Comparative Transcriptome Analysis of Bombyx mori (Lepidoptera) Larval Hemolymph in Response to Autographa californica Nucleopolyhedrovirus in Differentially Resistant Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Silkworm, AcMNPV, and Sample Preparation
2.2. Library Construction, Illumina Sequencing, and Read Assembly
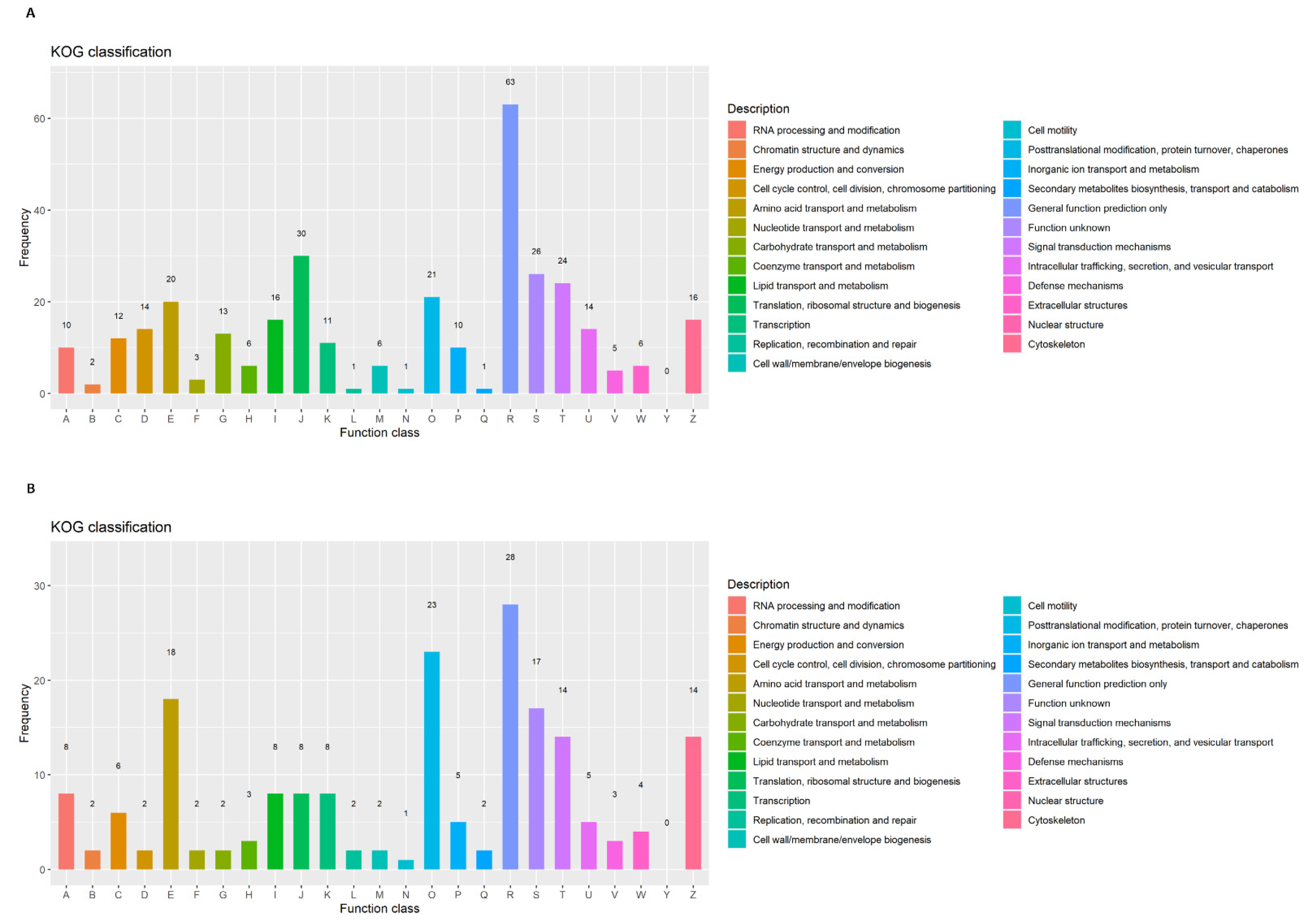
2.3. Functional Annotation and Enrichment Analysis
2.4. Identification of Differentially Expressed Genes (DEGs)
2.5. RNA Extraction, First-Strand cDNA Synthesis, and Real-Time Quantitative PCR (RT-qPCR)
2.6. Construction of pIZT-mCherry-BmTex261 Overexpression Vector
2.7. Synthesis of siRNA
2.8. BmN Cell Culture and Transfection
2.9. Statistical Analysis
3. Results
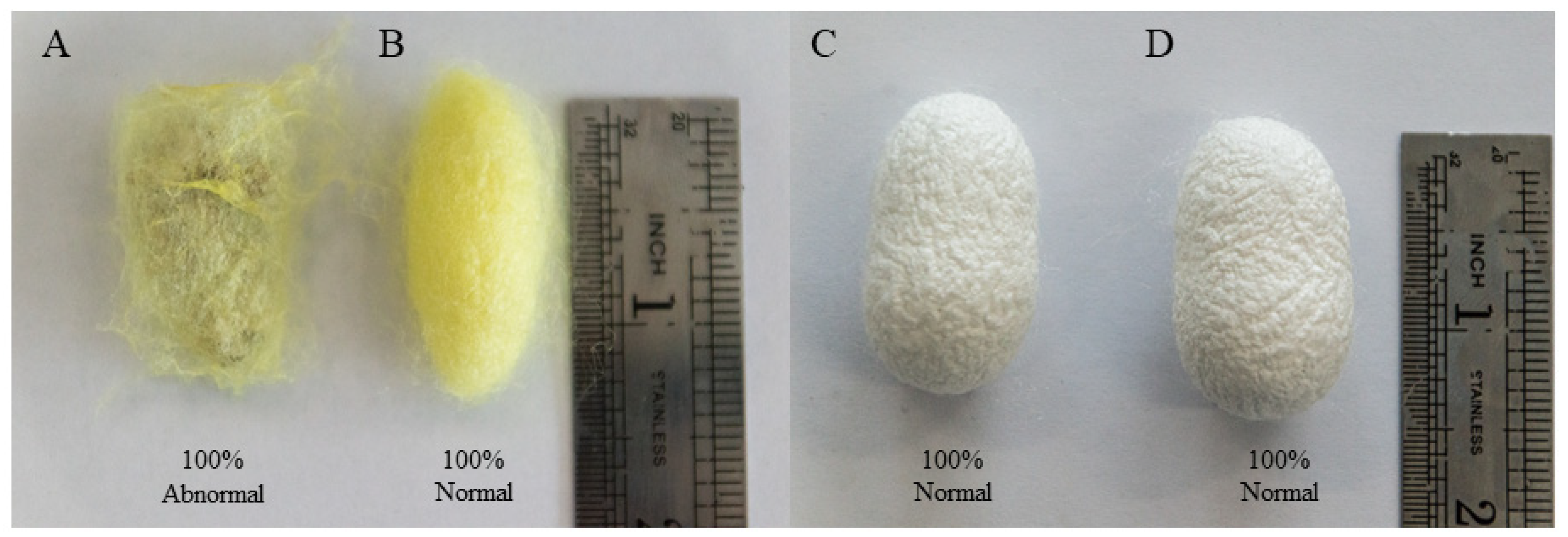
3.1. Analysis of AcMNPV Infection in Differentially Resistant Strains
3.2. Overview of the Transcriptome of Silkworm Hemolymph
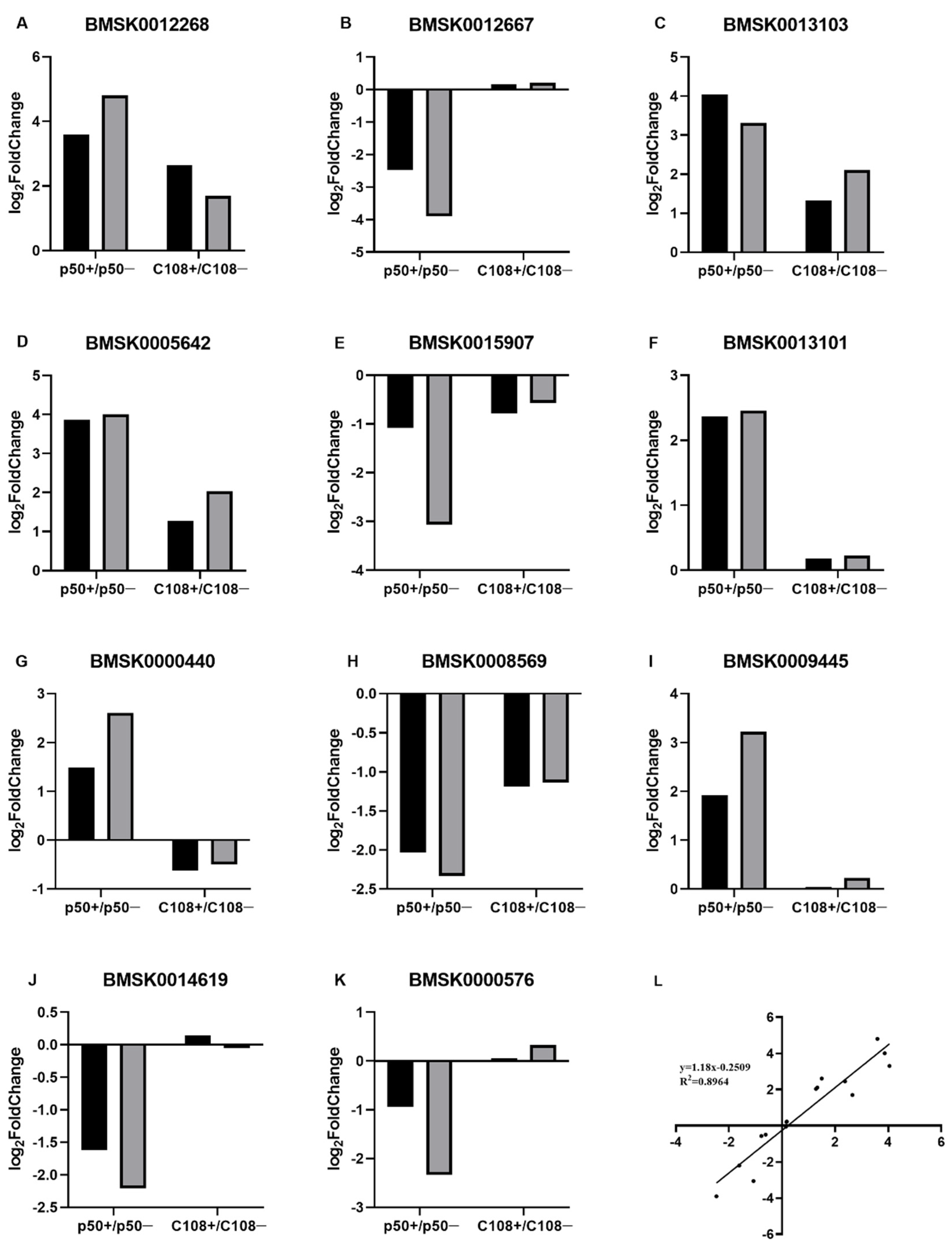
3.3. RT-qPCR Validation of Differentially Expressed Transcripts
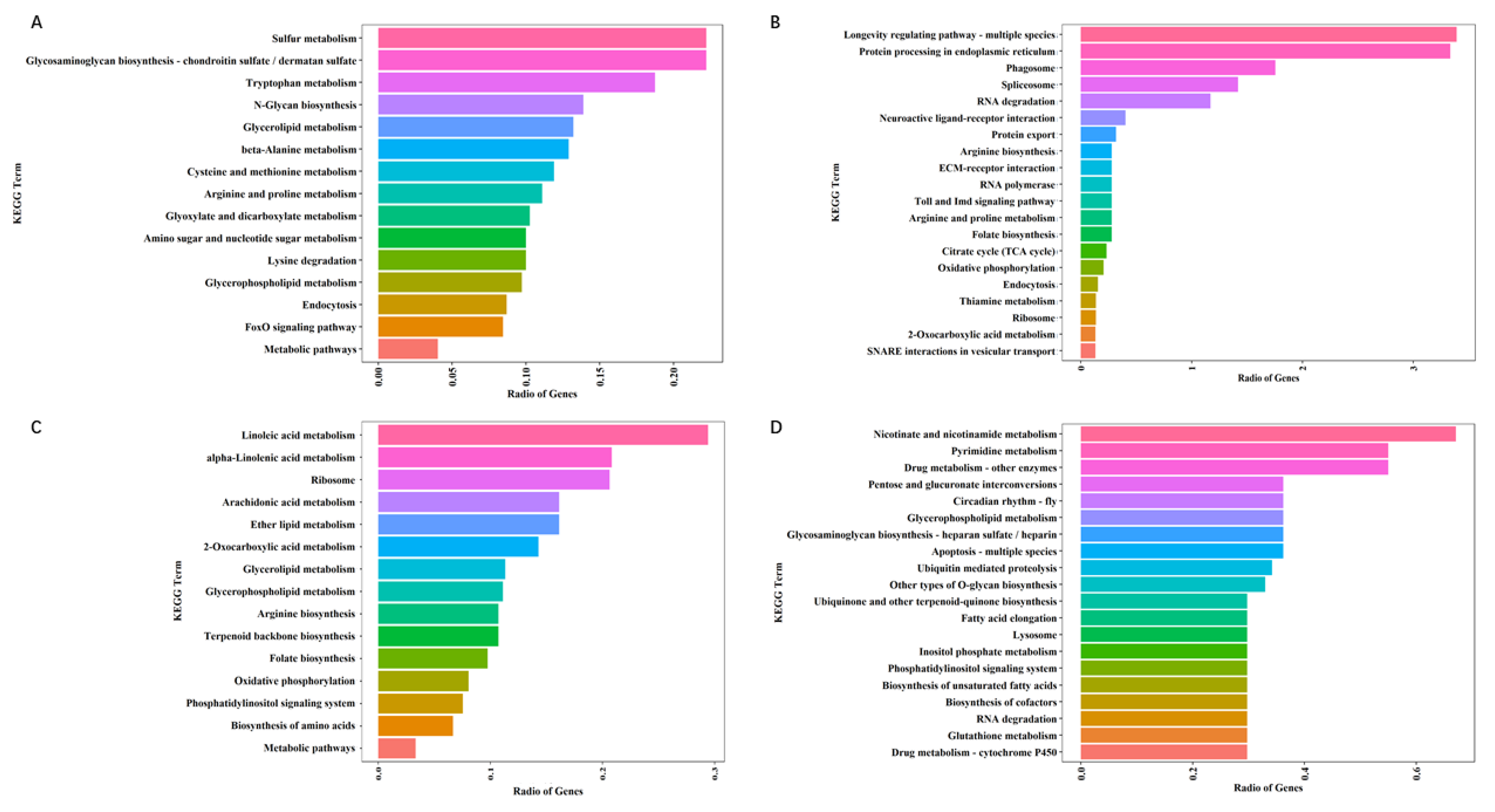
3.4. Detection and Enrichment Analysis of DEGs
3.5. DEGs Involved in Metabolic and Apoptosis Pathways Showed Significant Responses to AcMNPV Infection
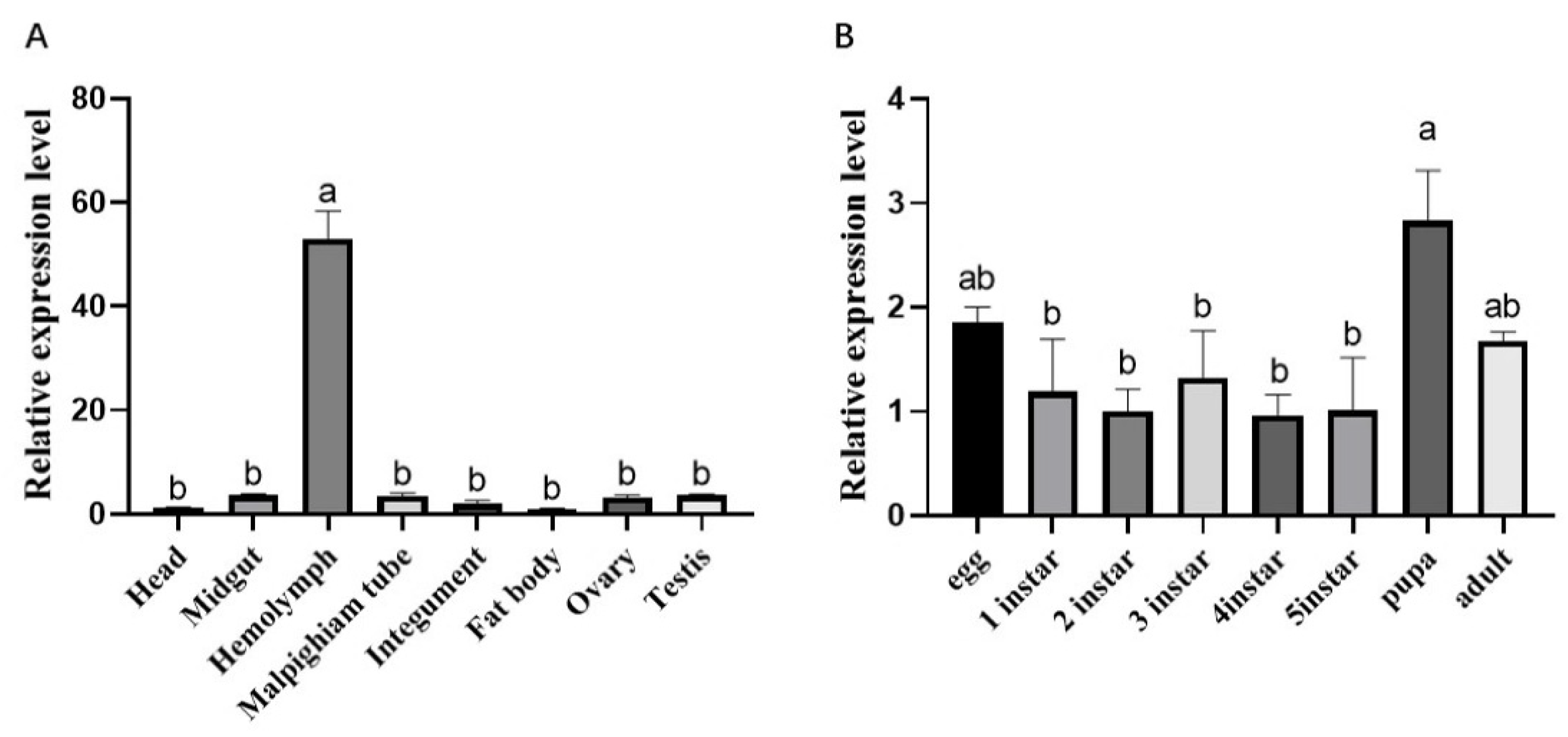
3.6. Spatiotemporal Expression Pattern of B. mori Testis Expressed Genes 261 (BmTex261)
3.7. Response Analysis of BmTex261 to AcMNPV Infection
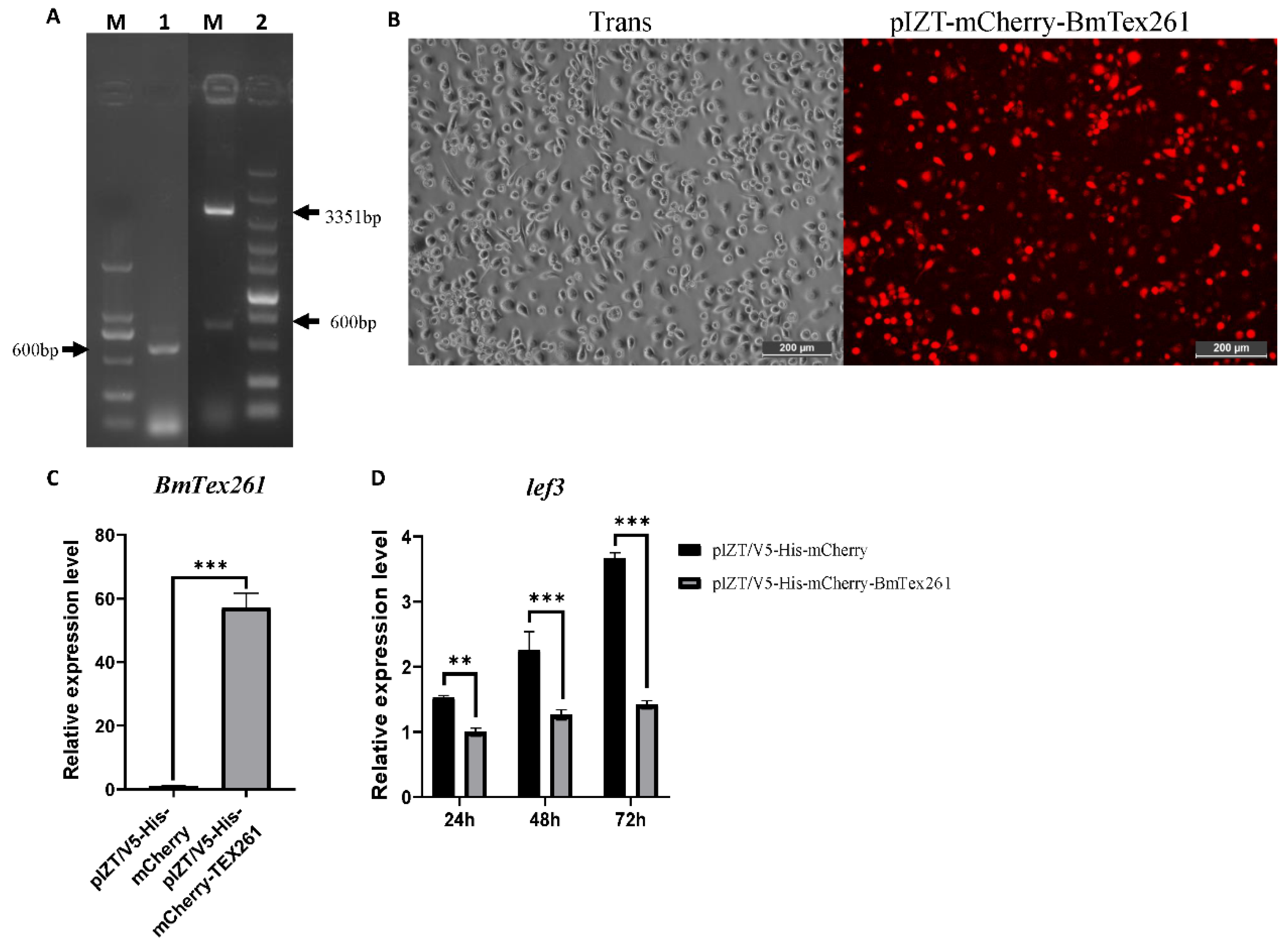
3.8. Overexpression of BmTex261 Inhibited AcMNPV Infection in BmN Cells
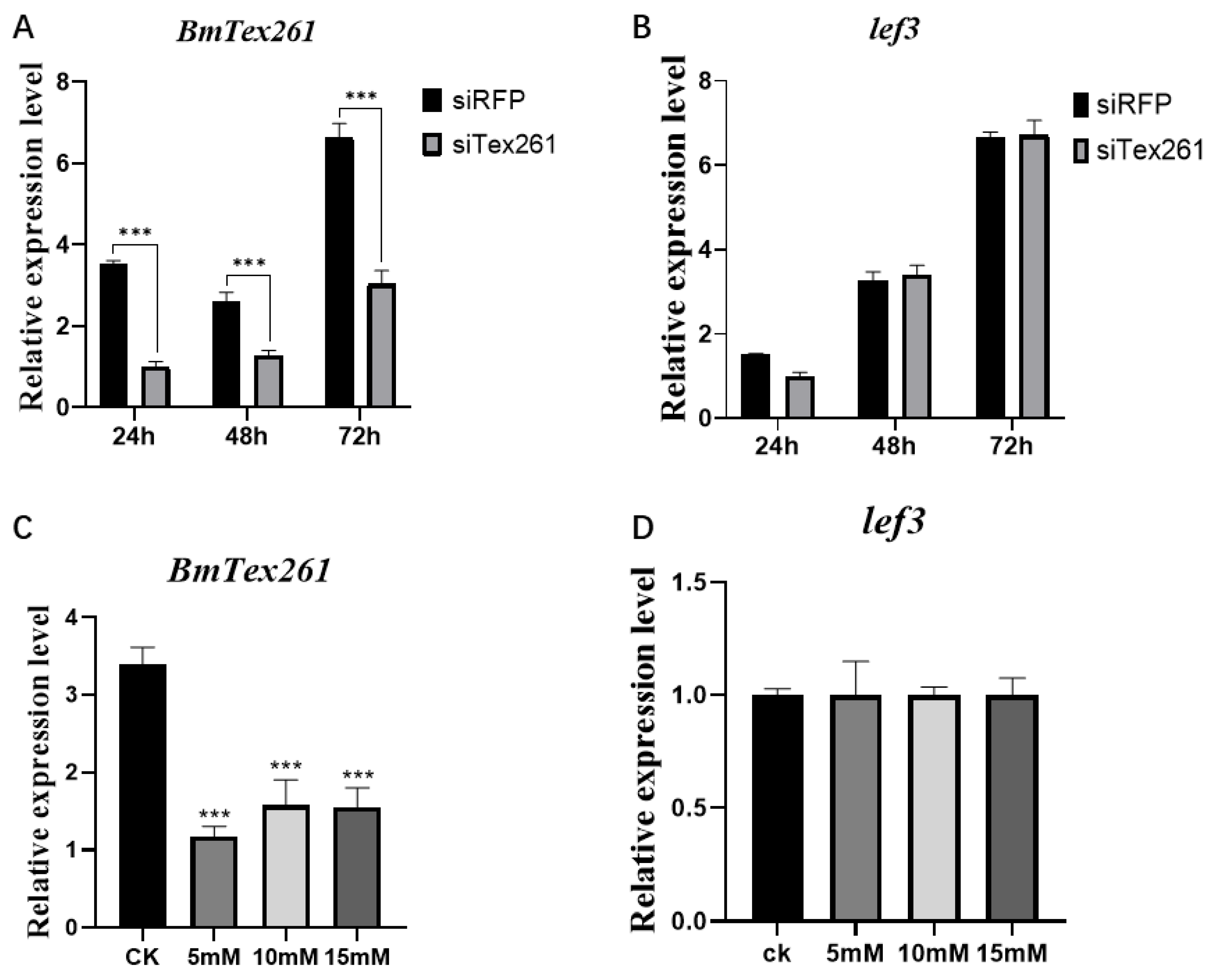
3.9. Knockdown BmTex261 Has No Effect on AcMNPV Infection in BmN Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nerome, K.; Yamaguchi, R.; Fuke, N.; Izzati, U.Z.; Maegawa, K.; Sugita, S.; Kawasaki, K.; Kuroda, K.; Nerome, R. Development of a Japanese encephalitis virus genotype V virus-like particle vaccine in silkworms. J. Gen. Virol. 2018, 99, 897–907. [Google Scholar] [CrossRef]
- Dash, R.; Mandal, M.; Ghosh, S.K.; Kundu, S.C. Silk sericin protein of tropical tasar silkworm inhibits UVB-induced apoptosis in human skin keratinocytes. Mol. Cell. Biochem. 2008, 311, 111–119. [Google Scholar] [CrossRef]
- Nourmohammadi, J.; Roshanfar, F.; Farokhi, M.; Nazarpak, M.H. Silk fibroin/kappa-carrageenan composite scaffolds with enhanced biomimetic mineralization for bone regeneration applications. Mater. Sci. Eng. C 2017, 76, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Yang, M. Silk-based biomaterials. Microsc. Res. Tech. 2017, 80, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Blissard, G.W.; Rohrmann, G.F. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 1990, 35, 127–155. [Google Scholar] [CrossRef] [PubMed]
