Possible Intraindividual Evolution of SARS-CoV-2 in Nasopharyngeal and Anal Swab in an Octogenarian: A Case Report
Abstract
Introduction
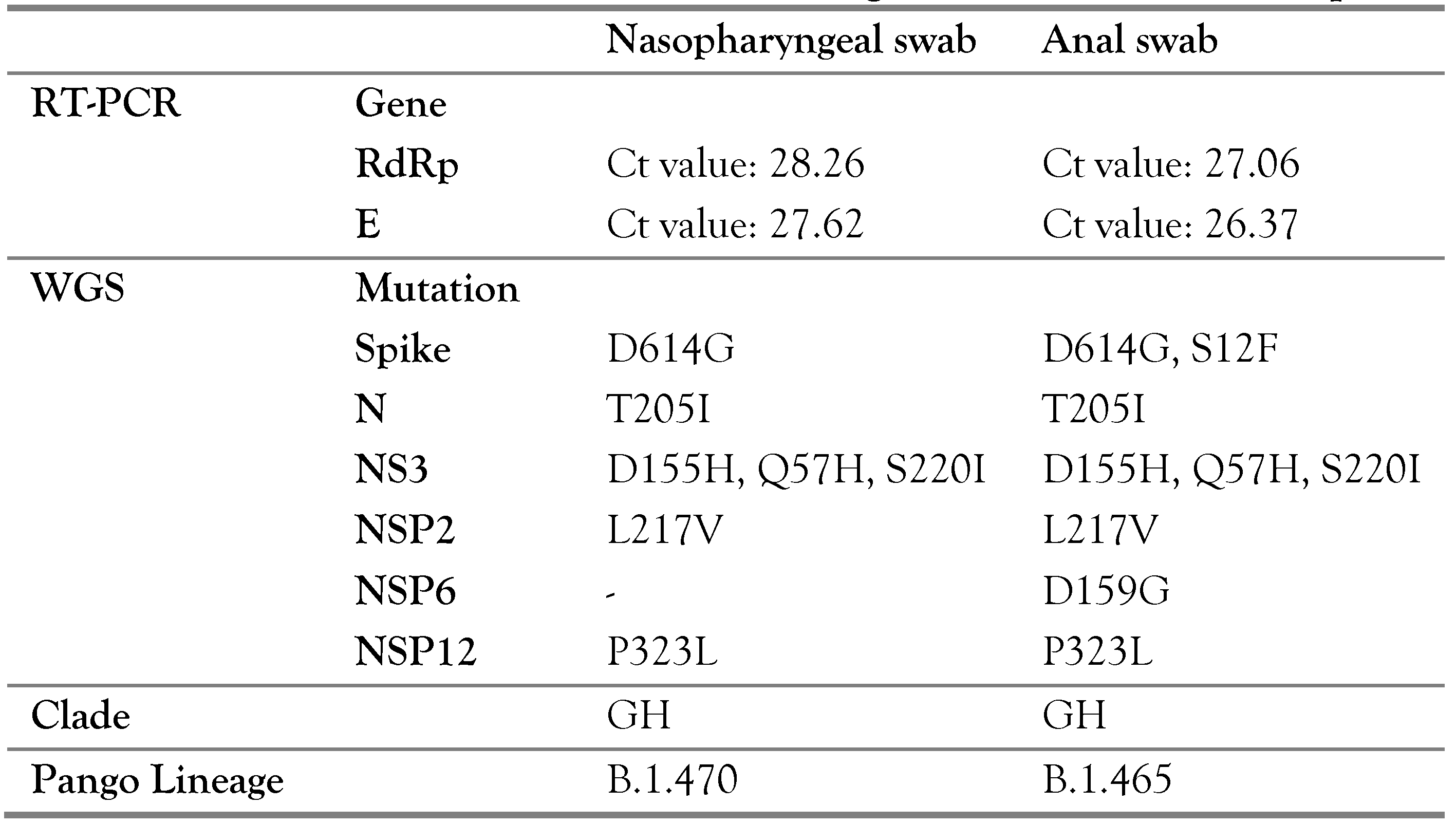
Case Report
Discussion
Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Consent
References
- Dhama, K.; Patel, S.K.; Kumar, R.; et al. Geriatric population during COVID-19 pandemic: Problems, considerations, exigencies and beyond. Front Public Health 2020, 8, 574198. [Google Scholar] [CrossRef] [PubMed]
- Perrotta, F.; Corbi, G.; Mazzeo, G.; et al. COVID-19 and the elderly: Insights into pathogenesis and clinical decision-making. Aging Clin Exp Res. 2020, 32, 1599–1608. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Chen, Y.; Lin, R.; Han, K. Clinical features of COVID-19 in elderly patients: A comparison with young and middle-aged patients. J Infect. 2020, 80, e14–e18. [Google Scholar] [CrossRef] [PubMed]
- Bwire, G.M.; Majigo, M.V.; Njiro, B.J.; Mawazo, A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis. J Med Virol. 2021, 93, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Parasa, S.; Desai, M.; Thoguluva Chandrasekar, V.; et al. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019, a systematic review and meta-analysis. JAMA Netw Open 2020, 3, e2011335. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Wu, C.; Li, X.; et al. Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020, 71, 2255–2258. [Google Scholar] [CrossRef] [PubMed]
- Papoutsis, A.; Borody, T.; Dolai, S.; et al. Detection of SARS-CoV-2 from patient fecal samples by whole genome sequencing. Gut Pathog. 2021, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ren, L.; Zhang, L.; et al. High anal swab viral load predisposes adverse clinical outcomes in severe COVID-19 patients. Emerg Microbes Infect. 2020, 9, 2707–2714. [Google Scholar] [CrossRef] [PubMed]
