The Regulation and Role of piRNAs and PIWI Proteins in Cancer
Abstract
:1. Introduction
2. PIWI Proteins in Diverse Cancers
3. Oncogenic piRNAs and Tumor-Suppressor piRNAs
4. The Role of PIWI in Cancer in a piRNA-Independent Manner
5. Databases of piRNA Expression in Cancers
6. piRNAs as Non-Invasive Biomarkers for Cancer
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Appendix A
| Cancer | Expression in Cancer; Mechanism | Sample Tested | Role; Mechanism | Ref. |
|---|---|---|---|---|
| Endometrial cancer | Up; Estrogen-ERα signaling and hypomethylation of PIWIL1 promoter | Patients, Ishikawa, RL95–2 cell lines | Oncogenic; Inducing epithelial-mesenchymal transition | [32,33] |
| Lung adenocarcinoma | Up; Hypomethylation of PIWIL1 promoter | Patients, A549, H1299 cell lines | Oncogenic | [34] |
| Glioblastoma | Up; Downregulation of miR-154-5p that targets PIWIL1 mRNA | Patients, glioma stem cell lines (4121, 3832, 387, 3359) & glioblastoma cell lines (U251, U87, A172, LN229, SNB19, LN308) | Oncogenic | [36] |
| Esophageal squamous cell carcinoma | Up (cytoplasmic); Unknown | Patients, Kyse140 cell line | Oncogenic | [37] |
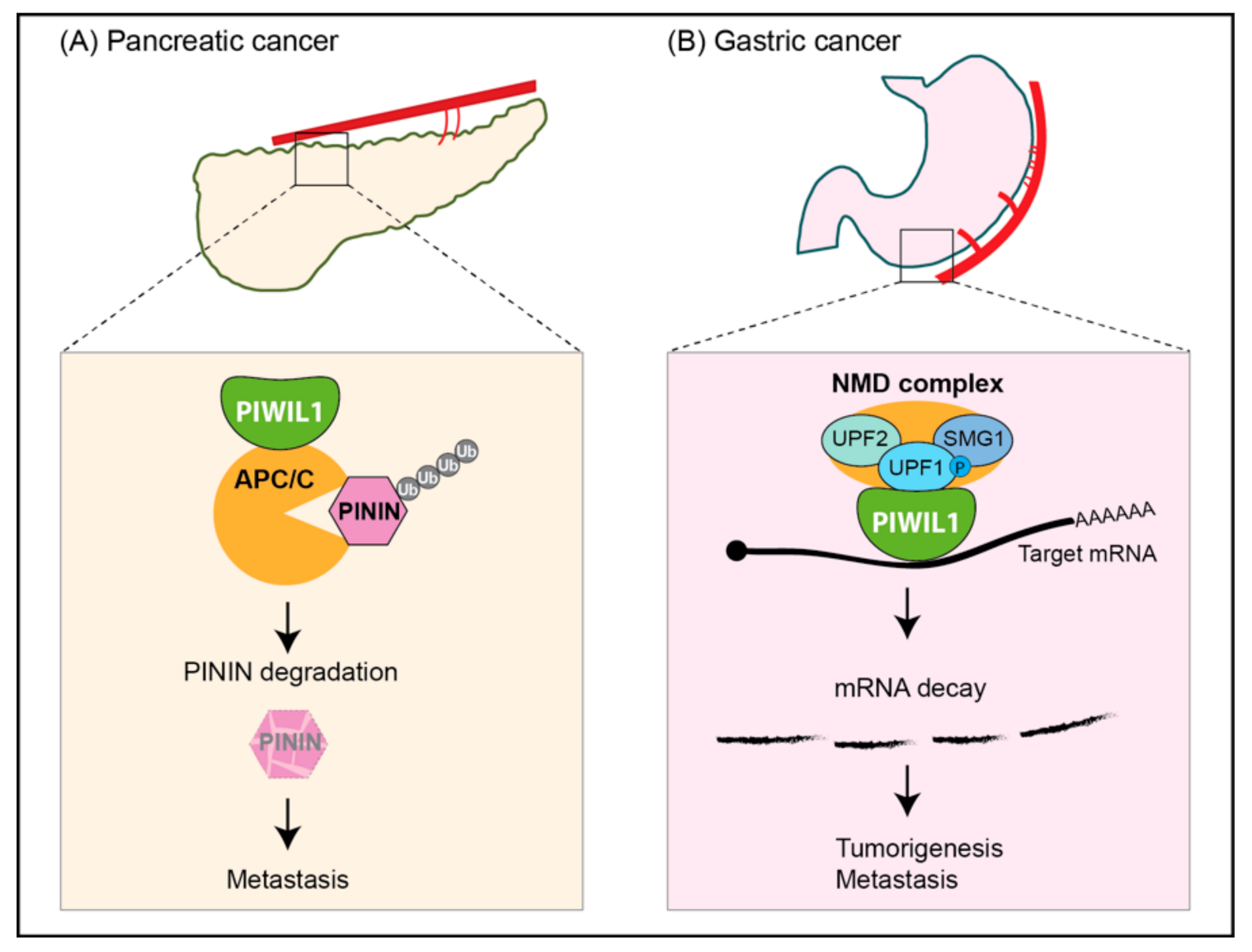
| Pancreatic ductal adenocarcinomas | Up; Unknown | Patients, BxPC-3, AsPC-1 cell lines | Oncogenic; piRNA-independent function; acting with APC/C to degrade PININ | [59] |
| Gastric cancer | Up; Unknown | Patients, AGS, HGC-27, N87, SNU-1/5/16 cell lines | Oncogenic; piRNA-independent function; acting with NMD components to likely degrade tumor suppressors mRNAs | [60] |
| Cancer | piRNA | Expression in Cancer | Samples Tested | Role | Mechanism | Ref. |
|---|---|---|---|---|---|---|
| Breast cancer | piR-36026 (DQ597960) | Up | Patients, MCF7 cell line | Oncogenic | Targeting SERINA1 and LRAT mRNA | [41,42] |
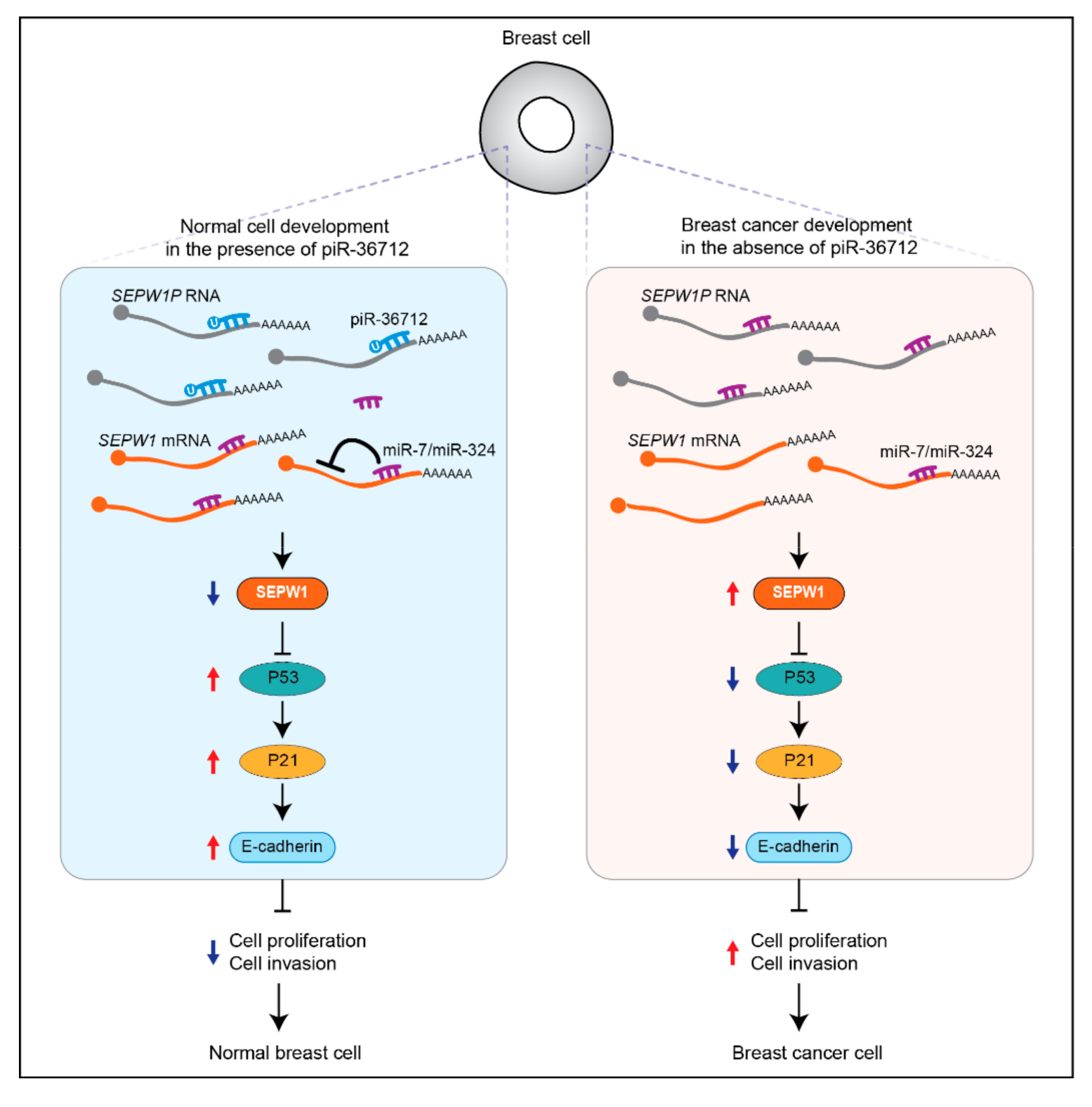
| piR-36712 | Down | Patients | Tumor suppressing | Increasing P53 expression by targeting SEPW1P RNA | [43] | |
| Colorectal cancer | piR-54265 | Up | Patients (tissue & blood), HCT116, LoVo, SW480, SW620, HT-29, DLD-1 cell lines | Oncogenic | Activating STAT3 signaling by interacting with PIWIL2 | [44,45,46] |
| piR-1245 | Up | Patients, HCT116, SW480 cell lines | Oncogenic | Likely regulating a panel of tumor suppressor genes mRNA | [47] | |
| piR-823 | Up | Patients, HCT15/116, DLD-1 cell lines | Oncogenic | Enhancing the transcriptional activity of HSF1 by promoting its phosphorylation | [49] | |
| piR-24000 | Up | Patients | Oncogenic | Unknown | [48] | |