- Mori, H. Transovarian transmission of a foreign gene in the silkworm, Bombyx mori, by Autographa californica nuclear polyhedrosis virus. Biotechnology 1995, 13, 1005–1007. [Google Scholar] [CrossRef]
- Rahman, M.; Gopinathan, K.P. Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res. 2004, 101, 109–118. [Google Scholar] [CrossRef]
- Katsuma, S.; Mita, K.; Shimada, T. ERK- and JNK-Dependent Signaling Pathways Contribute to Bombyx mori Nucleopolyhedrovirus Infection. J. Virol. 2007, 81, 13700–13709. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Baculovirus Molecular Biology, 2rd ed.; Bethesda: Rockville, MD, USA, 2013. [Google Scholar]
- Slack, J.M.; Lawrence, S. Evidence for proteolytic cleavage of the baculovirus occlusion-derived virion envelope protein P74. J. Gen. Virol. 2005, 86, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Sparks, W.O.; Harrison, R.L.; Bonning, B.C. Autographa californica multiple nucleopolyhedrovirus ODV-E56 is a per os infectivity factor, but is not essential for binding and fusion of occlusion-derived virus to the host midgut. Virology 2011, 409, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Ayres, M.D.; Howard, S.C.; Kuzio, J.; Lopez-Ferber, M.; Possee, R.D. The Complete DNA Sequence of Autographa californica Nuclear Polyhedrosis Virus. Virology 1994, 202, 586–605. [Google Scholar] [CrossRef] [PubMed]
- Gomi, S.; Majima, K.; Maeda, S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 1999, 80, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.-Y.; Tang, X.-D.; Lv, Z.-Y.; Wang, X.; Tian, C.-H.; Xu, Y.P.; Zhang, C.-X. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 2009, 94, 138–145. [Google Scholar] [CrossRef]
- Xue, J.; Qiao, N.; Zhang, W.; Cheng, R.; Zhang, X.-Q.; Bao, Y.-Y.; Xu, Y.-P.; Gu, L.-Z.; Han, J.-D.J.; Zhang, C.-X. Dynamic Interactions between Bombyx mori Nucleopolyhedrovirus and Its Host Cells Revealed by Transcriptome Analysis. J. Virol. 2012, 86, 7345–7359. [Google Scholar] [CrossRef] [PubMed]
- Yin, H. Comprehensive analysis of lncRNA-mRNA regulatory network in BmNPV infected cells treated with Hsp90 inhibitor. Mol. Immunol. 2020, 127, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Analysis of lncRNA-mediated gene regulatory network of Bombyx mori in response to BmNPV infection. J. Invertebr. Pathol. 2020, 170, 107323. [Google Scholar] [CrossRef]
