Abstract
Introduction: The aim of this study was to assess the clinical performance of different automated immunoassays available in Europe to detect anti-SARS-CoV-2 antibodies; an ELISA assay and a CLIA. The second goal was to estimate the seroprevalence of SARS-CoV-2 antibodies among healthcare workers in Evros area during the first pandemic wave of COVID-19. Methods: The study included serum samples from 101 patients with confirmed COVID-19 by RT-PCR and 208 negative patients. Furthermore, it included 1036 healthcare workers (HWs) of the Evros Region, Northern Greece. The measurement of anti-SARS-CoV-2 antibodies was performed using the Abbott SARS-CoV-2 IgG and anti-SARS-CoV-2 ELISA IgG assay (Epitope Diagnostics, USA). Results: Of 101 confirmed COVID-19 patients, 82 were hospitalized and 19 were outpatients. Hospitalized patients had higher IgG levels in comparison to outpatients (6.46±2.2 vs. 3.52±1.52, p<0.001). Of 208 non−COVID-19 patients only 1 was positive in both ELISA and CLIA assay. SARS-CoV-2-IgG antibodies were detected in 6 HWs out of 1036 (0.58%) with mean S/CO-value of anti-SARS-CoV-2 IgG 3.12±1.3 (confidence interval 0.95), which was lower than in COVID-19 patients (3.12 vs. 5.9; p=0.016). The clinical evaluation of two immunoassays showed remarkably high true positivity rates in the confirmed COVID-19 patients. Sensitivities obtained with CLIA and ELISA methods were 99.02% vs. 97.09% and specificities 99.52% vs 99.05% respectively. Conclusions: We found an acceptable accordance between CLIA and ELISA assays in the confirmed COVID-19 patients. In all subjects included in this study in the past medical history, the information that was obtained included details about the presence of autoimmune diseases.
Keywords:
COVID-19; Anti-SARS-CoV-2 Antibodies; SARS-CoV-2; Diagnosis; RT-PCR; Global Health Problem Introduction
The novel coronavirus disease (COVID-19) due to the severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) was first reported as cluster of cases in Wuhan, China in December 2019 [1]. On 11 March 2020, the World Health Organization characterized the global health emergency of COVID-19 as a pandemic. As of 14 May 2021, over 160 million SARS-CoV-2 infections cases and over 3 million deaths have been reported globally [2].
Laboratory diagnosis is based on detection of viral RNA by real time reverse transcriptase-polymerase chain reaction (RT-PCR) in nasopharyngeal (NP) or oropharyngeal (OP) swabs and bronchoalveolar lavage (BAL) samples [3]. This method is considered as the gold standard method for screening and diagnosis [4]. The infected people develop specific antibodies to SARS-CoV-2 after the second week of infection. Antibodies measurement is an extremely helpful tool in assessing COVID’s course as well as response to vaccinations. In view of the vague course and outcome of the disease additional information by calculating the immune response could be of significant value. In contrast to the molecular (RT-PCR) tests designed to directly detect SARS-CoV-2 RNA during the acute phase of disease, serological tests measure antibodies that remain detectable after acute infection [5,6]. Moreover, development of new serological tests could be helpful as a complementary diagnostic tool and to increase the sensitivity of diagnostic tests. Different assays have recently been developed to for the detection of antibodies. Currently, automated tests such as chemiluminescence enzyme immunoassays (CLIA) and enzyme-linked immunosorbent assays (ELISA) or rapid detection tests, lateral flow immunoassays (LFIA) are available [6,7].
As previously reported, the main route of transmission for SARS-CoV-2 is person-to-person spread [8,9]. Healthcare workers (HWs) are at high risk for infection due to frequent and close contact to COVID-19 patients [9]. Understanding the risk for SARS-CoV-2 infection among these frontline healthcare personnel is fundamental for designing and executing the national planning to the COVID-19 pandemic.