| Glioma | piR-33221 (DQ593109) | Up | Glioma endothelial cells | Oncogenic | Acting with PIWIL1 to suppress BTB permeability via regulating MEG3/miR-330-5p/RUNX3 axis | [50] |
| Diffuse large B-cell lymphoma | piR-30473 | Up | Patients | Oncogenic | Stabilizing WTAP mRNA via m6A RNA methylation | [51,52] |
| Multiple myeloma | piR-823 | Up | Patients, Plasma cell, RPMI8226, ARH-77, U266, KM3 cell lines | Oncogenic | Inducing de novo DNA methylation | [53] |
| piR-004800 | Up | Patients (exosomes from bone marrow supernatant), RPMI8226, U266 cell lines | Oncogenic | Regulation of PI3K/Akt/mTOR pathway | [54] |
References
- Huang, X.; Wong, G. An old weapon with a new function: PIWI-interacting RNAs in neurodegenerative diseases. Transl. Neurodegener. 2021, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Pan, Y.; Fang, Y.; Zhang, J.; Xie, M.; Yang, F.; Yu, T.; Ma, P.; Li, W.; Shu, Y. The biogenesis and functions of piRNAs in human diseases. Mol. Ther. Nucleic Acids 2020, 21, 108–120. [Google Scholar] [CrossRef]
- Kim, K.W. PIWI proteins and piRNAs in the nervous system. Mol. Cells 2019, 42, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [Green Version]
- Weick, E.M.; Miska, E.A. piRNAs: From biogenesis to function. Development 2014, 141, 3458–3471. [Google Scholar] [CrossRef] [Green Version]
- Su, J.-F.; Concilla, A.; Zhang, D.-Z.; Zhao, F.; Shen, F.-F.; Zhang, H.; Zhou, F.-Y. PIWI-interacting RNAs: Mitochondria-based biogenesis and functions in cancer. Genes Dis. 2020, 8, 603–622. [Google Scholar] [CrossRef]
- Czech, B.; Munafo, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-guided genome defense: From biogenesis to silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef]
- Gamez, S.; Srivastav, S.; Akbari, O.S.; Lau, N.C. Diverse defenses: A perspective comparing Dipteran Piwi-piRNA pathways. Cells 2020, 9, 2180. [Google Scholar] [CrossRef]
- Parhad, S.S.; Theurkauf, W.E. Rapid evolution and conserved function of the piRNA pathway. Open Biol. 2019, 9, 180181. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.C.; Gu, W.; Shirayama, M.; Youngman, E.; Conte, D., Jr.; Mello, C.C. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 2012, 150, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, J.P.T.; Folkmann, A.; Bernard, L.; Lee, C.Y.; Seroussi, U.; Charlesworth, A.G.; Claycomb, J.M.; Seydoux, G. P Granules Protect RNA Interference Genes from Silencing by piRNAs. Dev. Cell 2019, 50, 716–728. [Google Scholar] [CrossRef]