- Matos, L.; Gonçalves, V.; Pinto, E.; Laranjeira, F.; Prata, M.J.; Jordan, P.; Desviat, L.R.; Pérez, B.; Alves, S. Functional analysis of splicing mutations in the IDS gene and the use of antisense oligonucleotides to exploit an alternative therapy for MPS II. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2015, 1852, 2712–2721. [Google Scholar] [CrossRef]
- Hu, X.; Zhu, M.; Wang, S.; Zhu, L.; Xue, R.; Cao, G.; Gong, C. Proteomics analysis of digestive juice from silkworm during Bombyx mori nucleopolyhedrovirus infection. Proteomics 2015, 15, 2691–2700. [Google Scholar] [CrossRef]
- Kang, L.; Shi, H.; Liu, X.; Zhang, C.; Yao, Q.; Wang, Y.; Chang, C.; Shi, J.; Cao, J.; Kong, J.; et al. Arginine kinase is highly expressed in a resistant strain of silkworm (Bombyx mori, Lepidoptera): Implication of its role in resistance to Bombyx mori nucleopolyhedrovirus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 158, 230–234. [Google Scholar] [CrossRef]
- Flipsen, J.; Martens, J.; Van Oers, M.; Vlak, J.; Van Lent, J. Passage of Autographa californica Nuclear Polyhedrosis Virus through the Midgut Epithelium of Spodoptera exigua Larvae. Virology 1995, 208, 328–335. [Google Scholar] [CrossRef]
- Washburn, J.O.; Chan, E.Y.; Volkman, L.E.; Aumiller, J.J.; Jarvis, D.L. Early Synthesis of Budded Virus Envelope Fusion Protein GP64 Enhances Autographa californica Multicapsid Nucleopolyhedrovirus Virulence in Orally Infected Heliothis virescens. J. Virol. 2003, 77, 280–290. [Google Scholar] [CrossRef]
- Si, Y.-H.; Fang, M.; Huang, Y.; Zheng, F.-L.; Li, T.; Hu, Z.; Wang, H.-Z. Construction and Characterization of aHelicoverpa armigeraNucleopolyhedrovirus Bacterial Artificial Chromosome with Deletion of Ecdysteroid UDP-Glucosyltransferase Gene. Biosci. Biotechnol. Biochem. 2007, 71, 2435–2441. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kiuchi, T.; Fujii, T.; Daimon, T.; Li, M.; Banno, Y.; Kikuta, S.; Kikawada, T.; Katsuma, S.; Shimada, T. Mutation of a novel ABC transporter gene is responsible for the failure to incorporate uric acid in the epidermis of ok mutants of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2013, 43, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, M.; Islam, I.; You, L.; Wang, Y.; Li, Z.; Ling, L.; Zeng, B.; Xu, J.; Huang, Y.; et al. Allelic-specific expression in relation to Bombyx mori resistance to Bt toxin. Insect Biochem. Mol. Biol. 2014, 54, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Braunagel, S.C. Identification of BV/ODV-C42, an Autographa californica nucleopolyhedrovirus orf101-encoded structural protein detected in infected-cell complexes with ODV-EC27 and p78/83. J. Virol. 