–22. [Google Scholar]
- Sultan, A.M.; Amer, G.F.; Nabiel, Y. Quinolone-resistant uropathogenic E. coli: is there a relation between qnr genes, gyrA gene target site mutation and biofilm formation? J Med Microbiol. 2021, 70, 10. [Google Scholar] [CrossRef]
- Ugwuanyi, F.C.; Ajayi, A.; Ojo, D.A.; Adeleye, A.I.; Smith, S.I. Evaluation of efflux pump activity and biofilm formation in multidrug resistant clinical isolates of Pseudomonas aeruginosa isolated from a Federal Medical Center in Nigeria. Ann Clin Microbiol Antimicrob. 2021, 20, 11. [Google Scholar] [CrossRef]
- Zaki, M.E.S.; Mahmoud, N.M.; Rizk, M.A. Molecular study of integrase gene I and integrase gene II in clinical isolates of Pseudomonas aeruginosa. Infect Disord Drug Targets. 2022, 22, 56–61. [Google Scholar] [CrossRef]
- Chen, J.; Su, Z.; Liu, Y.; et al. Identification and characterization of class 1 integrons among Pseudomonas aeruginosa isolates from patients in Zhenjiang, China. Int J Infect Dis. 2009, 13, 717–21. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, L.; Shirtliff, M.E.; Alam, M.J.; Yamasaki, S.; Shi, L. Occurrence and characteristics of class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in southern China. J Clin Microbiol. 2009, 47, 230–34. [Google Scholar] [CrossRef] [PubMed]
- Poonsuk, K.; Tribuddharat, C.; Chuanchuen, R. Class 1 integrons in Pseudomonas aeruginosa and Acinetobacter baumannii isolated from clinical isolates. Southeast Asian J Trop Med Public Health. 2012, 43, 376–84. [Google Scholar] [PubMed]
- Zahedani, S.S.; Tahmasebi, H.; Jahantigh, M. Coexistence of virulence factors and efflux pump genes in clinical isolates of Pseudomonas aeruginosa: analysis of biofilmforming strains from Iran. Int J Microbiol. 2021, 2021, 5557361. [Google Scholar] [CrossRef] [PubMed]
- Azizi, O.; Shakibaie, M.R.; Modarresi, F.; Shahcheraghi, F. Molecular detection of class-D OXA carbapenemase genes in biofilm and non-biofilm forming clinical isolates of Acinetobacter baumannii. Jundishapur J Microbiol. 2015, 8, e21042. [Google Scholar] [CrossRef]
- Stalder, T.; Cornwell, B.; Lacroix, J.; et al. Evolving populations in biofilms contain more persistent plasmids. Mol Biol Evol. 2020, 37, 1563–76. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2024.