- Shen, E.Z.; Chen, H.; Ozturk, A.R.; Tu, S.; Shirayama, M.; Tang, W.; Ding, Y.H.; Dai, S.Y.; Weng, Z.; Mello, C.C. Identification of piRNA binding sites reveals the Argonaute regulatory landscape of the C. elegans germline. Cell 2018, 172, 937–951.e918. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Tu, S.; Stubna, M.; Wu, W.S.; Huang, W.C.; Weng, Z.; Lee, H.C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 2018, 359, 587–592. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.W.; Tang, N.H.; Andrusiak, M.G.; Wu, Z.; Chisholm, A.D.; Jin, Y. A neuronal piRNA pathway inhibits axon regeneration in C. elegans. Neuron 2018, 97, 511–519. [Google Scholar] [CrossRef] [Green Version]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef] [Green Version]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Carmell, M.A.; Xuan, Z.; Zhang, M.Q.; Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002, 16, 2733–2742. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, T.; Shiohama, A.; Minoshima, S.; Shimizu, N. Identification of eight members of the Argonaute family in the human genome. Genomics 2003, 82, 323–330. [Google Scholar] [CrossRef]
- Juliano, C.; Wang, J.; Lin, H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 2011, 45, 447–469. [Google Scholar] [CrossRef] [Green Version]
- Ramat, A.; Simonelig, M. Functions of PIWI proteins in gene regulation: New arrows added to the piRNA quiver. Trends Genet. 2021, 37, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Kashima, M.; Agata, K.; Shibata, N. What is the role of PIWI family proteins in adult pluripotent stem cells? Insights from asexually reproducing animals, planarians. Dev. Growth Differ. 2020, 62, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Sousa-Victor, P.; Ayyaz, A.; Hayashi, R.; Qi, Y.; Madden, D.T.; Lunyak, V.V.; Jasper, H. Piwi is required to limit exhaustion of aging somatic stem cells. Cell Rep. 2017, 20, 2527–2537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, L.; Mu, S.; Sun, C.; Fan, F.; Yan, H.; Qin, Y.; Cui, G.; Wang, Y.; Guo, T.; Mei, H.; et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol. Cancer 2019, 18, 88. [Google Scholar] [CrossRef]
- Sturm, A.; Ivics, Z.; Vellai, T. The mechanism of ageing: Primary role of transposable elements in genome disintegration. Cell. Mol. Life Sci. 2015, 72, 1839–1847. [Google Scholar] [CrossRef] [Green Version]
- Wood, J.G.; Helfand, S.L. Chromatin structure and transposable elements in organismal aging. Front. Genet. 2013, 4, 274. [Google Scholar] [CrossRef] [Green Version]