2001, 75, 12331–12338. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y. Autographa californica multiple nucleopolyhedrovirus nucleocapsid protein BV/ODV-C42 mediates the nuclear entry of P78/83. J. Virol. 2008, 82, 4554–4561. [Google Scholar] [CrossRef]
- Bhatia, N.K.; Carrillo, E.; Durham, R.J.; Berka, V.; Jayaraman, V. Allosteric Changes in the NMDA Receptor Associated with Calcium-Dependent Inactivation. Biophys. J. 2020, 119, 2349–2359. [Google Scholar] [CrossRef]
- Li, K.; Wang, Y.; Bai, H.; Wang, Q.; Song, J.; Zhou, Y.; Wu, C.; Chen, X. The Putative Pocket Protein Binding Site of Autographa californica Nucleopolyhedrovirus BV/ODV-C42 Is Required for Virus-Induced Nuclear Actin Polymerization. J. Virol. 2010, 84, 7857–7868. [Google Scholar] [CrossRef]
- Vieira, H.L.A.; Pereira, A.C.P.; Carrondo, M.J.T.; Alves, P. Catalase effect on cell death for the improvement of recombinant protein production in baculovirus-insect cell system. Bioprocess Biosyst. Eng. 2006, 29, 409–414. [Google Scholar] [CrossRef]
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef]
- Miller, L.K. Baculovirus interaction with host apoptotic pathways. J. Cell Physiol. 1997, 173, 178–182. [Google Scholar] [CrossRef]
- Clem, R.J.; Fechheimer, M.; Miller, L.K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 1991, 254, 1388–1390. [Google Scholar] [CrossRef]
- Bump, N.J.; Hackett, M.; Hugunin, M.; Seshagiri, S.; Brady, K.; Chen, P.; Ferenz, C.; Franklin, S.; Ghayur, T.; Li, P.; et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science 1995, 269, 1885–1888. [Google Scholar] [CrossRef]
- Lopez-Fernandez, L.A.; Parraga, M.; del Mazo, J. Tex261, a novel gene presumably related but distinct from steroidogenic acute regulatory (StAR) gene, is regulated during the development of germ cells. Biochem. Biophys. Res. Commun. 1998, 242, 565–569. [Google Scholar] [CrossRef]
