Candida auris and Other Phylogenetically Related Species – A Mini-Review of the Literature
Abstract
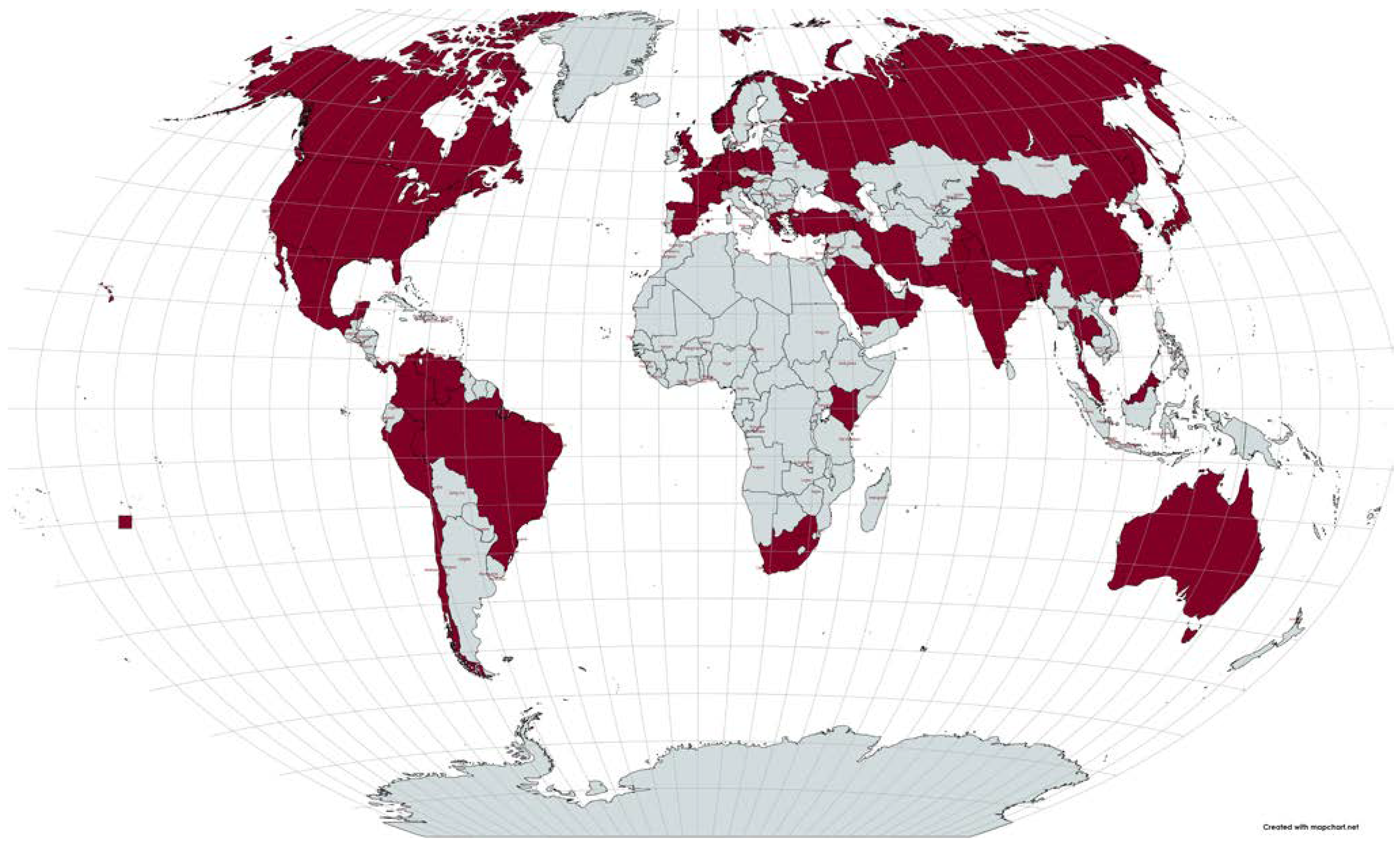
Introduction
Emergence
Virulence
Immune response
Laboratory identification
Resistance
Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Borman, A.M.; Johnson, E.M. Name changes for fungi of medical importance, 2018 to 2019. J Clin Microbiol. 2021, 59, e01811–20. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.U.; Al-Sweih, N.A.; Ahmad, S.; et al. Outbreak of fungemia among neonates caused by Candida haemulonii resistant to amphotericin b, itraconazole, and fluconazole. J Clin Microbiol. 2007, 45, 2025–2027. [Google Scholar] [CrossRef]
- Eyre, D.W.; Sheppard, A.E.; Madder, H.; et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018, 379, 1322–1331. [Google Scholar] [CrossRef]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Lee, W.G.; Shin, J.H.; Uh, Y.; et al. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 2011, 49, 3139–3142. [Google Scholar] [CrossRef] [PubMed]
- Desnos-Ollivier, M.; Fekkar, A.; Bretagne, S. Earliest case of Candida auris infection imported in 2007 in Europe from India prior to the 2009 description in Japan. J Mycol Med. 2021, 31, 101139. [Google Scholar] [CrossRef]
- Chybowska, A.D.; Childers, D.S.; Farrer, R.A. Nine things genomics can tell us about Candida auris. Front Genet. 2020, 11, 351. [Google Scholar] [CrossRef]
- Casadevall, A.; Kontoyiannis, D.P.; Robert, V. On the emergence of Candida auris: Climate change, azoles, swamps, and birds. mBio. 2019, 10, e01397–19. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef]
- Chow, N.A.; de Groot, T.; Badali, H.; Abastabar, M.; Chiller, T.M.; Meis, J.F. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019, 25, 1780–1781. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Berman, J.; Novikov, A.; et al. Multidrug- resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 2017, 23, 195–203. [Google Scholar] [CrossRef]
- Rossato, L.; Colombo, A.L. Candida auris: What have we learned about its mechanisms of pathogenicity? Front Microbiol. 2018, 9, 3081. [Google Scholar] [CrossRef] [PubMed]
- Allaw, F.; Kara Zahreddine, N.; Ibrahim, A.; et al. First Candida auris outbreak during a COVID-19 pandemic in a tertiary-care center in Lebanon. Pathogens. 2021, 10, 157. [Google Scholar] [CrossRef]
- Prestel, C.; Anderson, E.; Forsberg, K.; et al. Candida auris outbreak in a COVID-19 specialty care unit—Florida, July-August 2020. MMWR Morb Mortal Wkly Rep. 2021, 70, 56–57. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, J.N., Jr; Francisco, E.C.; Hagen, F.; et al. Emergence of Candida auris in Brazil in a COVID-19 intensive care unit. J Fungi (Basel). 2021, 7, 220. [Google Scholar] [CrossRef] [PubMed]