- Anwar, S.L.; Wulaningsih, W.; Lehmann, U. Transposable elements in human cancer: Causes and consequences of deregulation. Int. J. Mol. Sci. 2017, 18, 974. [Google Scholar] [CrossRef] [Green Version]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Watanabe, T.; Lin, H. Posttranscriptional regulation of gene expression by Piwi proteins and piRNAs. Mol. Cell 2014, 56, 18–27. [Google Scholar] [CrossRef] [Green Version]
- Moyano, M.; Stefani, G. piRNA involvement in genome stability and human cancer. J. Hematol. Oncol. 2015, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Mentis, A.A.; Dardiotis, E.; Romas, N.A.; Papavassiliou, A.G. PIWI family proteins as prognostic markers in cancer: A systematic review and meta-analysis. Cell. Mol. Life Sci. 2020, 77, 2289–2314. [Google Scholar] [CrossRef]
- Chen, Z.; Che, Q.; He, X.; Wang, F.; Wang, H.; Zhu, M.; Sun, J.; Wan, X. Stem cell protein Piwil1 endowed endometrial cancer cells with stem-like properties via inducing epithelial-mesenchymal transition. BMC Cancer 2015, 15, 811. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Yang, H.J.; Lin, Q.; Zhu, M.J.; Yu, Y.Y.; He, X.Y.; Wan, X.P. Estrogen-ERalpha signaling and DNA hypomethylation co-regulate expression of stem cell protein PIWIL1 in ERalpha-positive endometrial cancer cells. Cell Commun. Signal. 2020, 18, 84. [Google Scholar] [CrossRef]
- Xie, K.; Zhang, K.; Kong, J.; Wang, C.; Gu, Y.; Liang, C.; Jiang, T.; Qin, N.; Liu, J.; Guo, X.; et al. Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer Med. 2018, 7, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Yu, X.; Han, X.; Hao, J.; Zhao, J.; Bebek, G.; Bao, S.; Prayson, R.A.; Khalil, A.M.; Jankowsky, E.; et al. Piwil1 regulates glioma stem cell maintenance and glioblastoma progression. Cell Rep. 2021, 34, 108522. [Google Scholar] [CrossRef]
- Wang, X.; Sun, S.; Tong, X.; Ma, Q.; Di, H.; Fu, T.; Sun, Z.; Cai, Y.; Fan, W.; Wu, Q.; et al. MiRNA-154-5p inhibits cell proliferation and metastasis by targeting PIWIL1 in glioblastoma. Brain Res. 2017, 1676, 69–76. [Google Scholar] [CrossRef]
- He, W.; Wang, Z.; Wang, Q.; Fan, Q.; Shou, C.; Wang, J.; Giercksky, K.E.; Nesland, J.M.; Suo, Z. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer 2009, 9, 426. [Google Scholar] [CrossRef] [Green Version]
- Taubert, H.; Wach, S.; Jung, R.; Pugia, M.; Keck, B.; Bertz, S.; Nolte, E.; Stoehr, R.; Lehmann, J.; Ohlmann, C.H.; et al. Piwil 2 expression is correlated with disease-specific and progression-free survival of chemotherapy-treated bladder cancer patients. Mol. Med. 2015, 21, 371–380. [Google Scholar] [CrossRef]