- Taniura, H. Tex261 modulates the excitotoxic cell death induced by N-methyl-D-aspartate (NMDA) receptor activation. Biochem. Biophys. Res. Commun. 2007, 362, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Li, T. The haemolymph melanization response is related to defence against the AcMNPV infection in Bombyx mori. Arch. Insect Biochem. Physiol. 2021, 2021, e21764. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- The International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Mita, K.; Kasahara, M.; Sasaki, S.; Nagayasu, Y.; Yamada, T.; Kanamori, H.; Namiki, N.; Kitagawa, M.; Yamashita, H.; Yasukochi, Y.; et al. The Genome Sequence of Silkworm, Bombyx mori. DNA Res. 2004, 11, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Biology Analysis Group; Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.-Y.; Liu, B.; Zhao, P.; Zha, X.; Cheng, T.; et al. A Draft Sequence for the Genome of the Domesticated Silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, M.P.D.K.G.M.P.S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Natale, D.A.; Garkavtsev, I.V.; Tatusova, T.A.; Shankavaram, U.T.; Rao, B.S.; Kiryutin, B.; Galperin, M.; Fedorova, N.D.; Koonin, E.V. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Puri, B.K. Calcium Signaling and Gene Expression. Adv. Exp. Med. Biol. 2019, 1131, 537–545. [Google Scholar]
- Vessaro-Silva, A.S.; Neto, M.H.M.; Brancalhão, R.M.C.; Ribeiro, L.D.F.C.; Guimarães, A.T.B.; De Oliveira, C.M.T. Antioxidant Systems as a Response to Midgut Cellular of Bombyx mori Lineu, 1758 (Lepidoptera: Bombycidae) Infection for Baculoviruses. J. Econ. Èntomol. 2019, 112, 1089–1097. [Google Scholar] [CrossRef]
- Oliveira, J.H.M.; Talyuli, O.A.C.; Goncalves, R.L.S.; Paiva-Silva, G.O.; Sorgine, M.H.F.; Alvarenga, P.H.; Oliveira, P. Catalase protects Aedes aegypti from oxidative stress and increases midgut infection prevalence of Dengue but not Zika. PLOS Neglected Trop. Dis. 2017, 11, e0005525. [Google Scholar] [CrossRef] [PubMed]
- Zou, F.-M.; Lou, D.-S.; Zhu, Y.-H.; Wang, S.-P.; Jin, B.-R.; Gui, Z.-Z. Expression profiles of glutathione S-transferase genes in larval midgut of Bombyx mori exposed to insect hormones. Mol. Biol. Rep. 2010, 38, 639–647. [Google Scholar] [CrossRef]
- Zhuang, W.; Zhang, C.; Hao, F.; Sun, X. Baculoviral IAP Repeat Containing 6 (BIRC6) Is a Predictor of Prognosis in Prostate Cancer. Med Sci. Monit. 2018, 24, 839–845. [Google Scholar] [CrossRef]
- Orgueira, A.M.; López, M.C.; Raíndo, A.P.; Arias, J.D.; Rodríguez, B.A.; Pérez, L.B.; Vence, N.A.; Ángeles, B.L.; Blanco, A.A.; Valentín, P.M.; et al. Detection of Rare Germline Variants in the Genomes of Patients with B-Cell Neoplasms. Cancers 2021, 13, 1340. [Google Scholar] [CrossRef]