- Magnasco, L.; Mikulska, M.; Giacobbe, D.R.; et al. Spread of carbapenem-resistant gram-negatives and Candida auris during the COVID-19 pandemic in critically ill patients: One step back in antimicrobial stewardship? Microorganisms. 2021, 9, 95. [Google Scholar] [CrossRef]
- Schwartz, I.S.; Hammond, G.W. First reported case of multidrug-resistant Candida auris in Canada. Can Commun Dis Rep. 2017, 43, 150–153. [Google Scholar] [CrossRef]
- Villanueva-Lozano, H.; Treviño-Rangel, R.J.; González, G.M.; et al. Outbreak of Candida auris infection in a COVID-19 hospital in Mexico. Clin Microbiol Infect. 2021, 27, 813–816. [Google Scholar] [CrossRef]
- Paucar-Miranda, C.J.; Sandoval-Ahumada, R.E.; López- Martínez, R.L.; Terrel, *!!! REPLACE !!!*; et al. Primer reporte de Candida auris en Perú. An Fac Med. 2021, 82, 56–61. [Google Scholar]
- Barantsevich, N.E.; Orlova, O.E.; Shlyakhto, E.V.; et al. Emergence of Candida auris in Russia. J Hosp Infect. 2019, 102, 445–448. [Google Scholar] [CrossRef]
- Plachouras, D.; Lötsch, F.; Kohlenberg, A.; Monnet, D.L. ; Candida auris survey collaborative group. Candida auris: Epidemiological situation, laboratory capacity and preparedness in the European Union and European Economic Area*, January 2018 to May 2019. Euro Surveill. 2020, 25, 2000240. [Google Scholar] [CrossRef] [PubMed]
- Kurt, A.F.; Kuskucu, M.A. ; Balkan IiI; et al. Candida auris fungemia and a local spread taken under control with infection control measures: First report from Turkey. Indian J Med Microbiol. 2021, 39, 228–230. [Google Scholar] [CrossRef]
- Lone, S.A.; Ahmad, A. Candida auris-the growing menace to global health. Mycoses. 2019, 62, 620–637. [Google Scholar] [CrossRef]
- de Cássia Orlandi Sardi, J.; Silva, D.R.; Soares Mendes- Giannini, M.J.; Rosalen, P.L. Candida auris: Epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb Pathog. 2018, 125, 116–121. [Google Scholar] [CrossRef]
- Ruiz-Gaitán, A.; Moret, A.M.; Tasias-Pitarch, M.; et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018, 61, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Morales-López, S.E.; Parra-Giraldo, C.M.; Ceballos-Garzón, A.; et al. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis. 2017, 23, 162–164. [Google Scholar] [CrossRef]
- Vogelzang, E.H.; Weersink, A.J.L.; van Mansfeld, R. ; The first two cases of Candida auris in the Netherlands. J Fungi (Basel). 2019, 5, 91. [Google Scholar] [CrossRef]
- Kathuria, S.; Singh, P.K.; Sharma, C.; et al. Multidrug- resistant Candida auris misidentified as Candida haemulonii: Characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol. 2015, 53, 1823–1830. [Google Scholar] [CrossRef]