- Guo, B.; Li, D.; Du, L.; Zhu, X. piRNAs: Biogenesis and their potential roles in cancer. Cancer Metastasis Rev. 2020, 39, 567–575. [Google Scholar] [CrossRef]
- Weng, W.; Li, H.; Goel, A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 160–169. [Google Scholar] [CrossRef]
- Hashim, A.; Rizzo, F.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Santamaria, G.; Cordella, A.; Cantarella, C.; et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014, 5, 9901–9910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.J.; Moon, S.U.; Park, M.G.; Jung, W.Y.; Park, Y.K.; Song, S.K.; Ryu, J.G.; Lee, Y.S.; Heo, H.J.; Gu, H.N.; et al. Multiplex bioimaging of piRNA molecular pathway-regulated theragnostic effects in a single breast cancer cell using a piRNA molecular beacon. Biomaterials 2016, 101, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Ye, Y.; Li, M.; Pan, L.; Su, J.; et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mai, D.; Ding, P.; Tan, L.; Zhang, J.; Pan, Z.; Bai, R.; Li, C.; Li, M.; Zhou, Y.; Tan, W.; et al. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 2018, 8, 5213–5230. [Google Scholar] [CrossRef]
- Mai, D.; Zheng, Y.; Guo, H.; Ding, P.; Bai, R.; Li, M.; Ye, Y.; Zhang, J.; Huang, X.; Liu, D.; et al. Serum piRNA-54265 is a New Biomarker for early detection and clinical surveillance of Human Colorectal Cancer. Theranostics 2020, 10, 8468–8478. [Google Scholar] [CrossRef]
- Tosar, J.P.; Garcia-Silva, M.R.; Cayota, A. Circulating SNORD57 rather than piR-54265 is a promising biomarker for colorectal cancer: Common pitfalls in the study of somatic piRNAs in cancer. RNA 2021, 27, 403–410. [Google Scholar] [CrossRef]
- Weng, W.; Liu, N.; Toiyama, Y.; Kusunoki, M.; Nagasaka, T.; Fujiwara, T.; Wei, Q.; Qin, H.; Lin, H.; Ma, Y.; et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol. Cancer 2018, 17, 16. [Google Scholar] [CrossRef]
- Iyer, D.N.; Wan, T.M.; Man, J.H.; Sin, R.W.; Li, X.; Lo, O.S.; Foo, D.C.; Pang, R.W.; Law, W.L.; Ng, L. Small RNA profiling of piRNAs in colorectal cancer identifies consistent overexpression of piR-24000 that correlates clinically with an aggressive disease phenotype. Cancers 2020, 12, 188. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Jiang, X.Y.; Qi, W.; Ji, C.G.; Xie, X.L.; Zhang, D.X.; Cui, Z.J.; Wang, C.K.; Bai, Y.; Wang, J.; et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017, 108, 1746–1756. [Google Scholar] [CrossRef]
