Abstract
Clostridium beijerinckii is a relatively widely studied, yet non-model, bacterium. While 246 genome assemblies of its various strains are available currently, the diversity of the whole species has not been studied, and it has only been analyzed in part for a missing genome of the type strain. Here, we sequenced and assembled the complete genome of the type strain Clostridium beijerinckii DSM 791T, composed of a circular chromosome and a circular megaplasmid, and used it for a comparison with other genomes to evaluate diversity and capture the evolution of the whole species. We found that strains WB53 and HUN142 were misidentified and did not belong to the Clostridium beijerinckii species. Additionally, we filtered possibly misassembled genomes, and we used the remaining 237 high-quality genomes to define the pangenome of the whole species. By its functional annotation, we showed that the core genome contains genes responsible for basic metabolism, while the accessory genome has genes affecting final phenotype that may vary among different strains. We used the core genome to reconstruct the phylogeny of the species and showed its great diversity, which complicates the identification of particular strains, yet hides possibilities to reveal hitherto unreported phenotypic features and processes utilizable in biotechnology.
1. Introduction
Clostridium beijerinckii belongs to the group of authentic Clostridium spp., also referred to as Cluster I “Sensu stricto” [1]. As a Gram-positive, spore forming, rod shaped anaerobe capable of solventogenesis, C. beijerinckii represents an industrially relevant microorganism. From that perspective, butanol seems to be the main subject of research interest. Butanol is produced within either an acetone-butanol-ethanol (ABE) [2] or isopropanol-butanol-ethanol (IBE) [3] fermentation pathway that covers a bi-phasic process in which acid formation is later followed by the formation of solvents. Moreover, solventogenesis is usually coupled with sporulation. Nevertheless, both processes do not seem to be closely linked in all strains [4]. Thus, evolutionary and comparative studies of various clostridial strains are required to help reveal hidden aspects of the production of valuable chemicals by microbial cell factories [5].
Although the evolution and taxonomy of the clostridia were revisited several times [1,6,7,8], additional studies are needed as the clostridia still represent a polyphyletic group with uncertain phylogenetic affinities and reidentifications of particular strains and reclassifications of various clostridial species are quite common [9,10,11]. The most recent change affecting C. beijerinckii is the reclassification of C. diolis as C. beijerinckii [12]. These taxonomic readjustments were expected as the previous taxonomy was built upon phenotypic differences that do not necessarily reflect genetic heterogeneity [13]. A massive reduction in DNA sequencing costs over the past 20 years resulted in many genomes of non-model bacteria being sequenced. This applies also to C. beijerinckii strains, and, currently, there are 241 genome assemblies of various C. beijerinckii strains and five genome assemblies of three different C. diolis strains in the GenBank database (May 2021). Unfortunately, most genomes are assembled only in the form of draft assemblies, thus leading to gaps in knowledge. Although draft assemblies of type strains C. beijerinckii DSM 791T and C. diolis DSM 15410T were sufficient to reveal that C. bejerinckii and C. diolis are heterotypic synonyms [12], it was the complete genome assembly that helped to reveal hitherto unreported features of C. diolis DSM 15410T. A thorough analysis proved its ability to produce isopropanol [14] that was hidden for 18 years since the description of the species in 2002 [15]. There are also other strains of C. beijerinckii that are studied without the knowledge of a genome sequence; for example, the strain C. beijerinckii F-6, a butanol-tolerant hydrogen producer for which only a 16S RNA gene sequence is known [16,17].
Following reclassification there are now two type strains: C. beijerinckii DSM 791T (=ATCC 25752, E. McCoy A-67, L.S. McClung 1671, NCIMB 9362) and C. beijerinckii, formerly C. diolis, DSM 15410T (=DSM 5431, SH1, 88-273, ATCC BAA-557). Although the reclassification means the loss of type strain designation, this has not been universally agreed by various sources. Thus, the strain C. beijerinckii DSM15410T can be found as the type strain in the German Collection of Microorganisms and Cell Cultures (DSZM) catalogue and must be searched as C. diolis DSM 15410 in the GenBank database. Here, we refer to all C. beijerinckii/diolis strains as C. beijerinckii strains for two reasons. First, the species name C. beijerinckii was proposed several decades before C. diolis [15,18]. Second, the number of assemblies is considerably higher for the C. beijerickii species, and it contains several well-studied strains, e.g., C. beijerinckii NCIMB 8052 [19,20,21,22] (formerly C. acetobutylicum [23]), C. beijerinckii NRRL B-598 [24,25,26,27] (formerly C. pasteurianum [9]), and C. beijerinckii DSM 6423 [28,29]. Despite that, only a draft genome sequence of the type strain C. beijerinckii DSM 791T has been available until now. In this paper, we sequenced and assembled the complete genome of C. beijerinckii DSM 791T that contains a chromosome and a single megaplasmid. The presence of the plasmid has never been reported before as the type strain has not been studied in detail. Additionally, we performed its annotation, and included the classification of protein coding sequences (CDSs) into clusters of orthologous groups (COG), a prediction of the operon structure, and the identification of prophage DNA and CRISPR arrays. Eventually, we compared its main genomic features to the other type strain (DSM 15410T) and performed an extensive comparative study of all currently available C. beijerinckii genomes to define its pangenome. Although some reports comparing genomes of various C. beijerinckii strains have been published [9,12,14,17,30], they are limited to comparing only a few genomes simultaneously, and, sometimes, they have used only 16S rRNA gene sequences. This is the first report to show the diversity of the C. beijerinckii species and the first study to use the maximum information available, thanks to the definition of the core genome from 237 genomes.
2. Materials and Methods
2.1. Bacterial Strain
The strain Clostridium beijerinckii DSM 791T was obtained from the German public collection of microorganisms at the Leibnitz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.
2.2. Cultivation
The strain inoculum was prepared from a cryopreserved culture stored at −80 °C in 30% glycerol. Cells were cultivated at 37 °C in liquid tryptone-yeast extract-acetate (TYA) medium overnight and subsequently transferred on solidified TYA medium with agar. The selected colony was transferred into a liquid medium and cultivated for 24 h. Cells from 10 mL of suspension were pelleted by centrifugation (6000× g), washed with sterile demi water, pelleted again, and stored at −80 °C prior to DNA isolation. Up to the final centrifugation and wash step, all operations were performed under a nitrogen atmosphere in an anaerobic chamber Concept 400 (Ruskinn). The composition of TYA media was as follows: 20 g·L−1 glucose; 2 g·L−1 yeast extract; 6 g·L−1 tryptone; 0.5 g·L−1 KH2PO4; 3 g·L−1 ammonium acetate; 0.3 g·L−1 MgSO4.7H2O; 0.01 g·L−1 FeSO4; (agar 20 g·L−1); pH was adjusted to 6.8 prior to sterilization (121 °C, 20 min).