- Hindson, J. COVID-19: Faecal-oral transmission? Nat Rev Gastroenterol Hepatol. 2020, 17, 259. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Pongor, S.; Győrffy, B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int J Antimicrob Agents 2021, 57, 106272. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.; Choudhary, M.C.; Regan, J.; et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020, 383, 2291–2293. [Google Scholar] [CrossRef] [PubMed]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021, 592, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Avanzato, V.A.; Matson, M.J.; Seifert, S.N.; et al. Case study: Prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020, 183, 1901–1912. [Google Scholar] [CrossRef] [PubMed]

 |
© GERMS 2025.
Share and Cite
Zaini, J.; Putra, A.C.; Ridwanuloh, A.M.; Saniyyah, Z.; Haryanto, B.; Utomo, A.R.H.; Dharmayanthi, A.B.; Prasetyoputri, A.; Andriani, A.; Hariyatun, H.; et al. Possible Intraindividual Evolution of SARS-CoV-2 in Nasopharyngeal and Anal Swab in an Octogenarian: A Case Report. GERMS 2022, 12, 298-303. https://doi.org/10.18683/germs.2022.1332
Zaini J, Putra AC, Ridwanuloh AM, Saniyyah Z, Haryanto B, Utomo ARH, Dharmayanthi AB, Prasetyoputri A, Andriani A, Hariyatun H, et al. Possible Intraindividual Evolution of SARS-CoV-2 in Nasopharyngeal and Anal Swab in an Octogenarian: A Case Report. GERMS. 2022; 12(2):298-303. https://doi.org/10.18683/germs.2022.1332
Chicago/Turabian StyleZaini, Jamal, Andika Chandra Putra, Asep Muhamad Ridwanuloh, Zahrah Saniyyah, Budi Haryanto, Ahmad Rusdan Handoyo Utomo, Anik Budhi Dharmayanthi, Anggia Prasetyoputri, Ade Andriani, Hariyatun Hariyatun, and et al. 2022. "Possible Intraindividual Evolution of SARS-CoV-2 in Nasopharyngeal and Anal Swab in an Octogenarian: A Case Report" GERMS 12, no. 2: 298-303. https://doi.org/10.18683/germs.2022.1332
APA StyleZaini, J., Putra, A. C., Ridwanuloh, A. M., Saniyyah, Z., Haryanto, B., Utomo, A. R. H., Dharmayanthi, A. B., Prasetyoputri, A., Andriani, A., Hariyatun, H., Nuryana, I., Iryanto, S. B., Saputra, S., Wardiana, A., & Ningrum, R. A. (2022). Possible Intraindividual Evolution of SARS-CoV-2 in Nasopharyngeal and Anal Swab in an Octogenarian: A Case Report. GERMS, 12(2), 298-303. https://doi.org/10.18683/germs.2022.1332