- Shen, S.; Yu, H.; Liu, X.; Liu, Y.; Zheng, J.; Wang, P.; Gong, W.; Chen, J.; Zhao, L.; Xue, Y. PIWIL1/piRNA-DQ593109 regulates the permeability of the Blood-Tumor Barrier via the MEG3/miR-330-5p/RUNX3 axis. Mol. Ther. Nucleic Acids 2018, 10, 412–425. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Fan, G.; Song, S.; Jiang, Y.; Qian, C.; Zhang, W.; Su, Q.; Xue, X.; Zhuang, W.; Li, B. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 2020, 137, 1603–1614. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Wu, Q.L.; Sun, C.Y.; Ai, L.S.; Deng, J.; Zhang, L.; Chen, L.; Chu, Z.B.; Tang, B.; Wang, K.; et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015, 29, 196–206. [Google Scholar] [CrossRef]
- Ma, H.; Wang, H.; Tian, F.; Zhong, Y.; Liu, Z.; Liao, A. PIWI-Interacting RNA-004800 is regulated by S1P receptor signaling pathway to keep myeloma cell survival. Front. Oncol. 2020, 10, 438. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Hong, J.; Hong, M.; Wang, Y.; Yu, T.; Zang, S.; Wu, Q. piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene 2019, 38, 5227–5238. [Google Scholar] [CrossRef]
- Xu, J.; Camfield, R.; Gorski, S.M. The interplay between exosomes and autophagy—Partners in crime. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [Green Version]
- Colletti, M.; Ceglie, D.; Di Giannatale, A.; Nazio, F. Autophagy and Exosomes Relationship in Cancer: Friends or Foes? Front. Cell Dev. Biol. 2020, 8, 614178. [Google Scholar] [CrossRef]
- Genzor, P.; Cordts, S.C.; Bokil, N.V.; Haase, A.D. Aberrant expression of select piRNA-pathway genes does not reactivate piRNA silencing in cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 11111–11112. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Yuan, P.; Rao, M.; Jin, C.H.; Tang, W.; Rong, Y.F.; Hu, Y.P.; Zhang, F.; Wei, T.; Yin, Q.; et al. piRNA-independent function of PIWIL1 as a co-activator for anaphase promoting complex/cyclosome to drive pancreatic cancer metastasis. Nat. Cell Biol. 2020, 22, 425–438. [Google Scholar] [CrossRef]
- Shi, S.; Yang, Z.Z.; Liu, S.; Yang, F.; Lin, H. PIWIL1 promotes gastric cancer via a piRNA-independent mechanism. Proc. Natl. Acad. Sci. USA 2020, 117, 22390–22401. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, P.; Lu, Y.; Li, Y.; Zheng, Y.; Kan, Y.; Chen, R.; He, S. piRBase: A comprehensive database of piRNA sequences. Nucleic Acids Res. 2019, 47, D175–D180. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Si, X.; Skogerbo, G.; Wang, J.; Cui, D.; Li, Y.; Sun, X.; Liu, L.; Sun, B.; Chen, R.; et al. piRBase: A web resource assisting piRNA functional study. Database (Oxford) 2014, 2014, bau110. [Google Scholar] [CrossRef] [Green Version]