- Xiong, C.; Wang, Z.; Wang, G.; Zhang, C.; Jin, S.; Jiang, G.; Bai, D. Identification of CDC20 as an immune infiltration-correlated prognostic biomarker in hepatocellular carcinoma. Investig. New Drugs 2021, 1–15. [Google Scholar] [CrossRef]
- Wang, L.; Wang, M.; Hu, C.; Li, P.; Qiao, Y.; Xia, Y.; Liu, L.; Jiang, X. Protein salvador homolog 1 acts as a tumor suppressor and is modulated by hypermethylation in pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 62953–62961. [Google Scholar] [CrossRef] [PubMed]
- Swevers, L.; Cherbas, L.; Cherbas, P.; Iatrou, K. Bombyx EcR (BmEcR) and Bombyx USP (BmCF1) combine to form a functional ecdysone receptor. Insect Biochem. Mol. Biol. 1996, 26, 217–221. [Google Scholar] [CrossRef]
- Yao, T.-P.; Segraves, W.A.; Oro, A.E.; McKeown, M.; Evans, R. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 1992, 71, 63–72. [Google Scholar] [CrossRef]
- Ryu, S.H.; Lee, S.Y.; Lee, K.; Rhee, S.G. Catalytic properties of inositol trisphosphate kinase: Activation by Ca2+ and calmodulin. FASEB J. 1987, 1, 388–393. [Google Scholar] [CrossRef] [PubMed]


| No. | Primer Names | Forward Primer (5′-3′) | Reverse Prime (5′-3′) |
|---|---|---|---|
| 1 | BMSK0012268 | CAGGCGATGAAGCTGGAGAA | GCGGACTTCCTCGTTTACCT |
| 2 | BMSK0012667 | GGCGAAGCAAAATGGCAGAA | ATTTGACGCGCTTATCGTGC |
| 3 | BMSK0013103 | CCAACTCAGCTAGACGATGCC | GATGCCAAGTTCCCGAAGATAG |
| 4 | BMSK0005642 | AACTCTGGCCGCTAAGTTCA | TCAGCTGCTCGTCCAATTCC |
| 5 | BMSK0015907 | AAAGACCAACGGAACTGCGA | CCTGTGAATTCGGTCCCCTC |
| 6 | BMSK0013101 | ACCGCACGGGAACTAGGA | CCAAGCCTAGATGCTCGTTGT |
| 7 | BMSK0000440 | GCAGTTCCGGTGAAGAGACA | AAGAAGGAGGTGGGAAGGGA |
| 8 | BMSK0008569 | AAAACACGCCCGATTCACAC | CGCGACTGTAAGTGGGAGAA |
| 9 | BMSK0009445 | TGCTACAGACGAGACTACCC | TGGATCTGTTCGCCCCTT |
| 10 | BMSK0014619 | CCGACATTGTTTGCCGTTGT | GCACTTCTGGTTGATGATGCC |
| 11 | BMSK0000576 | TAAACAAGGTCGGTCACGCA | GCCGTTTTGAACTGTGGCTT |
| 12 | BmTex261 | CGTGTTGCCAACGACAGAAG | CGCTTTCTTGTTCCGGTGAG |
| 13 | BmGAPDH | CCGCGTCCCTGTTGCTAAT | CTGCCTCCTTGACCTTTTGC |
| 14 | lef3 | CAAACGCGTTGCTTCGTACA | TGCTCGAGTCGGAAGAGGTA |
| 15 | BmTex261 KX | GGGGTACCATGTTATTCTTGTATTTATTGAGTTATT | GCTCTAGAGAACGCTTTCTTGTTCCG |
| Primer Names | Sequences (5′-3′) |
|---|---|
| BmTex261-1 Olig-1 | GATCACTAATACGACTCACTATAGGGAAGTCATCACGTATGCTGTATTT |
| BmTex261-1 Olig-2 | AAATACAGCATACGTGATGACTTCCCTATAGTGAGTCGTATTAGTGATC |
| BmTex261-1 Olig-3 | AAAAGTCATCACGTATGCTGTATCCCTATAGTGAGTCGTATTAGTGATC |
| BmTex261-1 Olig-4 | GATCACTAATACGACTCACTATAGGGATACAGCATACGTGATGACTTTT |
| BmTex261-2 Olig-1 | GATCACTAATACGACTCACTATAGGGAACGTTCTGACGGATTATCTGTT |