The aim of this study was to assess the clinical performance of different assays available in Europe (CE marked) to detect SARS-CoV-2 antibodies: immunoassays targeting two different proteins: qualitative ELISA assay (Epitope Diagnostics USA – EDI) and semiquantitative chemiluminescent microparticle immunoassay (Abbott, USA). The second goal was to estimate the seroprevalence of SARS-CoV-2 antibodies (Abs) among HWs in Evros area, Northern Greece, during the first pandemic wave of COVID-19 and explore risk factors among HWs with positive results.
Methods
Study design
This study was performed at the University General Hospital of Alexandroupolis and Democritus University of Thrace in Alexandroupolis, Greece. The study protocol was approved by the local committee of ethics and deontology in accordance with the Declaration of Helsinki (Number 17240/5-6-2020).
Study population
The study groups consisted of 101 patients with confirmed COVID-19 by qualitative RT real-time PCR (qRT-PCR) and 208 COVID-19 negative patients. More specifically, the COVID-19 positive patients consisted of 82 hospitalized and 19 outpatients. The criteria for admission to the hospital were: oxygen saturation ≤92%, respiratory distress, severe critical condition, multiorgan involvement, and comorbidities (diabetes mellitus, cardiovascular diseases, transplanted, and immunocompromised). The 208 COVID-19 negative patients were 31 with fever of unknown origin (FUO) and 177 patients with samples obtained before the COVID-19 pandemic. These 177 patients were distributed as following: 81 patients with systemic lupus erythematosus (SLE), 43 with rheumatoid arthritis (RA) and 53 patients with other autoimmune disorders like systemic sclerosis (SSc) and Sjogren's syndrome, ANCA-associated vasculitis (AAV), ankylosing spondylarthritis (AS) and antiphospholipid syndrome (APS). Furthermore, HWs of the University General Hospital of Alexandroupolis (Hospital 1), as well as workers from General Hospital of Didymoteicho (Hospital 2), Healthcare Centers of Alexandroupolis (HC 1), Orestiada (HC 2) Sufli, (HC 3), Dikaia (HC 4) and Samothraki (HC 5) (Figure 1), as high-risk group were also recruited in this study. HWs who were ill or quarantined during the study period were excluded. Healthcare workers were divided into two groups. A high-risk group included workers who had regular direct contact with adult patients with COVID-19 at the emergency department, intensive care unit (ICU), and a specialized inpatient COVID-19 unit. A low-risk group included healthcare workers from Healthcare Centers, as well as other departments and the administrative staff of the two hospitals (Alexandroupolis and Didymoteicho).
Figure 1.
Location of all the participating centers. HC – health center.
All clinical and epidemiological data for COVID-19 positive patients are included in the present study. The period of sampling was June - July 2020.
IgG testing
CLIA assay
Detection of anti-SARS-CoV-2 antibodies was performed using the Abbott Architect i1000SR instrument (Abbott Diagnostics Division, USA) using the Abbott SARS-CoV-2 IgG kit after FDA notification and following manufacturer’s instructions. The assay is a chemiluminescent microparticle immunoassay for qualitative detection of anti-SARS-CoV-2 Abs type IgG against the CoV-2 nucleoprotein (Np) in human serum. Qualitative results are reported as an index value, and results <1.4 were non-reactive or negative and ≥1.4 were positive or reactive in accordance with the Abbott-determined positivity cutoff of 1.40.