2.3. DNA Extraction and Sequencing
For long-read sequencing, genomic high molecular weight DNA was extracted using the MagAttract HMW DNAKit (Qiagene, Venlo. NL). The extracted DNA purity and proper length were checked using the NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent 4200 TapeStation (Agilent technologies, Santa Clara, CA, USA), respectively. Library preparation for Oxford Nanopore sequencing was performed using the Ligation sequencing 1D Kit (Oxford Nanopore Technologies, Oxford, UK). The library was sequenced using the R9.4.1 flowcell and the MinION platform (Oxford Nanopore Technologies).
For short-read sequencing, genomic DNA was purified using the GenElute Bacterial Genomic DNA Kit (SIGMA-ALDRICH, St. Louis, MI, USA). The extracted DNA purity was checked by NanoDrop (Thermo Fisher Scientific). The sequencing library was prepared using the KAPA HyperPlus kit and was carried out using the Miseq Reagent Kit v2 (500 cycles) and the Illumina MiSeq platform (Illumina, San Diego, CA, USA).
2.4. Genome Assembly
Basecalling of raw nanopore squiggles was performed with Guppy v3.4.4 and the quality of reads was checked with MinIONQC [31]. Similarly, the initial quality assessment of Illumina raw reads was conducted through a combination of FastQC v0.11.5 and MultiQC v1.7 [32]. The adapter and quality trimming was performed using Trimmomatic v1.36 [33]. Initial genome assemblies were constructed using long reads with a Flye v2.8.1 assembler [34] and short reads with a plasmidSPAdes v3.11.1 assembler [35]. Contigs from both assemblies were compared with NUCmer v3.1 [36] and selected contigs were further polished. The first step of polishing was done by four rounds of mapping long reads with minimap2 v2.17 [37] and polishing with racon v1.4.20 [38]. The second step consisted of two rounds of polishing with medaka v1.2.5, again in combination with long reads. Finally, the third step of polishing was done by two rounds of mapping short reads with BWA v0.7.17 [39] and polishing with pilon v1.24 [40], while handling files of mapped reads with SAMtools v1.7 [41]. Resulting contigs were manually examined for circularity by concatenating their ends and mapping short and long reads with BWA and minimap2, respectively. Missing or duplicated bases were manually added or trimmed. Eventually, replication origins of contigs were predicted. Chromosomal replication origin (oriC) was predicted with Ori-finder [42] and the sequence was rearranged according to its position, so the dnaA gene is the first gene in the chromosomal sequence. Similarly, replication origin in plasmid (oriV) was predicted using BLAST [43] searches against the database of replication origins DoriC [44] and the sequence was rearranged according to its position, so the repB gene is the first gene in the plasmid sequence.
2.5. Genome Annotation and Analysis
A genome annotation was added by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [45]. An operon prediction was completed using the Operon-mapper [46]. The functional annotation of the protein coding genes was performed by assigning clusters of orthologous group (COG) categories from the eggNOG database with the eggNOG-mapper [47]. Circular genome maps of a chromosome and a megaplasmid were prepared with a DNAplotter [48] integrated in Artemis [49]. The genome was searched for prophage DNA with PhiSpy v4.2.12 [50] and for clustered regularly interspaced short palindromic repeats (CRISPR) arrays with CRISPRDetect v2.3 [51].
Entrez was used to search the GenBank database for C. beijerinckii/diolis genomes [52]. A circular graph showing whole-genome alignments was produced with BRIG v0.95 [53]. Nucleotide sequences of reference genes from the strain C. beijerinckii DSM 791T were localized in other strains using BLAST [43] searches and a 90% sequence similarity in Ridom SeqSphere+ v7.6.1. Outliers were detected with R package graphics [54] using the boxplot function and setting a whiskers range as 10 times the interquartile range. Digital DNA to DNA hybridization (dDDH) values were calculated using the type strain genome server (TYGS) [55]. The core and accessory genomes and unique genes were identified with BPGA v1.3.0 [56] using amino acid sequences and a 90% sequence similarity. All sequences of unique genes and reference sequences of pangenome genes were uploaded to FAIRDOMHub [57]. The phylogenetic tree was reconstructed using the concatenated core genome sequences with the neighbor-joining method in BPGA. A reduction of leaf nodes in the tree was done by collapsing branches where the whole length was shorter than 1% of the longest branch in the tree. A final visualization of trees was conducted through Evolview v3 [58].
3. Results
3.1. Genome Sequencing and Assembly
Oxford Nanopore sequencing produced 690,277 reads with a N50 length of 21,578 bp. Moreover, 700 reads exceeded the length of 100 kbp, while five were even longer than 200 kbp. The Illumina sequencing produced more than 2.1 million additional paired reads of 250 bp in length. The final genome assembly consisted of two circular contigs. While the first one represented a circular chromosome of length 5,876,902 bp, the second corresponded to a 73,345 bp long megaplasmid. Both sequences have been deposited at the DDBJ/EMBL/GenBank under accession numbers CP073653 for the chromosome and CP073654 for the megaplasmid. Coverage of the assembly after the filtering steps was approximately 1063× and the assembly was reconstructed with the contribution of more than 2.1 million paired Illumina reads (more than 99% of all Illumina reads and more than 99% of all Illumina sequenced bases after quality trimming) and more than 640,000 of Oxford Nanopore reads (almost 90% of all Oxford Nanopore reads and more than 93% of all Oxford Nanopore sequenced bases). Average coverage, considering only short reads, was 88× for chromosome and 192× for plasmid.
3.2. The Characteristics of the C. beijerinckii DSM 791 Genome
The guanine–cytosine (GC) content of the genome was calculated as 29.87%. While the GC content of the chromosome was almost 29.90%, the GC content of the plasmid was slightly lower, reaching only 27.84%, see Table 1. The complete genome contained 5279 annotated open reading frames (ORFs) divided into 3291 operons, see Table 1 for separate statistics for chromosome and plasmid. The majority of ORFs consisted of protein coding genes, but 134 pseudogenes were also found. The sequences of 60 pseudogenes were found to be incomplete, 56 were frameshifted, 34 contained internal stops, and 13 suffered from multiple problems. The positions of particular features within the chromosome and plasmid are shown in Figure 1. Additionally, protein coding genes and pseudogenes were assigned COG categories. Unfortunately, 687 CDSs were not assigned any COG and an additional 990 CDSs were assigned to group S with an unknown function. Nevertheless, the remaining 3454 CDSs (out of all 5131 protein coding genes and pseudogenes) were divided into the remaining COG categories, see Supplementary Table S1.