- van Uden, N.; Kolipinski, M.C. Torulopsis haemulonii nov. spec. a yeast from the Atlantic ocean. Antonie van Leeuwenhoek. 1962, 28, 78–80. [Google Scholar] [CrossRef]
- Cendejas-Bueno, E.; Kolecka, A.; Alastruey-Izquierdo, A.; et al. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii Group I), C. duobushaemulonii sp. nov. (C. haemulonii Group II), and C. haemulonii var. vulnera var. nov.: Three multiresistant human pathogenic yeasts. J Clin Microbiol. 2012, 50, 3641–3651. [Google Scholar] [CrossRef] [PubMed]
- Sugita, T.; Takashima, M.; Poonwan, N.; Mekha, N. Candida pseudohaemulonii sp. nov., an amphotericin b- and azole- resistant yeast species, isolated from the blood of a patient from Thailand. Microbiol Immunol. 2006, 50, 469–473. [Google Scholar] [CrossRef]
- Lima, S.L.; Francisco, E.C.; de Almeida Júnior, J.N.; et al. Increasing prevalence of multidrug-resistant Candida haemulonii species complex among all yeast cultures collected by a reference laboratory over the past 11 years. J Fungi (Basel). 2020, 6, 110. [Google Scholar] [CrossRef] [PubMed]
- Gade, L.; Muñoz, J.F.; Sheth, M.; et al. Understanding the emergence of multidrug-resistant Candida: Using whole- genome sequencing to describe the population structure of Candida haemulonii species complex. Front Genet. 2020, 11, 554. [Google Scholar] [CrossRef]
- Kim, M.N.; Shin, J.H.; Sung, H.; et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: Identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 2009, 48, e57–61. [Google Scholar] [CrossRef]
- Ruan, S.Y.; Kuo, Y.W.; Huang, C.T.; Hsiue, H.C.; Hsueh, P.R. Infections due to Candida haemulonii: Species identification, antifungal susceptibility and outcomes. Int J Antimicrob Agents. 2010, 35, 85–88. [Google Scholar] [CrossRef]
- Pérez-Lazo, G.; Morales-Moreno, A.; Soto-Febres, F.; Hidalgo, J.A.; Neyra, E.; Bustamante, B. Liver abscess caused by Candida haemulonii var. vulnera. First case report in Peru. Rev Iberoam Micol. 2021, 38, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Frías-De-León, M.G.; Martínez-Herrera, E.; Acosta- Altamirano, G.; Arenas, R.; Rodríguez-Cerdeira, C. Superficial candidosis by Candida duobushaemulonii: An emerging microorganism. Infect Genet Evol. 2019, 75, 103960. [Google Scholar] [CrossRef]
- Kumar, A.; Prakash, A.; Singh, A.; et al. Candida haemulonii species complex: An emerging species in India and its genetic diversity assessed with multilocus sequence and amplified fragment-length polymorphism analyses. Emerg Microbes Infect. 2016, 5, e49. [Google Scholar] [CrossRef] [PubMed]
- Saluja, P.; Prasad, G.S. Candida ruelliae sp. nov. , a novel yeast species isolated from flowers of Ruellia sp. (Acanthaceae). FEMS Yeast Res. 2008, 8, 660–666. [Google Scholar] [CrossRef]