ELISA
The Epitope Diagnostics (EDI), anti-SARS-CoV-2 ELISA IgG assay (Epitope Diagnostics, USA) was used according to the manufacturer’s guidelines on the DS2® system, an automated microplate technology (Dynex Technologies GmbH, Germany). The microplate wells are coated with recombinant S1 structural protein, and the assay detects anti-SARS-CoV-2 IgG against the viral recombinant protein. ELISA positivity cutoff was calculated according to manufacturer's instructions. More specifically, positive cutoff (PC) was calculated using the following formula: PC = 1.1 X (x NC-Negative Control + 0.18), where x refers to the number of negative control’s runs. The PC value was 0.46 in this study, with x equal to 3 for all our specimens. qRT-PCR nucleic acid SARS-CoV-2 testing was performed in HWs with titer of IgG >0.9S/CO according to CLIA assay. Moreover, 10% of HWs with negative titer of IgG were also tested for nucleic acid SARS-CoV-2 by molecular methods. Viral RNA from the nasopharyngeal samples (NPS) was extracted by using the Viral 1RNA/DNA Nucleospin Virus kit, (Macherey-Nagel, Germany). 200 μL of NPS sample were used for RNA extraction. The qRT-PCR assay was performed by RT real time PCR kit (Primerdesign Ltd, UK) and run by SaCycler-96 Real Time PCR System (Sacace Biotechnologies, Italy). For each run, positive and negative controls as well as internal control for extraction were included.
Statistical analysis
Continuous variables were presented as mean ± standard deviation (SD) for normally distributed data. The counting data were expressed by rate (%). The independent sample T-test was used to detect the data between two groups. The p value <0.05 indicated a statistically significant difference. All statistical analyses were performed using Origin Pro 8 (OriginLab Northampton, USA).
Results
From 101 qRT-PCR confirmed COVID-19 patients, 82 were hospitalized and 19 were outpatients, based on disease severity.
Demographic, clinical and laboratory data of qRT-PCR confirmed COVID-19 patients is shown in Table 1. Hospitalized patients had statistically significant higher IgG levels in comparison to outpatients (6.46±2.2 vs 3.52±1.52, p<0.001) (Figure 2A).

Table 1.
Demographical, clinical and laboratory data of patients with COVID-19.
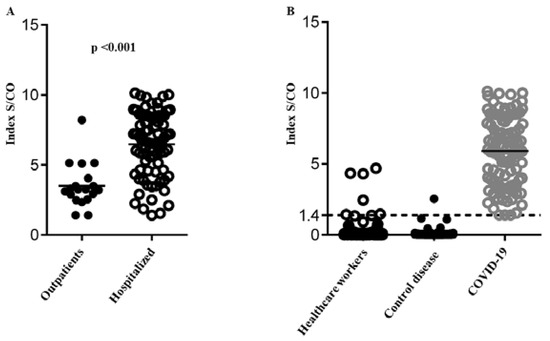
Figure 2.
Mean S/CO-value of anti-SARS-CoV-2 IgGs. 2A: IgG level of hospitalized patients in comparison to outpatients. 2B: IgG level of health workers, control group – patients with autoimmune disorders and patients – COVID-19 patients.
From 208 non-COVID-19 patients, none was positive by the qRT-PCR test and only 1 was positive by both ELISA and CLIA antibody assay. This patient suffered from SLE and the blood sample had been collected during the exacerbation of SLE.
Sensitivity and specificity of two SARS-CoV-2 IgG assays
The clinical evaluation of two immunoassays showed remarkably high true positivity rates in the confirmed COVID-19 patients. Sensitivities and specificities obtained by the two methods are summarized in Table 2. Specifically, only one false positive result was detected by CLIA assay, which corresponded to a pre-pandemic specimen from a patient with SLE. Two false positive results were observed with IgG ELISA: one sample from a patient with SLE and one with negative qRT-PCR result and negative IgG result with CLIA assay.

Table 2.
Performance of CLIA and ELISA assays.