Table 1.
Genome features of Clostridium beijerinckii DSM 791T.
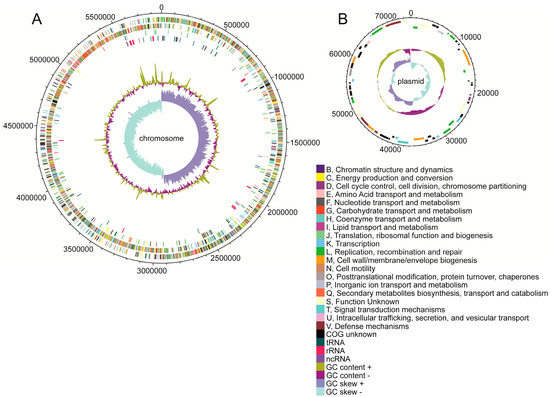
Figure 1.
Circular maps of C. beijerinckii DSM 791 (A) chromosome and (B) plasmid. The outermost and the second outermost circles represent CDSs on the forward and reverse strands, respectively. The third circle represents pseudogenes, and the colors represent the COG functional classification. The fourth circle represents RNA genes, while the colors distinguish between tRNA, rRNA, and ncRNA. The inner shaded area represents (from outside in) GC content and GC skew, plotted using a 10-kb window with a step of 200 bp while the colors distinguish between above and below average values.
ORFs were searched for prophage genes, which resulted in 10 identified prophages of lengths ranging from 6583 bp to 42,566 bp, see Supplementary Table S2. All prophages were located on the chromosome. While the lowest number of genes in a prophage was eight, two prophages consisted of 51 genes. The cumulative length of prophages was 246,958 bp, which is less than 4.2% of the genome.
Only two CRISPR arrays were found (see Supplementary Table S3). Both arrays were extremely short, only 172 bp and 153 bp long, with two spacer units. Moreover, no cas or cas-like genes were found in their neighborhoods.
3.3. Diversity of C. beijerinckii Strains
A search for C. beijerijcki/diolis genomes in the GenBank database obtained 246 genome assemblies. After deduplication of multiple assemblies for the same strains, 242 assemblies were preserved, from which 11 represented complete genomes or complete chromosomal sequences, see Supplementary Table S4.
A comparison of complete chromosomal sequences to the reference, C. beijerinckii DSM 791T chromosome, is shown in Figure 2. While the majority of genomes mapped to the reference with almost 100% identity in whole length, there were two genomes with lower similarities. While some parts of the DSM 6423 genome mapped with 95% identities, the majority of the WB53 genome mapped with considerably lower identities, not exceeding 90%. Those results were further supported by dDDH analysis that showed dDDH values between particular strains and the reference ranging from 71.9% to 86.4%, except for strains DSM 6423 and WB53 where the dDDH value to the strain DSM 791T was 67.4% and 20.3%, see Supplementary Table S5.
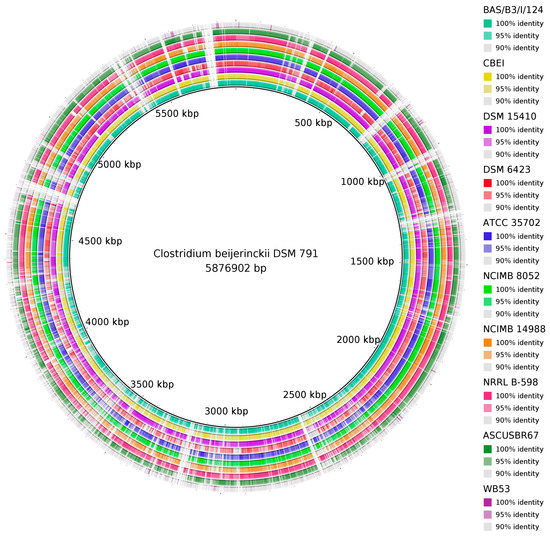
Figure 2.
Whole-genome comparison of the type strain C. beijerinckii DSM 791T to other C. beijerinckii complete genomes with the visualization of genomes percentage identity.
A whole-genome comparison was further applied to the whole dataset of 242 genomes. Genes of the reference strain C. beijerinckii DSM 791T were searched in other genomes, using nucleotide BLAST and a 90% sequence similarity, to find missing genes or genes with missing stop codon, see Supplementary Table S6. The median value of missing or corrupted genes was 993 and seven outliers were detected. While three outliers exceeded the upper fence, four outliers lay below the lower fence. Those exceeding the limit were genomes of strains ASCUSBR67, HUN142, and WB53, with 2599, 1811, and 4778 missing or corrupted genes, respectively. The dDDH value between the type strain and the strain HUN142 was 48.5%. Outliers below the lower fence were genomes of strains DJ079, DJ317, NBRC 109359, and NCTC13035, with 116, 96, 125, and 144 missing or corrupted genes, respectively.
3.4. Pangenome of C. beijerinckii
After removing unannotated genomes and genomes of strains that do not belong to the C. beijerinckii species, the remaining 237 genomes were used to define the core genome. In total, 2308 genes were present in all genomes, and formed the core genome. Additionally, 12,202 genes were found in at least two genomes and formed the accessory genome. Together, they presented the pangenome of C. beijerinckii containing 14,510 genes. Moreover, 5929 genes were unique, i.e., they were found in only one genome. The number of unique genes for particular strains ranged from zero to 516 (found in the strain DJ015), see Supplementary Table S7. While the median value of unique genes was three, type strains DSM 791T and DSM 15410T had one and 148 unique genes. In addition, genes exclusively absent were also counted. Such genes were found in every strain except for one. The number of exclusively missing genes ranged from zero to 86 (missing in the strain DJ032). The median value was zero and type strains DSM 791T and DSM 15410T were missing six genes and one gene, respectively.
A functional annotation of genes showed a different composition of the core genome and the accessory genome (see Figure 3). The core genome contained a larger proportion of genes connected to metabolism and energy production and conversion, except for genes in group (Q) “Secondary metabolites biosynthesis, transport and catabolism”, where the accessory genome and unique genes had higher relative abundances. Similarly, genes connected to repair and defense mechanisms were more abundant in the accessory genome and among unique genes. While only 6.28% of genes were not assigned any COG in the core genome, their abundance in the accessory genome and among unique genes exceeded 35%, see Supplementary Table S8.