- Larkin, E.; Hager, C.; Chandra, J.; et al. The emerging pathogen Candida auris: Growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob Agents Chemother. 2017, 61, e02396–16. [Google Scholar] [CrossRef]
- Chatterjee, S.; Alampalli, S.V.; Nageshan, R.K.; Chettiar, S.T.; Joshi, S.; Tatu, U.S. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics. 2015, 16, 686. [Google Scholar] [CrossRef]
- Muñoz, J.E.; Ramirez, L.M.; Dias, L.D.S.; et al. Pathogenicity levels of Colombian strains of Candida auris and Brazilian strains of Candida haemulonii species complex in both murine and Galleria mellonella experimental models. J Fungi (Basel). 2020, 6, 104. [Google Scholar] [CrossRef]
- Romera, D.; Aguilera-Correa, J.J.; Gadea, I.; Viñuela- Sandoval, L.; García-Rodríguez, J.; Esteban, J. Candida auris: A comparison between planktonic and biofilm susceptibility to antifungal drugs. J Med Microbiol. 2019, 68, 1353–1358. [Google Scholar] [CrossRef]
- Nett, J.E. Candida auris: An emerging pathogen “incognito”? PLoS Pathog. 2019, 15, e1007638. [Google Scholar] [CrossRef] [PubMed]
- Day, A.M.; McNiff, M.M.; da Silva Dantas, A.; Gow, N.A.R.; Quinn, J. Hog1 regulates stress tolerance and virulence in the emerging fungal pathogen Candida auris. mSphere. 2018, 3, e00506–18. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Arias, M.J.; Hernández-Chávez, M.J.; García- Carnero, L.C.; et al. Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect Drug Resist. 2019, 12, 783–794. [Google Scholar] [CrossRef]
- Bruno, M.; Kersten, S.; Bain, J.M.; et al. Transcriptional and functional insights into the host immune response against the emerging fungal pathogen Candida auris. Nat Microbiol. 2020, 5, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.; Heyl, J.; Babu, B.A. Candida auris in the age of resistance. Cureus. 2020, 12, e10334. [Google Scholar] [CrossRef]
- Jackson, B.R.; Chow, N.; Forsberg, K.; et al. On the origins of a species: What might explain the rise of Candida auris? J Fungi (Basel). 2019, 5, 58. [Google Scholar] [CrossRef]
- Kumar, A.; Sachu, A.; Mohan, K.; Vinod, V.; Dinesh, K.; Karim, S. Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal’s medium. Rev Iberoam Micol. 2017, 34, 109–111. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Prakash, A.; Meletiadis, J.; Sharma, C.; Chowdhary, A. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother. 2017, 61, e00485–17. [Google Scholar] [CrossRef] [PubMed]
- Rudramurthy, S.M.; Chakrabarti, A.; Paul, R.A.; et al. Candida auris candidaemia in Indian ICUs: Analysis of risk factors. J Antimicrob Chemother. 2017, 72, 1794–801. [Google Scholar] [CrossRef] [PubMed]
