Introduction
In the modern world, healthcare associated infections (HAIs) continue to be major health problems of a notable financial burden since they have considerable association with wide variety of morbidities and mortalities [
1].
Staphylococcus aureus is considered as a leading bacterial cause of HAIs responsible for bacteremia, pneumonia, skin or soft tissue infections and devices-associated infections [
2]. Interestingly,
S. aureus was found to be the second leading cause of HAIs in United States [
3]. However, it has been reported that up to 60% of nosocomial
S. aureus infections were attributed to methicillin resistant isolates [
4].
Methicillin resistant
S. aureus (MRSA) is considered as a common resistant pathogen to antibiotics particularly in certain settings and countries. Over the last decade, the rates of MRSA infection have increased owing to antibiotics abuse and continuous bacterial evolution. Furthermore, infections attributed to MRSA strains are more challenging to eradicate because of their resistance to many antimicrobials [
5].
Infection control guidelines have been established to control HAIs through the proper use of antiseptics such as chlorhexidine aiming for diminishing bacterial load from the infected or colonized body surfaces [
2]. Chlorhexidine is a known cationic topical antiseptic agent that is available in different forms such as acetate, gluconate or hydrochloride salts [
6]. Following adherence to bacterial cell wall, chlorhexidine molecules act through damaging the cell wall and cytoplasmic membrane leading to intra-cellular components leakage and subsequent bacterial death [
1,
6].
Chlorhexidine is one of the commonly used antiseptics in healthcare facilities because of its safety and tolerability as it can be tolerated by human tissue with a good safety record. It is also characterized by its broad-spectrum activity and residual effect which is the prolonged antimicrobial activity that extends after the product application. Furthermore, chlorhexidine has a wide range of applications in clinical practice including hand disinfection, skin disinfection prior to surgeries and invasive procedures such as vascular catheters insertion, MRSA decolonization and bathing patients in intensive care units (ICUs) [
6]. However, the wide use of chlorhexidine in healthcare settings could lead to reduction of its susceptibility among different bacterial isolates including
S. aureus [
2,
6,
7]. Furthermore, various HAIs and outbreaks have been attributed to chlorhexidine resistance [
7,
8].
Phenotypic susceptibility to chlorhexidine is usually tested by identifying the minimum inhibitory concentration (MIC) values [
1,
6]. Staphylococcal isolates with MIC≥4 mg/L are considered to have reduced chlorhexidine susceptibility, in other words chlorhexidine tolerance [
2,
6]. Chlorhexidine MIC values actually identify chlorhexidine tolerance instead of resistance. Chlorhexidine resistance indicates that the bacteria are able to survive at the in-use concentrations of chlorhexidine. On the other hand, when the bacteria are inhibited at lower concentrations, as in MIC tests, it should be defined as tolerance to chlorhexidine (MIC ≥4 mg/L). Therefore, the favored name for phenotypic chlorhexidine resistance is “chlorhexidine tolerance” or “reduced chlorhexidine susceptibility”, while genotypic resistance can be identified by the existence of chlorhexidine resistance genes [
6].
Chlorhexidine resistance is mainly caused by multidrug efflux pumps which can carry various antibiotics and antiseptic agents outside the bacterial cells [
2,
6]. In one report, chlorhexidine was effluxed in 96% of bloodstream
S. aureus isolates which highlighted the significance of efflux pumps [
9]. Genetic determinants responsible for the efflux-mediated antiseptic resistance include
qac (quaternary ammonium compound) and
smr (staphylococcal multidrug resistance) resistance genes [
6,
7].
Chlorhexidine resistance among staphylococcal isolates is frequently caused by
qacA and
qacB genes. These genes cause chlorhexidine resistance by encoding a proton motive force-dependent efflux pump. It is hard to differentiate between
qacA and
qacB genes by PCR because they are very similar and closely related. Hence, their PCR products are identified as
qacA/B gene negative or positive [
1]. In
S. aureus isolates, the
qacA/B gene is located in chromosomes and on plasmids [
7]. Both QacA and QacB proteins belong to the major facilitator superfamily of transport proteins. These large proteins have 14 transmembrane α-helical segments which can export chlorhexidine out of the bacterial cells [
1]. The
qacA/B gene was previously reported as the most frequent biocidal resistance gene among
S. aureus isolates [
7].