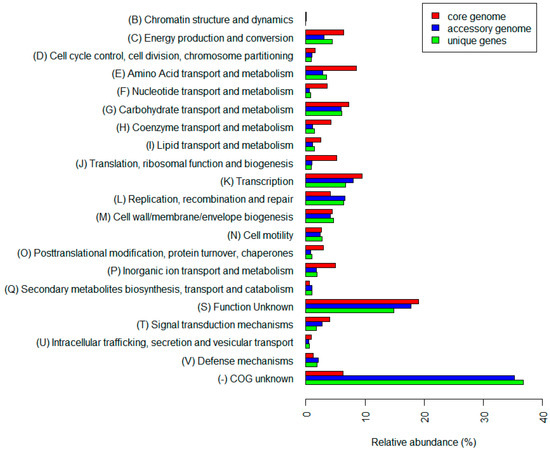
Figure 3.
Relative abundances of genes in particular clusters of orthologous groups. Color coding distinguishes between gene from core (red) and accessory (blue) genomes and genes unique for particular strains (green).
Amino acid sequences of unique genes for particular Clostridium beijerinckii strains as well as reference sequences of genes present in the Clostridium beijerinckii pangenome were uploaded to the FAIRDOMHub under the project “Clostridium beijerinckii pan-genome” https://fairdomhub.org/projects/242 (accessed on 1 June 2021).
3.5. Phylogeny
Finally, the phylogenetic tree of C. beijerinckii strains was reconstructed using the core genome (see Figure 4). Evolutionary closely related strains were collapsed into 16 clusters. While most clusters contained only units of strains, cluster 1 covered 145 strains, see Supplementary Table S9. The complete tree is showed in Supplementary Figure S2. While the type strain DSM 791T had four closely related strains and formed a cluster, the other type strain DSM 15410T formed an individual leaf node for which the strain NRRL B-598 was the closest strain.
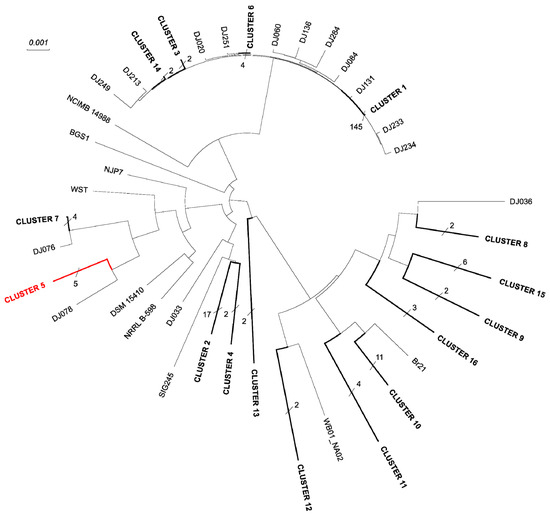
Figure 4.
Phylogenetic reconstruction of Clostridium beijerinckii species. The phylogeny was reconstructed using 2308 genes of the core genome, present in every strain. Branches that were shorter than 1% of the maximum branch length were collapsed. The number of strains hidden under collapsed branches is written to each collapsed branch and strains are listed in Supplementary Table S9. The branch containing the type strain C. beijerinckii DSM 791T is highlighted in red.
4. Discussion
The number of exceedingly long reads presented ideal data for a complete genome assembly that allowed for the identification of possible plasmids, while a number of high-quality short reads were ideal for polishing the final assembly. The presence of the megaplasmid was evident from the initial Oxford Nanopore assemblies by Flye as two long circular contigs, regardless of whatever input parameters were produced. However, the length of the shorter contig varied considerably between assemblies that were produced with different input parameters. Thus, the length of plasmid was initially predicted by comparing Flye contigs to contigs produced by plasmidSPAdes. The final confirmation was found in the different coverage of chromosome and plasmid in the final assembly as the read coverage is one possible way to distinguish between chromosome and plasmid [35]. Since almost all high-quality Illumina reads mapped unambiguously to the final genome assembly, the proposed assembly seems to be correct and of a high quality. Although the prediction of replication origin oriC in the chromosome was unambiguous, the prediction of replication origin oriV in the plasmid was complicated as no standardized algorithm is currently available [44]. The first from two putative replication initiators was frameshifted and the second, while having a complete coding sequence, had a noticeably short upstream intergenic region. As both genes were located relatively close to each other, the start of the sequence was set according to the first one.
The calculated GC content matched our presumptions as clostridia form a low GC content group of Gram-positive bacteria. While the GC content of the chromosome was the same as for other strains with complete genomes, e.g., NRRL B-598 [27], NCIMB 8052 [20], DSM 15410T [14], the GC content of the plasmid was slightly lower. Another difference between the chromosome and the plasmid was found in abundances of genes in particular COG categories, see Supplementary Table S1. While the plasmid carried zero genes from group (C) “Energy production and conversion”, the same group contained 6.28% genes of the chromosome. Similarly, the chromosome carried larger percentage of genes in groups (E) “Amino Acid transport and metabolism”, (F) “Nucleotide transport and metabolism”, (G) “Carbohydrate transport and metabolism”, (H) “Coenzyme transport and metabolism”, (I) “Lipid transport and metabolism”, and (J) “Translation, ribosomal function and biogenesis”, i.e., all groups directly or indirectly connected to metabolism. Thus, the plasmid in the strain C. beijerinckii DSM 791T does not contribute to the production of metabolites utilizable in industrial biotechnology. In other clostridia, plasmids sometimes carry genes necessary for solventogenesis. For example, sol operon in C. acetobutylicum ATCC 824 is located on a plasmid that can be lost during the degeneration process, resulting in a loss of ability to produce solvents [59]. On the contrary, the plasmid in C. beijerinckii DSM 791T carried a larger percentage of genes in groups (L) “Replication, recombination and repair” and (V) “Defense mechanisms”, which means that the plasmid possessed a machinery capable of protecting the cell. The latter group contained five genes from which three belonged to the cluster involved in the production of the bacteriocin circularin A. The cluster contained genes cfgR (KEC93_26215), cfgK (KEC93_26220), cfg02 (KEC93_26225), cfg01 (KEC93_26230), cirA (KEC93_26235), cirB (KEC93_26240), cirC (KEC93_26245), cirD (KEC93_26250), cirG (KEC93_26255), cirH (KEC93_26260), and cirI (KEC93_26265). Such a structure of the gene cluster has been already experimentally proved for C. beijerinckii DSM 791T. However, it was not revealed that it is carried by a plasmid [60]. In our sequence, gene cirE, which is necessary for bacteriocin production, was not annotated by PGAP. This was probably due to its short length of only 150 bp. Nevertheless, the gene is present in the sequence between cirD and cirG as its nucleotide sequence had 100% similarity to that already sequenced and experimentally proved by Kemperman et al. [60] (GenBank accession no. AJ566621.1). Circularin A has a wide activity range that inhibits C. tyrobutyricum, lactococci, enterococci, and some Lactobacillus strains. It is also highly resistant to digestion by sequence-specific endoproteinases [61]. For potential industrial use, the ability of the strain to kill other bacteria is highly advantageous, as contamination of fermentation processes by bacteria presents a compelling problem that makes the fermentation more expensive [62]. Moreover, the strain C. beijerinckii DSM 791T has already been proved to be a robust 1,3-propanediol producer [63].