- Xin, J.; Du, M.; Jiang, X.; Wu, Y.; Ben, S.; Zheng, R.; Chu, H.; Li, S.; Zhang, Z.; Wang, M. Systematic evaluation of the effects of genetic variants on PIWI-interacting RNA expression across 33 cancer types. Nucleic Acids Res. 2021, 49, 90–97. [Google Scholar] [CrossRef]
- Zhang, W.; Yao, G.; Wang, J.; Yang, M.; Wang, J.; Zhang, H.; Li, W. ncRPheno: A comprehensive database platform for identification and validation of disease related noncoding RNAs. RNA Biol. 2020, 17, 943–955. [Google Scholar] [CrossRef]
- Chen, S.; Ben, S.; Xin, J.; Li, S.; Zheng, R.; Wang, H.; Fan, L.; Du, M.; Zhang, Z.; Wang, M. The biogenesis and biological function of PIWI-interacting RNA in cancer. J. Hematol. Oncol. 2021, 14, 93. [Google Scholar] [CrossRef]
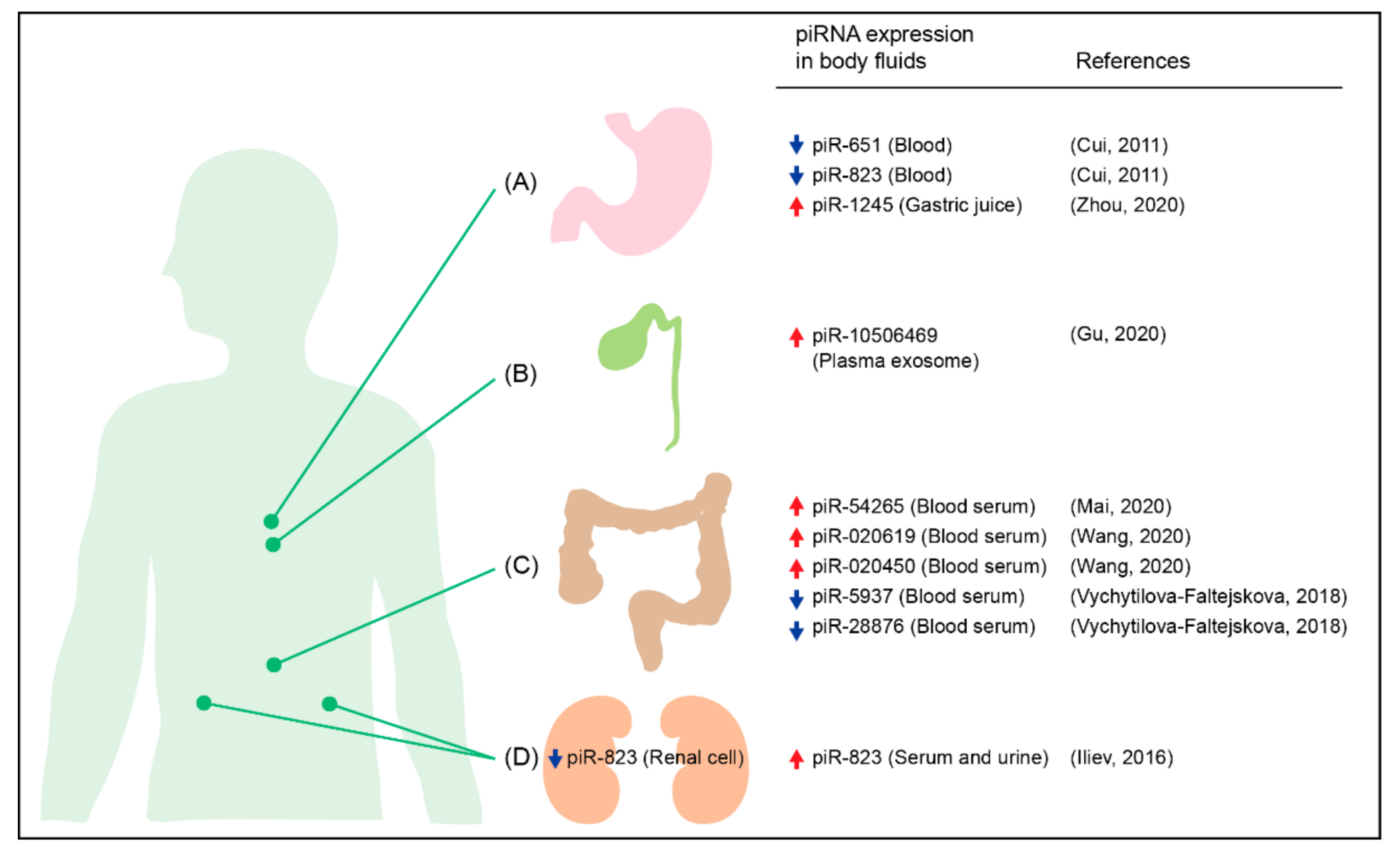
- Cui, L.; Lou, Y.; Zhang, X.; Zhou, H.; Deng, H.; Song, H.; Yu, X.; Xiao, B.; Wang, W.; Guo, J. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin. Biochem. 2011, 44, 1050–1057. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J.; Meng, A.; Zhang, L.; Wang, M.; Fan, H.; Peng, W.; Lu, J. Gastric juice piR-1245: A promising prognostic biomarker for gastric cancer. J. Clin. Lab. Anal. 2020, 34, e23131. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Wang, C.; Deng, H.; Qing, C.; Liu, R.; Liu, S.; Xue, X. Exosomal piRNA profiling revealed unique circulating piRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim. Biophys. Sin. (Shanghai) 2020, 52, 475–484. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Stitkovcova, K.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Kala, Z.; Svoboda, M.; Kiss, I.; Vyzula, R.; et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1019–1028. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yang, H.; Ma, D.; Mu, Y.; Tan, X.; Hao, Q.; Feng, L.; Liang, J.; Xin, W.; Chen, Y.; et al. Serum PIWI-interacting RNAs piR-020619 and piR-020450 are promising novel biomarkers for early detection of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2020, 29, 990–998. [Google Scholar] [CrossRef] [Green Version]
- Iliev, R.; Fedorko, M.; Machackova, T.; Mlcochova, H.; Svoboda, M.; Pacik, D.; Dolezel, J.; Stanik, M.; Slaby, O. Expression levels of PIWI-interacting RNA, piR-823, are deregulated in tumor tissue, blood serum and urine of patients with renal cell carcinoma. Anticancer Res. 2016, 36, 6419–6423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, Y.; Taketomi, Y.; Murakami, M.; Tsujimoto, M.; Yanoshita, R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol. Pharm. Bull. 2013, 36, 66–75. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Cheng, Y.; Lu, Q.; Wei, J.; Yang, H.; Gu, M. Detection of stably expressed piRNAs in human blood. Int. J. Clin. Exp. Med. 2015, 8, 13353–13358. [Google Scholar] [PubMed]