The
smr gene is another antiseptic resistance gene that was initially located on a plasmid recovered from a
S. aureus isolate. The protein encoded by
smr gene belongs to the small multidrug resistance family of transport proteins and causes an efflux-mediated antiseptic resistance. The
smr gene was found to be associated with reduced susceptibility toward chlorhexidine among
S. aureus isolates [
7].
The extensive usage of chlorhexidine in different healthcare settings raised an alarm about the development of acquired bacterial resistance toward this antiseptic agent [
7]. Previous studies have demonstrated reduced chlorhexidine susceptibility among
S. aureus isolates and reported different rates for antiseptic resistance genes [
6,
7]. To our knowledge, none of these studies were conducted in our locality or investigated the possible risk factors for such reduced susceptibility. Antiseptic resistance can compromise the efficiency of prevention strategies for HAIs. Therefore, the aim of our study was to determine chlorhexidine susceptibility among clinical
S. aureus isolates and to detect
qacA/B and
smr antiseptic resistance genes among these isolates. Furthermore, we aimed to identify possible risk factors for the reduction of chlorhexidine susceptibility among
S.
aureus isolates.
Results
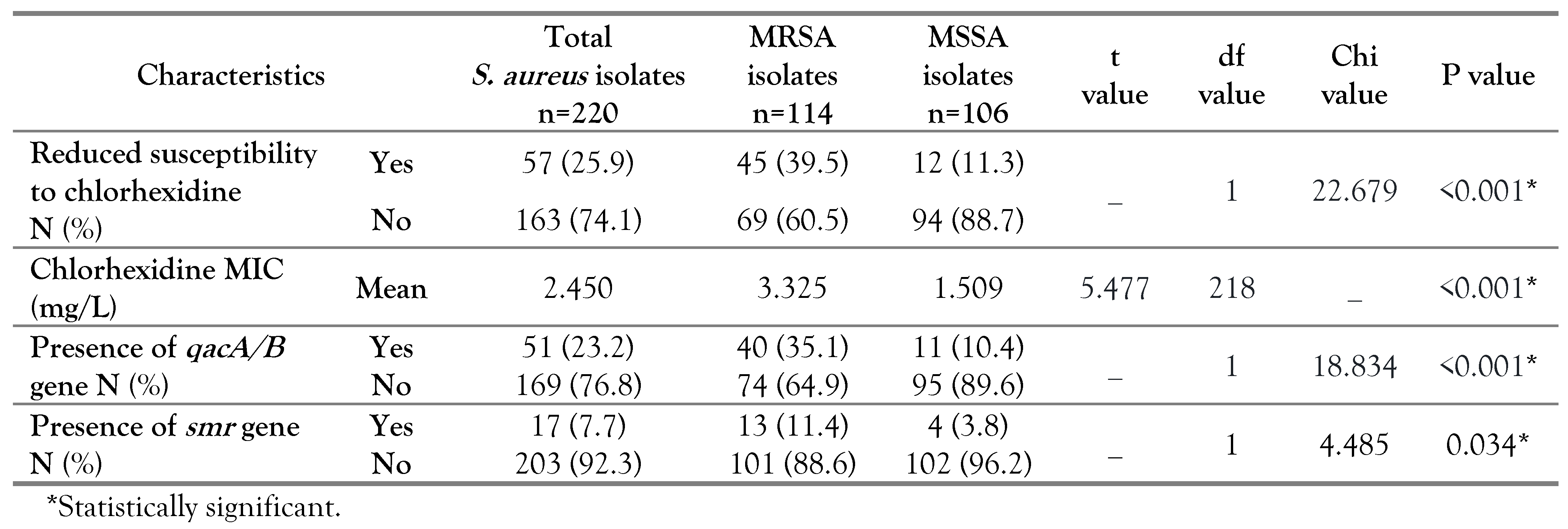
In our study, 220 S. aureus isolates were identified from the collected samples including 123 (55.9%) blood, 39 (17.7%) endotracheal aspirates, 30 (13.6%) wound drainages and 28 (12.7%) urine. The study subjects included 119 (54.1%) females and 101 (45.9%) males with mean age of 44±18 years.Out of the identified 220 S. aureus isolates, 114 (51.8%) were MRSA and 106 (48.2%) were methicillin susceptible S. aureus (MSSA). All MRSA strains were positive for cefoxitin disk diffusion test as well as mecA gene detection.