The number and cumulative length of putative prophages were approximately half compared with the strain DSM 15410T [14]. However, this prediction is questionable as two additional tools, PHASTER [64] and Prophage Hunter [65], predicted six and 30 prophages, respectively (data not shown). Although bacteriophage infection presents a serious problem for ABE fermentation, it has been addressed by only a few studies to date. Its analysis is not trivial and usually requires a separate study [66,67]. Thus, the question of phages in C. beijerinckii DSM 791T remains open. Unlike the strain DSM 15410T [14], CRISPR arrays in the strain DSM 791T had no cas or cas like genes in their neighborhoods. This suggests that while a culture of the strain DSM 791T might be resistant to other bacteria thanks to bacteriocin production, it might be defenseless against phage contaminations, as a CRISPR-associated system (Cas) forms somewhat of a bacterial immune system that provides protection from foreign genetic material [68].
Clostridium beijerinckii is a widely studied species, as the number of genome assemblies available suggests. Nevertheless, only a small fraction of them present high-quality complete genomes. While this analysis showed that the species is extremely diverse, and different strains may contain different genes, it is evident that some strains have been misidentified. This applies primarily to the strain that was WB53 isolated from a woodchip bioreactor [69]. All analyses, whole-genome alignment, dDDH, and a search for reference genes, presented strong evidence that the strain WB53 does not belong to the C. beijerinckii species and was omitted from following analyses to define the core genome. Similarly, the remaining two outliers arising from an analysis of the reference genes were discarded. Although the dDDH value for the strain ASCUSBR67 exceeded 70%, suggesting that it belongs to the C. beijerinckii species [70], its genome lacks the annotation that was needed since amino acid sequences were used in the following analysis. Unlike WB53, the strain ASCUSBR67 had a majority of corrupted, not missing genes (see Supplementary Table S4). This means that its assembly needs polishing by high-quality NGS data for further analyses, including genome annotation [71]. Yet, it is likely that the strain belongs to C. beijerinckii. The last outlier, the genome of the strain HUN142, had most of the reference genes missing. Together with a dDDH value below 70%, we can conclude that this strain does not belong to C. beijerinckii. It should be noted that when calculating the dDDH value, the d6 formula was used for complete genomes as it preserves the maximum amount of information, while the d4 formula was used for draft genomes as a more robust way to compare incomplete data [72]. Genomes of outliers below the lower fence in a reference genes’ analysis were preserved for following analyses as these represented strains that are evolutionary extremely close to the type strain C. beijerinckii DSM 791T.
Similar to ASCUSBR67, the study discarded other unannotated genomes for which amino acid sequences of protein coding genes are unavailable. Those were genomes of strains WB and G117. On the other hand, the study preserved the genome of the strain DSM 6423 [29], thedDDH value of which was below the 70% threshold. First, when considering the confidence interval, a dDDH value of 70% was reached. Second, its number of missing or corrupted reference genes did not differ substantially from other strains. Since clostridia present a diverse group of organisms, this study contends that following only a dDDH value for their delineation may be cumbersome.
It is not surprising that the core genome had a larger proportion of annotated genes as it contains housekeeping genes that maintain basic cellular functions; additionally, their orthologues are known as they are present in other related species [73]. The core genome can be used to improve a phylogenetic analysis and to correctly assign bacterial species [74]. This is highly convenient for clostridia where the reidentification of strains is still quite common. Thus, the use of the core genome defined within this study is suggested as a supplement to a dDDH analysis when assigning new strains to the C. beijerinckii species. From a biotechnological point of view, the core genome is not of the main interest as it contains primarily critical functions, the alteration or deletion of which are often not possible. This is the reason why the core genome lacks genes responsible for various biotechnologically relevant phenotypic manifestations, as the production of solvents varies among strains. The core genome contains master regulators, for example spo0A (Gene3468# in the core genome) that orchestrates except for sporulation also solvent production. On the other hand, particular genes coding enzymes necessary to form the final products, e.g., adh (Gene3611# in the accessory genome) that is responsible for isopropanol production or dhaT (Gene1153# in the accessory genome) that is responsible for 1,3-propandiol production, are parts of the accessory genome. Therefore, they are not present in every strain. Nevertheless, some of the enzymes still present parts of the core genome, e.g., adh (Gene4950# in the core genome) that is responsible for ethanol production or bdh (Gene1521# in the core genome) that is responsible for butanol production. This was expected as C. beijerinckii covers ABE and IBE fermentation strains, meaning that while the ability to produce acetone or isopropanol varies among strains, ethanol and butanol are produced by all strains. Nevertheless, other metabolic regulations may suppress their production, for example ethanol production is negligible in the strain C. beijerinckii NRRL B-598 under standard cultivation conditions [24]. Additionally, solvent production may be interrupted during the so-called acid crash phenomenon induced by cultivation conditions or genome mutations [75,76,77].
The core genome can find additional utilization in the identification of versatile reference genes for RT-qPCR in C. beijerinckii, as such genes should be present in every strain. However, experimental validation is always needed, as a reference gene-coding peptidase T (Gene4499# in the core genome), proposed as a reference gene for the strain NCIMB 8052 [20], was later proved to be not utilizable for the strain NRRL B-598 [78], where genes greA (Gene2564# in the core genome), zmp (Gene1191# in the core genome) and others performed better.
The fact that approximately one third of the accessory genome and unique genes was not assigned any COG suggests that orthologues for these genes are not present in more studied species. These genes might provide desirable phenotypic features, but advances in the field of functional annotation of non-model organisms are required to reveal these hidden properties. The accessory genome also contains several sequences of hypothetical proteins, the sequences of which are formed from repetitive subsequences. Even though these sequences were found in two or more genomes, they are probably not real proteins, and they might prevent the accessory genome from being used in BLAST searches. We suggest the use of the corrected accessory genome for BLAST searches where 76 sequences with a single kind of amino acid forming more than 25% of a sequence are discarded. The corrected Fasta file was uploaded to the FAIRDOMHub.