- Ji, L.; Chen, X. Regulation of small RNA stability: Methylation and beyond. Cell Res. 2012, 22, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Kurth, H.M.; Mochizuki, K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 2009, 15, 675–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimta, A.A.; Sigurjonsson, O.E.; Gulei, D.; Tomuleasa, C. The malignant role of exosomes as nanocarriers of rare RNA species. Int. J. Mol. Sci 2020, 21, 5866. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; Kalluri, R. Exosomes as a Multicomponent Biomarker Platform in Cancer. Trends Cancer 2020, 6, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Vourekas, A.; Zheng, Q.; Alexiou, P.; Maragkakis, M.; Kirino, Y.; Gregory, B.D.; Mourelatos, Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 2012, 19, 773–781. [Google Scholar] [CrossRef] [Green Version]
- Pon, J.R.; Marra, M.A. Driver and passenger mutations in cancer. Annu. Rev. Pathol. 2015, 10, 25–50. [Google Scholar] [CrossRef]
- Brase, J.C.; Wuttig, D.; Kuner, R.; Sultmann, H. Serum microRNAs as non-invasive biomarkers for cancer. Mol. Cancer 2010, 9, 306. [Google Scholar] [CrossRef] [Green Version]
- Rapisuwon, S.; Vietsch, E.E.; Wellstein, A. Circulating biomarkers to monitor cancer progression and treatment. Comput. Struct. Biotechnol. J. 2016, 14, 211–222. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, H.; Park, K.H.; Lee, Y.; Jeong, A.; Choi, S.; Kim, K.W. The Regulation and Role of piRNAs and PIWI Proteins in Cancer. Processes 2021, 9, 1208. https://doi.org/10.3390/pr9071208
Jeong H, Park KH, Lee Y, Jeong A, Choi S, Kim KW. The Regulation and Role of piRNAs and PIWI Proteins in Cancer. Processes. 2021; 9(7):1208. https://doi.org/10.3390/pr9071208
Chicago/Turabian StyleJeong, Hyeseon, Kyung Hwan Park, Yuri Lee, Ayoung Jeong, Sooji Choi, and Kyung Won Kim. 2021. "The Regulation and Role of piRNAs and PIWI Proteins in Cancer" Processes 9, no. 7: 1208. https://doi.org/10.3390/pr9071208
APA StyleJeong, H., Park, K. H., Lee, Y., Jeong, A., Choi, S., & Kim, K. W. (2021). The Regulation and Role of piRNAs and PIWI Proteins in Cancer. Processes, 9(7), 1208. https://doi.org/10.3390/pr9071208