According to MIC results, 57 out of 220
S. aureus isolates had reduced susceptibility to chlorhexidine with a percentage of 25.9%. The percentage of chlorhexidine tolerant isolates among MRSA and MSSA were 39.5% (45/114) and 11.3% (12/106) respectively, with a statistical significance (p<0.001). The MRSA strains had a range of MIC between 0.5 and 16 mg/L, whereas the MIC values for MSSA ranged from 0.5-8 mg/L. In parallel, the mean chlorhexidine MIC among the MRSA (3.325 mg/L) was significantly higher than MSSA isolates (1.509 mg/L), with p<0.001 as shown in
Table 1.
In our study, the molecular analysis revealed that 23.2% and 7.7% of
S. aureus strains harbored the
qacA/B and
smr genes, respectively. The percentage of
qacA/B gene among MRSA (35.1%) was significantly higher than in MSSA isolates (10.4%), p<0.001. Likewise, the MRSA had a higher percentage of
smr gene (11.4%) than the MSSA isolates (3.8%) which was statistically significant (p=0.034) (
Table 1). A significant association was found between
qacA/B gene existence and reduced chlorhexidine susceptibility as 94.1% of the
S. aureus isolates harboring this gene were chlorhexidine tolerant, while 5.9% were chlorhexidine susceptible (p<0.001). Also, an association was noted between the existence of