SARS-CoV-2 IgG Abs in healthcare workers
Overall, 1036 healthcare workers (HWs) were enrolled in the study (58.36% participation rate) – Table 3. The mean age of participants was 45.6±2.7 years, and 787 (75.9%) were women. Only 13 HWs had a history of a travel abroad in the past 3 months – Table 4. SARS-CoV-2-IgG antibodies were detected in 6 HWs out of 1036 (0.58%) with mean S/CO-value of anti-SARS-CoV-2 IgG 3.12±1.3 (confidence interval 0.95), which was lower than the mean S/CO-value of COVID-19 patients (3.12 vs. 5.9; p=0.016) – Figure 2B. The overall mean S/CO-value of anti-SARS-CoV-2 IgG in HWs was 0.065 (0.008-1.305) and did not differ significantly between high and low risk groups. Among subjects with positive IgGs, 4/6 (66.6%) reported to have suffered from COVID-19 associated symptoms before 1st of May 2020. Serum samples of HWs for the prevalence of anti-SARS-CoV-2 Abs were tested with CLIA and ELISA. There was no big discrepancy with both techniques. Unfortunately, a RT-PCR was not obtained at that time, because their symptoms were underestimated, during the disease, confirming the saying that HWs are the most disobedient patients, and the samples were obtained in convalescence. The most prevalent symptoms were headache, sore throat, cough, rhinitis, and muscle/joint pain. Only two subjects reported fever, while diarrhea or nausea/vomiting were not reported.

Table 3.
Participation rate.

Table 4.
Demographical data of health workers from the study population.
PCR of a nasopharyngeal swab was performed in 5 out of the 6 antibody-positive cases and was negative. Furthermore, in 10% of HWs that were negative for anti-SARS-CoV-2 antibodies, qRT-PCR for SARS-CoV-2 RNA was also negative. In 10 participants (0.97%) with negative qRT-PCR, the mean S/CO-value of anti-SARS-CoV-2 IgG was found to be between 0.5 to
1.37 by CLIA assay, that is characterized as borderline. The borderline result of antibodies was confirmed from a second sample, that was obtained after 1 week (0.905±0.31 vs. 0.833±0.34, p=0.634).
Discussion
The study was performed after the first wave of SARS-CoV-2, and before the second wave in Greece, from June 2020 to August 2020.
The serological assays used in clinical laboratories to evaluate infection, complementary to qRT-PCR, are ELISA and CLIA methods. Various CE marked assays are available in European Countries [10]. Given the low sensitivity, the antibody-based rapid test is not adequate for public health measures such as community screenings [11,12]. In this article, we evaluated two different CE marked commercial immunoassays for the detection of anti-SARS-CoV-2 antibodies in human serum. CLIA assay is a fully automated random-access test. In contrast, ELISA assay was performed on a 96 microplate technology requiring high handling. Overall, we found an acceptable accordance between the Abbott CLIA assay and the EDI ELISAs (IgG) in the confirmed COVID-19 patients. In addition, Kontou et al., in their meta-analysis report that ELISA-and CLIA-based methods have high sensitivity (90%-94%) in comparison to LFIA methods [13]. Furthermore, Coste et al., in their recent article tried to evaluate 17 SARS-CoV-2 serological tests. Between others, ELISA kit (Epitope Diagnostic) and CLIA kit (Abbott) were recommended with a combined sensitivity above 90% and specificity above 98% [14].
Anti-SARS-CoV-2 Abs in pre-pandemic sera from a patient suffering from SLE were also detected. This result indicates that there were cross reactions with non-SARS population to detect SARS-CoV-2 antibody by ELISA and CLIA in patients with autoimmune diseases. Previous studies reported that autoantibodies and specific antibodies against pathogens' antigens may cross-react. Our result is in line with the results reported by Wang et al., during the epidemic outbreak of SARS-CoV-1, in 2004, which verified that in patients with autoimmune diseases autoantibodies cross-react with viral antigens (coronavirus) [15]. Durlik-Popińska et al. demonstrated that antibodies isolated from RA patients' sera reacted stronger with the collagen. Additionally, cross-reactivity among Proteus mirabilis lipopolysaccharides was observed [16]. Moreover, Singh et al., who have generated two monoclonal antibodies (mAbs) against Epstein Barr’s viral nuclear antigen I (EBNA-1), demonstrated that these Abs cross-react with double-stranded DNA (dsDNA) [17].