Although genes coding bacteriocin circularin A have been described only for the type strain C. beijerinckii DSM 791T [60], they are present also in other strains, as the type strain has only one unique gene. Nevertheless, the presence of the whole cluster of all genes was not found in any other genome, nor in the closely related genomes that formed cluster 5 in the phylogenetic tree. Apart from the type strain, the cluster contained strains NBRC 10935, NCTC13035, DJ317, and DJ079, which were found to be closely related in the preceding analysis of reference genes as four outliers below the lower fence.
Various strains of C. beijerinckii are extremely diverse, except for one large cluster containing evolutionary closely related strains. The majority of them (DJ strains) were uploaded to the GenBank at the same time by the DOE Joint Genome Institute. Unfortunately, the study describing these strains is missing. However, not all of the DJ strains clustered together and some of them even formed particular leaf nodes. From the type strain, the evolutionary-distant strain DSM 6423 was clustered with an additional five strains into cluster 15. Although both type strains have different genome characteristics, they are evolutionary closer to each other than to DSM 6423. The closest neighbor to the IBE fermenting type strain DSM 15410T is the strain NRRL B-598, a typical ABE fermentation representative. This is surprising because evolutionary-distant DSM 6423 is also an IBE strain. The absence of a distinguished cluster of IBE strains is further supported by the position of the individual leaf node with the strain C. beijerinckii BGS1, which is another isopropanol producer [30]. These relations only confirm that the evolution of Clostridium beijerinckii is not trivial, and novel strains should be identified using the whole core genome rather than particular genes or a dDDH analysis.
5. Conclusions
In this study, we sequenced and assembled the first complete genome sequence of the type strain Clostridium beijerinckii DSM 791T. We discovered that the genome of the type strain is composed of a circular chromosome and a circular megaplasmid that carries a complete cluster of genes to produce bacteriocin circularin A, which is unique among C. beijerinckii strains. We used the genome sequence for whole-genome comparisons, and we found out that at least two strains currently assigned as C. beijerinckii, WB53 and HUN142, do not belong to the species. Moreover, we proved that some of the genome assemblies, e.g., the genome sequence of the strain C. beijerinckii ASCUSBR67, are of lower quality and should be polished before they can be used for comparative analyses. By collecting 237 genomes that met the quality criteria, we defined for the first time the pangenome of the Clostridium beijerinckii species. We used the core genome to reconstruct a phylogeny of the whole species, using the maximum sequence information available. As we demonstrated, phylogeny of the species is not trivial, and we suggest use of the core genome when performing comparative analysis and identification of novel strains. The accessory genome contained a large percentage of genes with unknown function. This means, therefore, that many unique properties of the C. beijerinckii species might be still unreported.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pr9071196/s1, Supplementary tables and figures: Figure S1: Boxplot of sum of missing and corrupted genes, Figure S2: Complete phylogenetic tree of Clostridium beijerinckii strains, Table S1: List of COG categories and number of genes present in each COG in C. beijerinckii DSM 791T genome, Table S2: Phage DNA within the C. beijerinckii DSM 791T genome, Table S3: CRISPR arrays in the C. beijerinckii DSM 791T genome, Table S4: List of C. beijerinckii genomes used for analysis, Table S5: Result of dDDH analysis for complete C. beijerinckii genomes, Table S6: Numbers of genes in reference genome that are missing in particular strains, Table S7: List of unique and uniquely missing genes in C. beijerinckii strains, Table S8: List of COG categories and number of genes present in each COG in C. beijerinckii core and accessory genomes and unique genes, Table S9: List of strains in each of collapsed branches in Figure 4.
Author Contributions
Conceptualization, K.S., M.L., P.P. and H.S.; methodology, K.S., M.N., M.B. and B.B., formal analysis, K.S. and M.N.; investigation, M.B. and B.B.; resources, K.S., M.L. and P.P.; data curation, K.S.; writing—original draft preparation, K.S., M.B. and B.B.; writing—review and editing, K.S.; visualization, K.S., M.N. and H.S.; supervision, K.S. and P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The genome assembly referred in this paper is the version GCA_018223745.1. Sequences of the chromosome and the plasmid were uploaded to GenBank under accession numbers CP073653.1 and CP073654.1. The whole-genome sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under the project accession number PRJNA724001. Sequences of pangenome and unique gene were uploaded to FAIRDOMHub under the project “Clostridium beijerinckii pan-genome” https://fairdomhub.org/projects/242 (accessed on 1 June 2021) under the Creative Commons Attribution-NonCommercial 4.0 license.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cruz-Morales, P.; Orellana, C.; Moutafis, G.; Moonen, G.; Rincon, G.; Nielsen, L.; Marcellin, E. Revisiting the Evolution and Taxonomy of Clostridia, a Phylogenomic Update. Genome Biol. Evol. 2019, 11, 2035–2044. [Google Scholar] [CrossRef]
- Schiel-Bengelsdorf, B.; Montoya, J.; Linder, S.; Dürre, P. Butanol fermentation. Environ. Technol. 2013, 34, 1691–1710. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.F.D.S.; Filho, F.M.; Filho, R.M.; Mariano, A.P. Acetone-free biobutanol production: Past and recent advances in the Isopropanol-Butanol-Ethanol (IBE) fermentation. Bioresour. Technol. 2019, 287, 121425. [Google Scholar] [CrossRef]
- Patakova, P.; Kolek, J.; Sedlar, K.; Koscova, P.; Branská, B.; Kupkova, K.; Paulova, L.; Provaznik, I. Comparative analysis of high butanol tolerance and production in clostridia. Biotechnol. Adv. 2018, 36, 721–738. [Google Scholar] [CrossRef]