smr gene and reduced chlorhexidine susceptibility (p<0.001) (
Table 2).
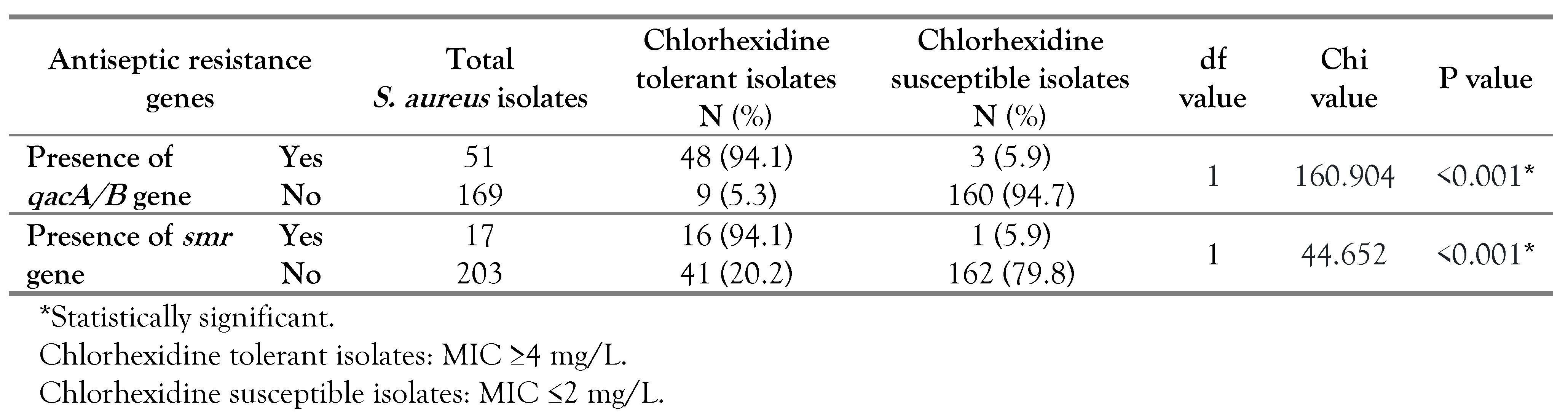
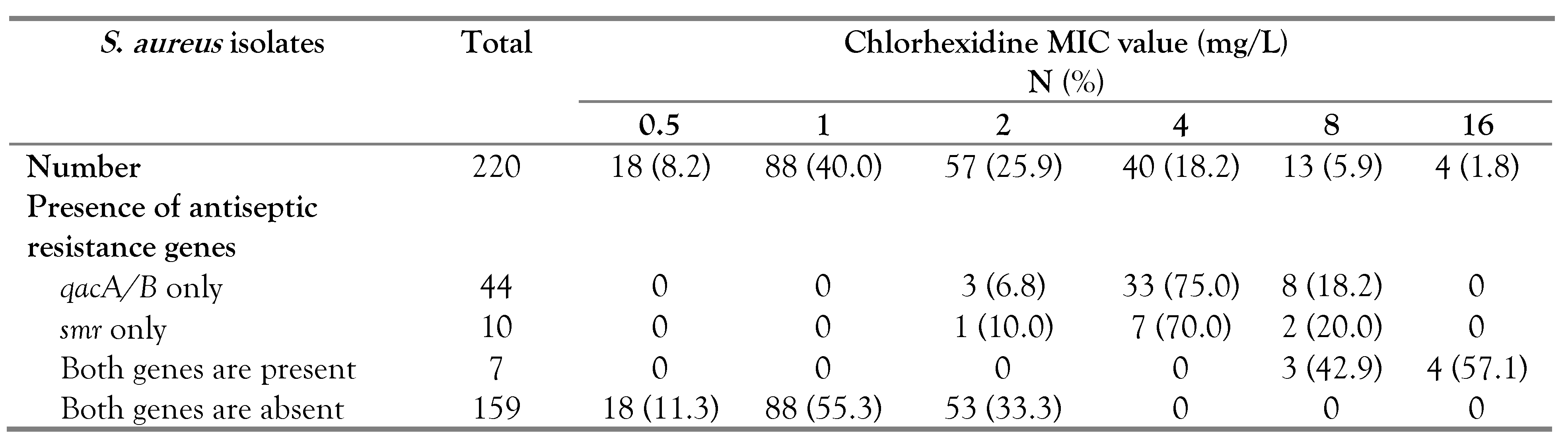
The antiseptic resistance genes were identified among 61 (27.7%) out of the 220
S. aureus isolates; 44 had the
qacA/B gene, 10 had the
smr gene and 7 harbored both genes (5 MRSA and 2 MSSA). Our results showed that all 57
S. aureus isolates with reduced susceptibility to chlorhexidine had one or both of the antiseptic resistance genes; 41 isolates carried the
qacA/B gene, 9 isolates had the
smr gene and 7 isolates harbored both genes (
Table 3). On the other hand, among the 163 chlorhexidine susceptible isolates, three had the
qacA/B and one carried the
smr gene (
Table 3). Isolates harboring antiseptic resistance genes displayed a wider range of chlorhexidine MIC (2-16 mg/L) compared to others lacking these genes (0.5-2 mg/L). It should be noted that
S. aureus isolates carrying either
qacA/B or
smr gene exhibited chlorhexidine MIC range of 2-8 mg/L, while isolates harboring both genes displayed higher chlorhexidine MIC values up to 16 mg/L with a chlorhexidine MIC range of 8-16 mg/L (
Table 3).