The seroprevalence of SARS-CoV-2 in HWs in this study was 0.58% (6/1036). This result is in line with previous studies which reported than the prevalence of IgG antibodies in HWs was between 0.5% and 7.6% [18,19,20,21]. Moreover, Galanis et al., in their meta-analysis, reported that seroprevalence among studies ranged between 0% and 45.3% [22]. Mughal et al. reported that the prevalence in a high-risk group of HW who worked in ICU was 0.83% [18]. Stubblefield et al. showed that among 249 HWs who worked in hospital units with COVID-19 patients, 19 (7.6%) tested positive for SARS-CoV-2 antibodies. Moreover, this study indicated that 8 subjects (42%) were asymptomatic, suggesting asymptomatic healthcare personnel could be an important source of SARS-CoV-2 transmission. In addition, the seropositivity among HWs was more common in those who were not universally using personal protective equipment [21]. Furthermore, Chen et al. reported a high prevalence of seropositivity among the workers who were exposed to patients with laboratory confirmed COVID-19 [23]. The low level of seropositivity among the HWs of Evros area can be explained by the good epidemiological status in this area during the first pandemic wave. A limitation of the study is the relatively small group of patients with asymptomatic infection and positive RT-PCR. Moreover, the exclusion of HWs who were ill or quarantined during the study period may have led to an underestimation of SARS-CoV-2 seroprevalence.
Conclusions
In conclusion, in this study, we demonstrated that the two methods (CLIA and ELISA) for detecting anti-SARS-CoV-2 Abs are concordant. Secondly, a patient with high immune activity (autoimmune disease) had autoantibodies that can cross-react with SARS-CoV-2 antigens and falsely give the impression of positive Abs. Thirdly, HWs in Thrace region were found to have a remarkable low level of anti-SARS-CoV-2 antibodies, that is quite satisfying, reflecting the personal measures that they apply. Nevertheless, the presence of positive Abs in asymptomatic HWs reminds us about the many faces of this devious disease and reinforces about the need for strong and continuous personal measures by HWs, who can otherwise silently transmit the virus to vulnerable patients and colleagues.
Author Contributions
Conceptualization, methodology, and validation: TK. Laboratory investigation: SZ, KP, EL, EF, AG, TG, EGK. and IN. Clinical data curation: IM, NL, VP, PS, PP and DC. Writing – original draft preparation: TK, IM. Writing – review and editing: CT, DC, TK and IM. Supervision: GM and MP. All authors read and approved the final version of the manuscript.
Funding
None to declare.
Conflicts of interest
All authors – none to declare.
References
- Zhu, N.; Zhang, D.; Wang, W.; et al. A novel coronavirus from patients with pneumonia in china, 2019. N Engl J Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. 2021. Available online: https://covid19.who.int (accessed on 14 May 2021).