- Černáková, L.; Roudbary, M.; Brás, S.; Tafaj, S.; Rodrigues, C.F. Candida auris: A quick review on identification, current treatments, and challenges. Int J Mol Sci. 2021, 22, 4470. [Google Scholar] [CrossRef]
- Lockhart, S.R. Candida auris and multidrug resistance: Defining the new normal. Fungal Genet Biol. 2019, 131, 103243. [Google Scholar] [CrossRef]
- Li, J.; Coste, A.T.; Liechti, M.; Bachmann, D.; Sanglard, D.; Lamoth, F. Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob Agents Chemother. 2021, 65, e02663–20. [Google Scholar] [CrossRef]
- Mayr, E.M.; Ramírez-Zavala, B.; Krüger, I.; Morschhäuser, J. A zinc cluster transcription factor contributes to the intrinsic fluconazole resistance of Candida auris. mSphere. 2020, 5, e00279–20. [Google Scholar] [CrossRef] [PubMed]
- Rybak, J.M.; Muñoz, J.F.; Barker, K.S.; et al. Mutations in TAC1B: A novel genetic determinant of clinical fluconazole resistance in Candida auris. mBio. 2020, 11, e00365–20. [Google Scholar] [CrossRef]
- Rhodes, J.; Abdolrasouli, A.; Farrer, R.A.; et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018, 7, 43. [Google Scholar] [CrossRef]
- Carolus, H.; Pierson, S.; Muñoz, J.F.; et al. genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. mBio. 2021, 12, e03333–20. [Google Scholar] [CrossRef]
- Sherry, L.; Ramage, G.; Kean, R.; et al. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef]
- Ramos, R.; Caceres, D.H.; Perez, M.; et al. Emerging multidrug-resistant Candida duobushaemulonii infections in Panama hospitals: Importance of laboratory surveillance and accurate identification. J Clin Microbiol. 2018, 56, e00371–18. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2021.
Share and Cite
Ciurea, C.N.; Mare, A.D.; Kosovski, I.-B.; Toma, F.; Vintilă, C.; Man, A. Candida auris and Other Phylogenetically Related Species – A Mini-Review of the Literature. GERMS 2021, 11, 441-448. https://doi.org/10.18683/germs.2021.1281
Ciurea CN, Mare AD, Kosovski I-B, Toma F, Vintilă C, Man A. Candida auris and Other Phylogenetically Related Species – A Mini-Review of the Literature. GERMS. 2021; 11(3):441-448. https://doi.org/10.18683/germs.2021.1281
Chicago/Turabian StyleCiurea, Cristina Nicoleta, Anca Delia Mare, Irina-Bianca Kosovski, Felicia Toma, Camelia Vintilă, and Adrian Man. 2021. "Candida auris and Other Phylogenetically Related Species – A Mini-Review of the Literature" GERMS 11, no. 3: 441-448. https://doi.org/10.18683/germs.2021.1281
APA StyleCiurea, C. N., Mare, A. D., Kosovski, I.-B., Toma, F., Vintilă, C., & Man, A. (2021). Candida auris and Other Phylogenetically Related Species – A Mini-Review of the Literature. GERMS, 11(3), 441-448. https://doi.org/10.18683/germs.2021.1281