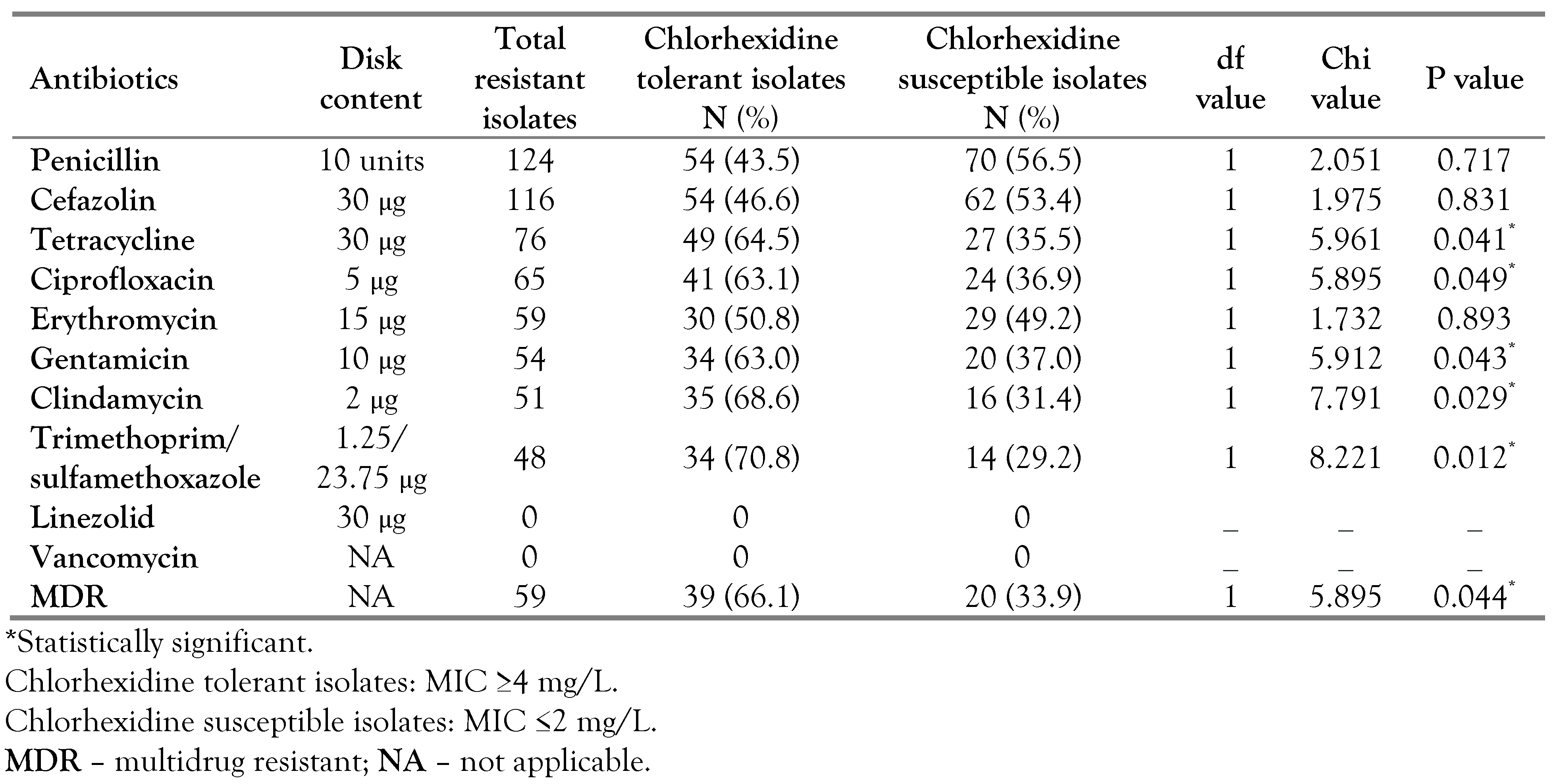
A significant association was found between
S. aureus isolates with reduced chlorhexidine susceptibility and resistance toward these antibiotics: tetracycline (p=0.041), ciprofloxacin (p=0.049), gentamicin (p=0.043), clindamycin (p=0.029) and trimethoprim/sulfamethoxazole (p=0.012). Of the 59 isolates showing MDR profiles, 66.1% were chlorhexidine tolerant and 33.9% were chlorhexidine susceptible showing a statistically significant difference (p=0.044). None of the tested
S. aureus strains demonstrated vancomycin or linezolid resistance (
Table 4).