- Charubin, K.; Bennett, R.K.; Fast, A.G.; Papoutsakis, E.T. Engineering Clostridium organisms as microbial cell-factories: Challenges & opportunities. Metab. Eng. 2018, 50, 173–191. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Song, Y.; Liu, C. Taxonomy—General Comments and Update on Taxonomy of Clostridia and Anaerobic cocci. Anaerobe 2002, 8, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Keis, S.; Bennett, C.F.; Ward, V.K.; Jones, D.T. Taxonomy and Phylogeny of Industrial Solvent-Producing Clostridia. Int. J. Syst. Bacteriol. 1995, 45, 693–705. [Google Scholar] [CrossRef]
- Johnson, J.L.; Francis, B.S. Taxonomy of the Clostridia: Ribosomal Ribonucleic Acid Homologies among the Species. J. Gen. Microbiol. 1975, 88, 229–244. [Google Scholar] [CrossRef]
- Sedlar, K.; Kolek, J.; Provaznik, I.; Patakova, P. Reclassification of non-type strain Clostridium pasteurianum NRRL B-598 as Clostridium beijerinckii NRRL B-598. J. Biotechnol. 2017, 244, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Lawson, P.A.; Rainey, F.A. Proposal to restrict the genus Clostridium Prazmowski to Clostridium butyricum and related species. Int. J. Syst. Evol. Microbiol. 2016, 66, 1009–1016. [Google Scholar] [CrossRef]
- Moon, C.D.; Pacheco, D.M.; Kelly, W.J.; Leahy, S.; Li, D.; Kopečný, J.; Attwood, G. Reclassification of Clostridium proteoclasticum as Butyrivibrio proteoclasticus comb. nov., a butyrate-producing ruminal bacterium. Int. J. Syst. Evol. Microbiol. 2008, 58, 2041–2045. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tanizawa, Y.; Sakamoto, M.; Nakamura, Y.; Ohkuma, M.; Tohno, M. Reclassification of Clostridium diolis Biebl and Spröer 2003 as a later heterotypic synonym of Clostridium beijerinckii Donker 1926 (Approved Lists 1980) emend. Keis et al. 2001. Int. J. Syst. Evol. Microbiol. 2020, 70, 2463–2466. [Google Scholar] [CrossRef]
- Ackermann, M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Genet. 2015, 13, 497–508. [Google Scholar] [CrossRef]
- Sedlar, K.; Vasylkivska, M.; Musilova, J.; Branska, B.; Provaznik, I.; Patakova, P. Phenotypic and genomic analysis of isopropanol and 1,3-propanediol producer Clostridium diolis DSM 15410. Genomics 2021, 113, 1109–1119. [Google Scholar] [CrossRef]
- Biebl, H.; Spröer, C. Taxonomy of the Glycerol Fermenting Clostridia and Description of Clostridium diolis sp. nov. Syst. Appl. Microbiol. 2002, 25, 491–497. [Google Scholar] [CrossRef]
- Wu, J.; Dong, L.; Zhou, C.; Liu, B.; Xing, D.; Feng, L.; Wu, X.; Wang, Q.; Cao, G. Enhanced butanol-hydrogen coproduction by Clostridium beijerinckii with biochar as cell’s carrier. Bioresour. Technol. 2019, 294, 122141. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Dong, L.; Zhou, C.; Liu, B.; Feng, L.; Wu, C.; Qi, Z.; Cao, G. Developing a coculture for enhanced butanol production by Clostridium beijerinckii and Saccharomyces cerevisiae. Bioresour. Technol. Rep. 2019, 6, 223–228. [Google Scholar] [CrossRef]
- Skerman, V.B.D.; Sneath, P.H.A.; McGowan, V. Approved Lists of Bacterial Names. Int. J. Syst. Evol. Microbiol. 1980, 30, 225–420. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Mao, Y.; Blaschek, H.P. Genome-wide dynamic transcriptional profiling in Clostridium beijerinckii NCIMB 8052 using single-nucleotide resolution RNA-Seq. BMC Genom. 2012, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Mao, Y.; Blaschek, H.P. Single-nucleotide resolution analysis of the transcriptome structure of Clostridium beijerinckii NCIMB 8052 using RNA-Seq. BMC Genom. 2011, 12, 479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ezeji, T.C. Transcriptional analysis of Clostridium beijerinckii NCIMB 8052 to elucidate role of furfural stress during acetone butanol ethanol fermentation. Biotechnol. Biofuels 2013, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Blaschek, H.P. Transcriptional Analysis of Clostridium beijerinckii NCIMB 8052 and the Hyper-Butanol-Producing Mutant BA101 during the Shift from Acidogenesis to Solventogenesis. Appl. Environ. Microbiol. 2008, 74, 7709–7714. [Google Scholar] [CrossRef]
- Wilkinson, S.R.; Young, M. Physical map of the Clostridium beijerinckii (formerly Clostridium acetobutylicum) NCIMB 8052 chromosome. J. Bacteriol. 1995, 177, 439–448. [Google Scholar] [CrossRef][Green Version]
- Kolek, J.; Sedlar, K.; Provaznik, I.; Patakova, P. Dam and Dcm methylations prevent gene transfer into Clostridium pasteurianum NRRL B-598: Development of methods for electrotransformation, conjugation, and sonoporation. Biotechnol. Biofuels 2016, 9, 1–14. [Google Scholar] [CrossRef]
- Sedlar, K.; Kolek, J.; Gruber, M.; Jureckova, K.; Branska, B.; Csaba, G.; Vasylkivska, M.; Zimmer, R.; Patakova, P.; Provaznik, I. A transcriptional response of Clostridium beijerinckii NRRL B-598 to a butanol shock. Biotechnol. Biofuels 2019, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Vasylkivska, M.; Branska, B.; Sedlar, K.; Jureckova, K.; Provaznik, I.; Patakova, P. Phenotypic and Genomic Analysis of Clostridium beijerinckii NRRL B-598 Mutants With Increased Butanol Tolerance. Front. Bioeng. Biotechnol. 2020, 8, 598392. [Google Scholar] [CrossRef]
- Sedlar, K.; Kolek, J.; Skutkova, H.; Branska, B.; Provaznik, I.; Patakova, P. Complete genome sequence of Clostridium pasteurianum NRRL B-598, a non-type strain producing butanol. J. Biotechnol. 2015, 214, 113–114. [Google Scholar] [CrossRef]
- Diallo, M.; Hocq, R.; Collas, F.; Chartier, G.; Wasels, F.; Wijaya, H.S.; Werten, M.W.; Wolbert, E.J.; Kengen, S.W.; van der Oost, J.; et al. Adaptation and application of a two-plasmid inducible CRISPR-Cas9 system in Clostridium beijerinckii. Methods 2020, 172, 51–60. [Google Scholar] [CrossRef] [PubMed]