| BmTex261-2 Olig-2 | AACAGATAATCCGTCAGAACGTTCCCTATAGTGAGTCGTATTAGTGATC |
| BmTex261-2 Olig-3 | AAAACGTTCTGACGGATTATCTGCCCTATAGTGAGTCGTATTAGTGATC |
| BmTex261-2 Olig-4 | GATCACTAATACGACTCACTATAGGGCAGATAATCCGTCAGAACGTTTT |
| RFP-Olig-1 | GATCACTAATACGACTCACTATAGGGGCACCCAGACCATGAGAATTT |
| RFP-Olig-2 | AAATTCTCATGGTCTGGGTGCCCCTATAGTGAGTCGTATTAGTGATC |
| RFP-Olig-3 | AAGCACCCAGACCATGAGAATCCCTATAGTGAGTCGTATTAGTGATC |
| RFP-Olig-4 | GATCACTAATACGACTCACTATAGGGATTCTCATGGTCTGGGTGCTT |
| p50− | p50+ | C108− | C108+ | |
|---|---|---|---|---|
| Total Reads | 44,728,956 | 46,036,772 | 46,693,816 | 43,864,330 |
| GC Content (%) | 48 | 46 | 48 | 48 |
| % ≥ Q30 (%) | 93.35 | 93.36 | 93.27 | 94.49 |
| Mapped Reads | 41,118,461 | 33,412,593 | 39,765,815 | 42,254,696 |
| Mapped Ratio (%) | 91.92 | 72.72 | 90.66 | 90.46 |
| Unique Mapped Reads | 34,225,042 | 30,271,861 | 37,902,000 | 39,895,987 |
| Unique Mapped Ratio (%) | 85.00 | 65.87 | 86.41 | 85.44 |
| Groups | DEGs | Ratio of Total Transcripts | Upregulation | Downregulation |
|---|---|---|---|---|
| p50+ vs. p50− | 679 | 6.73% | 301 (44.33%) | 378 (55.67%) |
| C108+ vs. C108− | 515 | 5.04% | 336 (65.24%) | 179 (34.76%) |
| Gene Name | Gene ID | p50− FRKM | p50+ FPKM | C108− FRKM | C108+ FRKM | p50+ vs. p50−Ratio | C108+ vs. C108−Ratio |
|---|---|---|---|---|---|---|---|
| Metabolic pathways | |||||||
| Catalase | BMSK0000352 | 101.20 | 24.49 | 64.09 | 85.41 | 0.24 | 0.75 |
| Mitochondrial aldehyde dehydrogenase | BMSK0006974 | 41.49 | 46.17 | 152.24 | 140.61 | 1.11 | 1.08 |
| Putative dopa decarboxylase protein | BMSK0002058 | 3.48 | 2.290 | 14.66 | 10.90 | 0.66 | 1.34 |
| Hydroxyacyl-coenzyme A dehydrogenase | BMSK0002060 | 52.09 | 17.74 | 35.47 | 36.35 | 0.34 | 0.98 |
| 3-hydroxyacyl-coa dehydrogenase | BMSK0001125 | 2.44 | 1.05 | 8.62 | 8.33 | 0.43 | 1.03 |
| Probable 2-oxoglutarate dehydrogenase E1 component | BMSK0003515 | 9.69 | 1.52 | 6.59 | 5.02 | 0.16 | 1.31 |
| Kynurenine formamidase | BMSK0008569 | 19.19 | 1.88 | 12.61 | 27.13 | 0.10 | 0.46 |
| Mitochondrial aldehyde dehydrogenase | BMSK0012254 | 338.85 | 59.03 | 97.36 | 121.13 | 0.17 | 0.80 |
| Tryptophan 2,3-dioxygenase | BMSK0008115 | 15.66 | 6.12 | 13.24 | 9.73 | 0.39 | 1.36 |
| Inositol-trisphosphate 3-kinase A | BMSK0005502 | 32.69 | 23.45 | 44.30 | 39.27 | 0.72 | 1.13 |
| Phosphatidylserine decarboxylase | BMSK0005994 | 43.74 | 10.12 | 28.99 | 32.13 | 0.23 | 0.90 |
| UDP-glucose 6-dehydrogenase | BMSK0014439 | 21.94 | 3.67 | 24.30 | 24.67 | 0.17 | 0.99 |