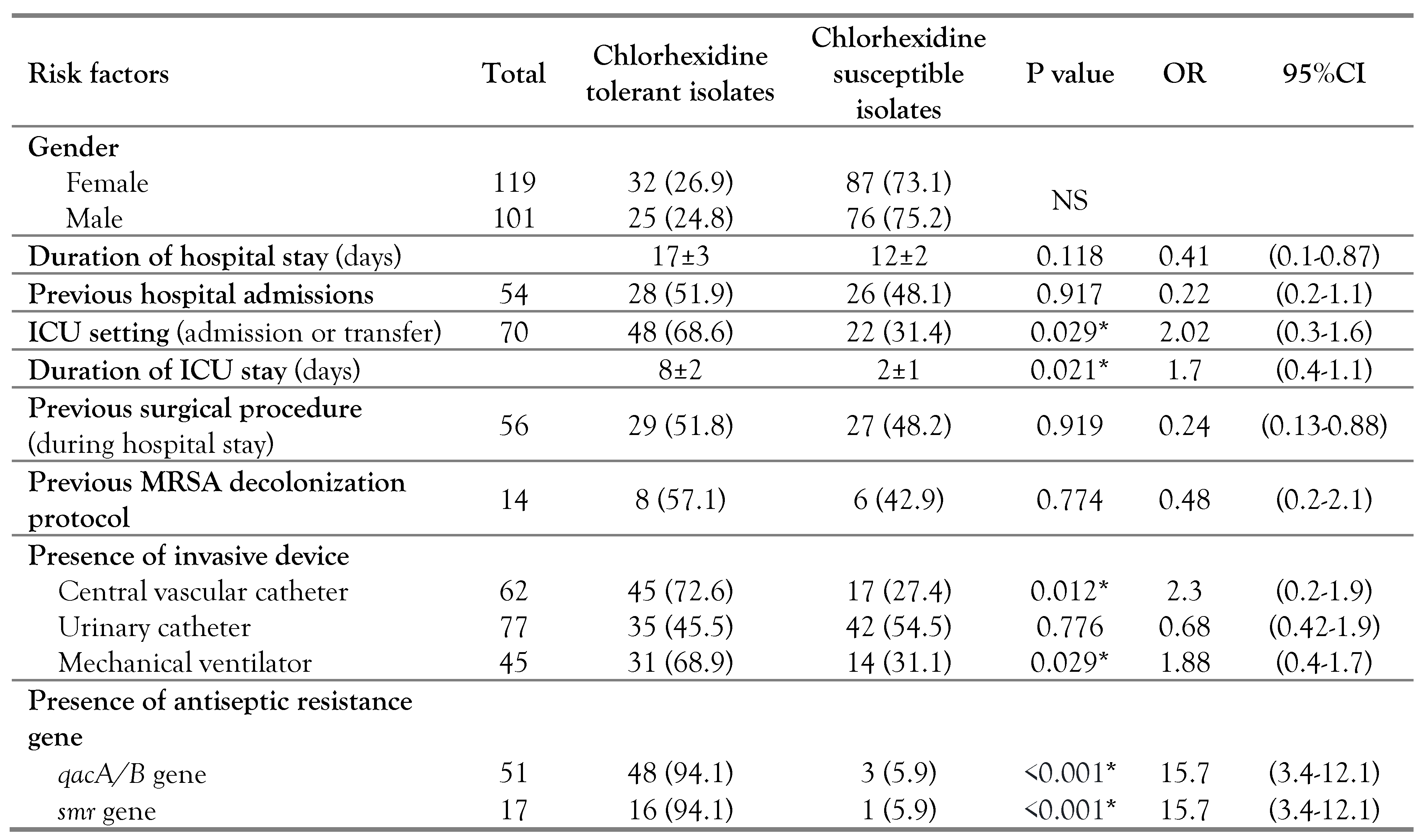
According to univariate analysis, the following were identified as risk factors for reduced susceptibility to chlorhexidine among
S. aureus isolates; ICU setting (p=0.029, OR=2.02, 95%CI: 0.3-1.6), prolonged ICU stay (p=0.021, OR=1.7, 95%CI: 0.4-1.1), presence of central vascular line (p=0.012, OR=2.3, 95%CI: 0.2-1.9), usage of mechanical ventilation (p=0.029, OR=1.88, 95%CI: 0.4-1.7) and acquisition of
qacA/B (p<0.001, OR=15.7, 95%CI: 3.4-12.1) or
smr gene (p<0.001, OR=15.7, 95%CI: 3.4-12.1). On the other hand, none of the following showed a significant association with reduced chlorhexidine susceptibility; gender, duration of hospital stay (p=0.118), previous hospital admissions (p=0.917), previous surgical procedure during hospital stay (p=0.919), previous MRSA decolonization protocol (p= 0.774) and the existence of urinary catheter (p=0.776) (
Table 5).