- Moore, A.J.; Nakahata, M.I.; Kalinich, C.C.; et al. The sensitivity of respiratory tract specimens for the detection of SARS-CoV-2: A protocol for a living systematic review and meta-analysis. medRxiv. 2020. [Google Scholar] [CrossRef]
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef]
- Egger, M.; Bundschuh, C.; Wiesinger, K.; et al. Comparison of the Elecsys® Anti-SARS-CoV-2 immunoassay with the EDITM enzyme linked immunosorbent assays for the detection of SARS-CoV-2 antibodies in human plasma. Clin Chim Acta. 2020, 509, 18–21. [Google Scholar] [CrossRef]
- Nicol, T.; Lefeuvre, C.; Serri, O.; et al. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech). J Clin Virol. 2020, 129, 104511. [Google Scholar] [CrossRef]
- Andrey, D.O.; Cohen, P.; Meyer, B.; et al. Diagnostic accuracy of Augurix COVID-19 IgG serology rapid test. Eur J Clin Invest. 2020, 50, e13357. [Google Scholar] [CrossRef]
- Iversen, K.; Bundgaard, H.; Hasselbalch, R.B.; et al. Risk of COVID-19 in health-care workers in Denmark: An observational cohort study. Lancet Infect Dis. 2020, 20, 1401–1408. [Google Scholar] [CrossRef]
- Koh, D. Occupational risks for COVID-19 infection. Occup Med (Lond). 2020, 70, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Kohmer, N.; Westhaus, S.; Rühl, C.; Ciesek, S.; Rabenau, H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J Clin Virol. 2020, 129, 104480. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Döhla, M.; Boesecke, C.; Schulte, B.; et al. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health 2020, 182, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Kontou, P.I.; Braliou, G.G.; Dimou, N.L.; Nikolopoulos, G.; Bagos, P.G. Antibody tests in detecting SARS-CoV-2 infection: A meta-analysis. Diagnostics (Basel). 2020, 10, 319. [Google Scholar] [CrossRef]
- Coste, A.T.; Jaton, K.; Papadimitriou-Olivgeris, M.; Greub, G.; Croxatto, A. Comparison of SARS-CoV-2 serological tests with different antigen targets. J Clin Virol. 2021, 134, 104690. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, S.; Shen, H.; et al. Cross-reaction of SARS-CoV antigen with autoantibodies in autoimmune diseases. Cell Mol Immunol. 2004, 1, 304–307. [Google Scholar]
- Durlik-Popińska, K.; Żarnowiec, P.; Lechowicz, Ł.; Gawęda, J.; Kaca, W. Antibodies isolated from rheumatoid arthritis patients against lysine-containing Proteus mirabilis O3 (S1959) lipopolysaccharide may react with collagen type I. Int J Mol Sci. 2020, 21, 9635. [Google Scholar] [CrossRef]
- Singh, D.; Oudit, O.; Hajtovic, S.; et al. Antibodies to an Epstein Barr Virus protein that cross-react with dsDNA have pathogenic potential. Mol Immunol 2021, 132, 41–52. [Google Scholar] [CrossRef]
- Mughal, M.S.; Kaur, I.P.; Patton, C.D.; Mikhail, N.H.; Vareechon, C.; Granet, K.M. The prevalence of severe acute respiratory coronavirus virus 2 (SARS-CoV-2) IgG antibodies in intensive care unit (ICU) healthcare personnel (HCP) and its implications-a single-center, prospective, pilot study. Infect Control Hosp Epidemiol 2021, 42, 638–639. [Google Scholar] [CrossRef]
- Schmidt, S.B.; Grüter, L.; Boltzmann, M.; Rollnik, J.D. Prevalence of serum IgG antibodies against SARS-CoV-2 among clinic staff. PLoS ONE. 2020, 15, e0235417. [Google Scholar] [CrossRef]
- Xu, X.; Sun, J.; Nie, S.; et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med 2020, 26, 1193–1195. [Google Scholar] [CrossRef]
- Stubblefield, W.B.; Talbot, H.K.; Feldstein, L.; et al. Seroprevalence of SARS-CoV-2 among frontline healthcare personnel during the first month of caring for COVID-19 patients - Nashville, Tennessee. Clin Infect Dis. 2021, 72, 1645–1648. [Google Scholar] [CrossRef] [PubMed]
- Galanis, P.; Vraka, I.; Fragkou, D.; Bilali, A.; Kaitelidou, D. Seroprevalence of SARS-CoV-2 antibodies and associated factors in healthcare workers: A systematic review and meta-analysis. J Hosp Infect. 2021, 108, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tong, X.; Wang, J.; et al. High SARS-CoV-2 antibody prevalence among healthcare workers exposed to COVID-19 patients. J Infect. 2020, 81, 420–426. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2021.