- De Gérando, H.M.; Wasels, F.; Bisson, A.; Clement, B.; Bidard, F.; Jourdier, E.; López-Contreras, A.M.; Ferreira, N.L. Genome and transcriptome of the natural isopropanol producer Clostridium beijerinckii DSM6423. BMC Genom. 2018, 19, 242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, T.; He, J. Characterization and genome analysis of a butanol–isopropanol-producing Clostridium beijerinckii strain BGS1. Biotechnol. Biofuels 2018, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Schalamun, M.; Kainer, D.; Wang, W.; Schwessinger, B. MinIONQC: Fast and simple quality control for MinION sequencing data. Bioinformatics 2019, 35, 523–525. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Antipov, D.; Hartwick, N.; Shen, M.; Rayko, M.; Lapidus, A.; Pevzner, P.A. plasmidSPAdes: Assembling plasmids from whole genome sequencing data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, C.-T. Ori-Finder: A web-based system for finding oriC s in unannotated bacterial genomes. BMC Bioinforma. 2008, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Luo, H.; Gao, F. DoriC 10.0: An updated database of replication origins in prokaryotic genomes including chromosomes and plasmids. Nucleic Acids Res. 2019, 47, D74–D77. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.V.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinform. 2008, 25, 119–120. [Google Scholar] [CrossRef]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.-A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinform. 2000, 16, 944–945. [Google Scholar] [CrossRef]
- Akhter, S.; Aziz, R.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
- Biswas, A.; Staals, R.; Morales, S.; Fineran, P.; Brown, C.M. CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genom. 2016, 17, 1–14. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Mizrachi, I.K. GenBank. Nucleic Acids Res. 2019, 48, D84–D86. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Murrell, P. R Graphics. Wiley Interdiscip. Rev. Comput. Stat. 2009, 1, 216–220. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed]
- Wolstencroft, K.; Krebs, O.; Snoep, J.L.; Stanford, N.J.; Bacall, F.; Golebiewski, M.; Kuzyakiv, R.; Nguyen, Q.; Owen, S.; Soiland-Reyes, S.; et al. FAIRDOMHub: A repository and collaboration environment for sharing systems biology research. Nucleic Acids Res. 2017, 45, D404–D407. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Cornillot, E.; Nair, R.V.; Papoutsakis, E.T.; Soucaille, P. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 1997, 179, 5442–5447. [Google Scholar] [CrossRef]
- Kemperman, R.; Jonker, M.; Nauta, A.; Kuipers, O.P.; Kok, J. Functional Analysis of the Gene Cluster Involved in Production of the Bacteriocin Circularin A by Clostridium beijerinckii ATCC 25752. Appl. Environ. Microbiol. 2003, 69, 6174–6178. [Google Scholar] [CrossRef]
- Kemperman, R.; Kuipers, A.; Karsens, H.; Nauta, A.; Kuipers, O.; Kok, J. Identification and Characterization of Two Novel Clostridial Bacteriocins, Circularin A and Closticin 574. Appl. Environ. Microbiol. 2003, 69, 1589–1597. [Google Scholar] [CrossRef]
- Thieme, N.; Panitz, J.C.; Held, C.; Lewandowski, B.; Schwarz, W.H.; Liebl, W.; Zverlov, V. Milling byproducts are an economically viable substrate for butanol production using clostridial ABE fermentation. Appl. Microbiol. Biotechnol. 2020, 104, 8679–8689. [Google Scholar] [CrossRef]
- Wischral, D.; Zhang, J.; Cheng, C.; Lin, M.; de Souza, L.M.G.; Pessoa, F.L.P.; Pereira, N.; Yang, S.-T. Production of 1,3-propanediol by Clostridium beijerinckii DSM 791 from crude glycerol and corn steep liquor: Process optimization and metabolic engineering. Bioresour. Technol. 2016, 212, 100–110. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Song, W.; Sun, H.-X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.; et al. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, W74–W80. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.E.; Liu, X.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Genome-directed analysis of prophage excision, host defence systems, and central fermentative metabolism in Clostridium pasteurianum. Sci. Rep. 2016, 6, 26228. [Google Scholar] [CrossRef]
- Schüler, M.; Stegmann, B.; Poehlein, A.; Daniel, R.; Dürre, P. Genome sequence analysis of the temperate bacteriophage TBP2 of the solvent producer Clostridium saccharoperbutylacetonicum N1-4 (HMT, ATCC 27021). FEMS Microbiol. Lett. 2020, 367. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-Mediated Adaptive Immune Systems in Bacteria and Archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.L.; Jang, J.; Venterea, R.T.; Feyereisen, G.W.; Ishii, S. Isolation and characterization of denitrifiers from woodchip bioreactors for bioaugmentation application. J. Appl. Microbiol. 2020, 129, 590–600. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Chen, Z.; Erickson, D.L.; Meng, J. Polishing the Oxford Nanopore long-read assemblies of bacterial pathogens with Illumina short reads to improve genomic analyses. Genomics 2021, 113, 1366–1377. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinforma. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Saha, J.; Saha, B.K.; Sarkar, M.P.; Roy, V.; Mandal, P.; Pal, A. Comparative Genomic Analysis of Soil Dwelling Bacteria Utilizing a Combinational Codon Usage and Molecular Phylogenetic Approach Accentuating on Key Housekeeping Genes. Front. Microbiol. 2019, 10, 2896. [Google Scholar] [CrossRef]
- Chung, M.; Munro, J.B.; Tettelin, H.; Hotopp, J.C.D. Using Core Genome Alignments to Assign Bacterial Species. mSystems 2018, 3, e00236-18. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Janssen, H.; Magis, A.; Wang, Y.; Lu, T.; Price, N.D.; Jin, Y.; Blaschek, H.P. Genomic, Transcriptional, and Phenotypic Analysis of the Glucose Derepressed Clostridium beijerinckii Mutant Exhibiting Acid Crash Phenotype. Biotechnol. J. 2017, 12, 1700182. [Google Scholar] [CrossRef]
- Patakova, P.; Branská, B.; Sedlar, K.; Vasylkivska, M.; Jureckova, K.; Kolek, J.; Koscova, P.; Provaznik, I. Acidogenesis, solventogenesis, metabolic stress response and life cycle changes in Clostridium beijerinckii NRRL B-598 at the transcriptomic level. Sci. Rep. 2019, 9, 1–21. [Google Scholar] [CrossRef]
- Branska, B.; Vasylkivska, M.; Raschmanova, H.; Jureckova, K.; Sedlar, K.; Provaznik, I.; Patakova, P. Changes in efflux pump activity of Clostridium beijerinckii throughout ABE fermentation. Appl. Microbiol. Biotechnol. 2021, 105, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Jureckova, K.; Raschmanova, H.; Kolek, J.; Vasylkivska, M.; Branska, B.; Patakova, P.; Provaznik, I.; Sedlar, K. Identification and Validation of Reference Genes in Clostridium beijerinckii NRRL B-598 for RT-qPCR Using RNA-Seq Data. Front. Microbiol. 2021, 12, 640054. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).