Discussion
The percentage of MRSA in our work with a value of 51.8% was found to be slightly higher than 48.9%, reported in an earlier study conducted in the same locality [
17]. Such observation highlighted a steady increment of MRSA infections in our community. In our study, the percentage of
S. aureus strains which had reduced chlorhexidine susceptibility, defined by MIC, was 25.9% that was almost two folds that been reported by Azrad et al. and by Hughes and Ferguson at values of 13.5% and 10% respectively [
2,
18]. The chlorhexidine tolerant isolates had a significantly higher frequency among MRSA than MSSA (p<0.001). In line with our findings, previous studies have reported that MSSA was more susceptible to chlorhexidine than MRSA [
2,
5]. In addition, MRSA had a significantly higher mean value of chlorhexidine MIC when compared to that of MSSA isolates (p<0.001) which was parallel to the reported results by Azrad and his colleagues [
2]. Such findings have raised a great concern because of the extensive use of chlorhexidine in MRSA decolonization protocols. However, the significance of such observation is not fully identified as the in-use concentrations of chlorhexidine were much higher than the MIC tested in vitro [
2,
6]. Nevertheless, Batra and his colleagues reported that usage of chlorhexidine-based protocol of decolonization in ICU has led to dissemination of MRSA strain with increased MIC value toward this antiseptic agent [
8]. These findings suggested that the low chlorhexidine susceptibility among MRSA could be associated with outbreaks inside healthcare settings.
In our study, the total percentage of
qacA/B gene among
S. aureus strains was 23.2%, which was higher than 13.15% reported by Azrad et al. [
2]. We have also found that 35.1% of the MRSA isolates harbored the
qacA/B gene. Similar to the findings of our study, the
qacA/B gene rates among MRSA isolates were reported at 33.9% in Japan and 37.7% in Korea [
14,
19]. Higher detection rates of that gene were reported in Turkey at 64.3% and 83% in China [
20,
21]. The
mecA gene, causing methicillin resistance, is positioned on staphylococcal cassette chromosome (SCC) that can integrate other plasmids carrying antiseptic resistance genes, and this can offer an explanation of our findings [
5,
16]. Sidhu and his colleagues have reported such genetic linkage between
qac genes and
mecA gene on the same mobile element [
22].
Out of
S. aureus isolates investigated in our study, 7.7% carried the
smr gene with a percentage of 11.4% among MRSA. Similar to the
qacA/B gene, the detection rates of the
smr gene among MRSA isolates varied as per different regions; 1.4% in Japan, 3.1% in Asia, 44.2% in Scotland, and 77.4% in China [
14,
16,
21,
23]. Such variations could be attributed to the geographical differences, clonal spread of tolerant strains, study design, applied infection control programs and most importantly the utilization pressure of chlorhexidine inside healthcare settings. Therefore, it is essential to continuously monitor the development of antiseptic resistance in different settings.