| Glutathione S-transferase delta 3 | BMSK0003600 | 29.77 | 13.67 | 33.11 | 24.70 | 0.46 | 1.34 |
| Apoptosis | |||||||
| baculoviral IAP repeat-containing protein 6 | BMSK0014897 | 6.73 | 2.42 | 7.44956 | 6.170963 | 0.36 | 1.21 |
| eukaryotic translation initiation factor 2-alpha kinase-like | BMSK0004001 | 12.72 | 6.04 | 12.1691 | 11.05708 | 0.47 | 1.10 |
| htra2 | BMSK0015308 | 7.03 | 2.32 | 3.154753 | 5.146924 | 0.33 | 0.61 |
| scaffold protein salvador | BMSK0014742 | 10.76 | 2.76 | 11.39895 | 15.83115 | 0.26 | 0.72 |
| Ultraspiracle 2 | BMSK0001870 | 5.92 | 2.48 | 4.06216 | 8.994249 | 0.42 | 0.45 |
| Cell division cycle protein 20 homolog | BMSK0013179 | 10.16 | 2.35 | 6.86 | 7.00 | 0.23 | 0.98 |
| Transcription factor E74 | BMSK0008350 | 5.72 | 1.96 | 11.45 | 14.80 | 0.34 | 0.77 |
| Mitogen-activated protein kinase kinase kinase 7-like | BMSK0001403 | 7.23 | 1.26 | 4.51 | 4.92 | 0.17 | 0.92 |
| Testis expressed genes 261 | BMSK0013995 | 2.66 | 1.56 | 2.36 | 2.05 | 0.59 | 0.87 |
| transcription factor kayak | BMSK0014876 | 33.23 | 23.73 | 49.37783 | 56.80742 | 0.71 | 0.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, X.-y.; Wang, X.-y.; Kong, Y.-h.; Zhao, C.-x.; Qin, S.; Sun, X.; Li, M.-w. Comparative Transcriptome Analysis of Bombyx mori (Lepidoptera) Larval Hemolymph in Response to Autographa californica Nucleopolyhedrovirus in Differentially Resistant Strains. Processes 2021, 9, 1401. https://doi.org/10.3390/pr9081401
Ding X-y, Wang X-y, Kong Y-h, Zhao C-x, Qin S, Sun X, Li M-w. Comparative Transcriptome Analysis of Bombyx mori (Lepidoptera) Larval Hemolymph in Response to Autographa californica Nucleopolyhedrovirus in Differentially Resistant Strains. Processes. 2021; 9(8):1401. https://doi.org/10.3390/pr9081401
Chicago/Turabian StyleDing, Xin-yi, Xue-yang Wang, Yun-hui Kong, Chun-xiao Zhao, Sheng Qin, Xia Sun, and Mu-wang Li. 2021. "Comparative Transcriptome Analysis of Bombyx mori (Lepidoptera) Larval Hemolymph in Response to Autographa californica Nucleopolyhedrovirus in Differentially Resistant Strains" Processes 9, no. 8: 1401. https://doi.org/10.3390/pr9081401
APA StyleDing, X.-y., Wang, X.-y., Kong, Y.-h., Zhao, C.-x., Qin, S., Sun, X., & Li, M.-w. (2021). Comparative Transcriptome Analysis of Bombyx mori (Lepidoptera) Larval Hemolymph in Response to Autographa californica Nucleopolyhedrovirus in Differentially Resistant Strains. Processes, 9(8), 1401. https://doi.org/10.3390/pr9081401