In our work, a significant association was reported between reduced chlorhexidine susceptibility and the existence of
qacA/B gene (p<0.001), and the same goes for
smr gene (p<0.001). Other studies have reported similar findings [
2,
5]. It is noteworthy that isolates harboring both genes displayed elevated MIC values compared to isolates carrying only one gene, which was in line with previous findings [
5]. The presence of antiseptic resistance genes does not necessarily induce tolerance to chlorhexidine as the gene could be silent and not expressed by the bacteria [
1,
2]. Therefore, it was not surprising to find that among chlorhexidine susceptible isolates, three isolates were
qacA/B gene-positive and one was
smr gene-positive.
In our study, resistance to tetracycline, ciprofloxacin, gentamicin, clindamycin and trimethoprim/sulfamethoxazole was significantly associated with chlorhexidine tolerant isolates. Such association between antiseptic resistance genes and antimicrobial resistance was previously reported [
1,
6]. We have also found that chlorhexidine tolerant strains displayed an MDR profile at a higher frequency than the susceptible strains (p=0.044), which was in line with previous studies [
1,
24]. These findings can be attributed to co-resistance and cross-resistance. Such cross-resistance occurs when one multidrug efflux pump carries chlorhexidine along with other antibiotics such as fluoroquinolones [
9]. On the other hand, the co-resistance is caused by the existence of different resistance genes on one genetic element [
2,
6]. The genetic link between the
qacA/B gene and resistance genes towards aminoglycosides and trimethoprim was reported among staphylococcal isolates [
1,
6]. In addition, the
qacA/B gene was detected in strains harboring SCCmec (types I–V) which carry resistance genes for tetracycline, aminoglycosides and lincosamides [
6]. These observations have raised an alarm that excessive exposure to antiseptics might result in selection of strains harboring both antiseptic and antibiotic resistance genes, which impose an additional challenge in controlling HAIs [
1]. It is also necessary to mention that the increasing utilization of antiseptics outside the healthcare settings, because of the pandemic of COVID-19, could exaggerate the problem of antiseptic resistance.
To the best of our knowledge, the current study is the first to explore possible risk factors for reduced chlorhexidine susceptibility among
S. aureus. The following risk factors were identified: ICU setting, prolonged duration of ICU stay, presence of central vascular catheter, mechanical ventilation and acquisition of
qacA/B or
smr gene. Our findings reflected the high selective pressure present in the ICU setting where chlorhexidine is frequently used [
6]. In addition, prolonged ICU stay is associated with repeated exposure to chlorhexidine building up more selective pressure over bacterial isolates. Vali and his colleagues have reported that recurrent exposure of MRSA to chlorhexidine has led to reduction in chlorhexidine susceptibility and induction of
qacA/B gene expression [
23]. Therefore, repeated chlorhexidine exposure in the ICU environment can lead to selection of chlorhexidine-tolerant isolates.
We also recognized the presence of central vascular catheter and mechanical ventilator as risk factors for reduced chlorhexidine susceptibility. In order to prevent catheter-associated bloodstream infections, chlorhexidine preparations are used for skin disinfection before the insertion and for the subsequent catheter care. Similarly, chlorhexidine can be used up to 6 times per day as an oral rinse to prevent ventilator-associated pneumonia [
6,
7]. Such high magnitude of chlorhexidine exposure can lead to selection of antiseptic tolerant organisms. In the UK, a significant correlation was reported between the magnitude of chlorhexidine exposure and the MIC values of clinical isolates [
25].
Chlorhexidine is an important antiseptic agent that is effective in preventing HAIs. To reduce selective pressure in healthcare settings, the unnecessary usage of chlorhexidine should be limited. A biocidal stewardship program should also be initiated to preserve the effectiveness of antiseptic agents. In addition, an ongoing surveillance system is necessary for early recognition of the antiseptic tolerant strains and for identification of possible risk factors. The limitation of our study includes lack of detection of other chlorhexidine resistance genes and relatively small sample size. Thus, further researches on a wider scale overcoming these limitations are needed.