Evaluation of Filter, Paramagnetic, and STAGETips Aided Workflows for Proteome Profiling of Symbiodiniaceae Dinoflagellate
Abstract
1. Introduction
2. Materials and Methods
2.1. Symbiodinium Culture and Maintenance
2.2. Sample Preparation Workflows for Proteome Profiling
2.2.1. Protein Extraction, Purification, Alkylation-Reduction, and Quantification
2.2.2. FASP, SP3, and STAGETips Based Clean-Up, Digestion, and Fractionation
2.3. Implementation of Outperformed SP3 Workflow in S. tridacnidorum Exposed to Low Light Conditions
2.4. LC-MS Analysis of Peptides
3. Statistical Analysis
4. Results and Discussion
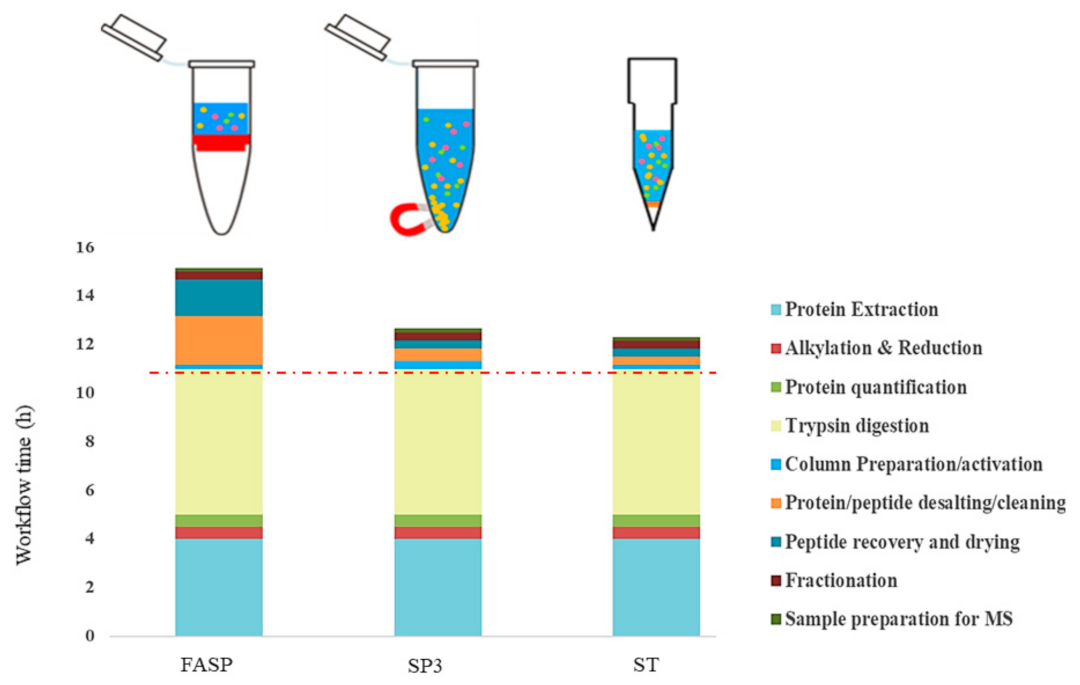
4.1. Sample Processing Timing Workflow for Protein Sample Preparation Methods
4.2. Differences in Protein and Peptides Identified, Their Quality Parameters, and Gene Ontology Analysis in Different Protein Sample Preparation Workflows
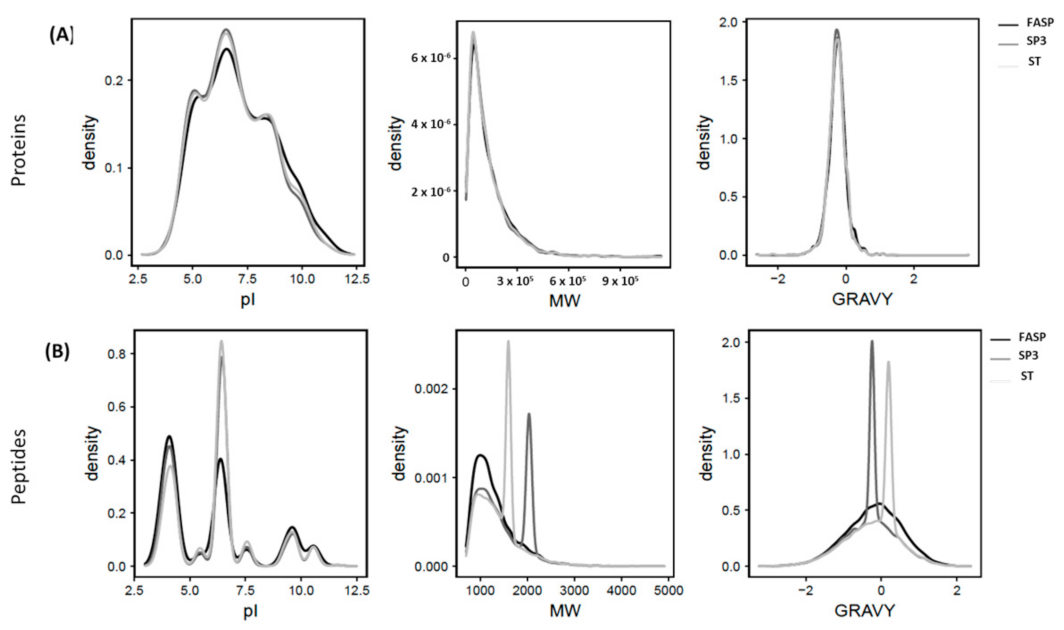
4.3. Physiochemical Properties of Proteins and Peptides Detected Different Protein Sample Preparation Workflows
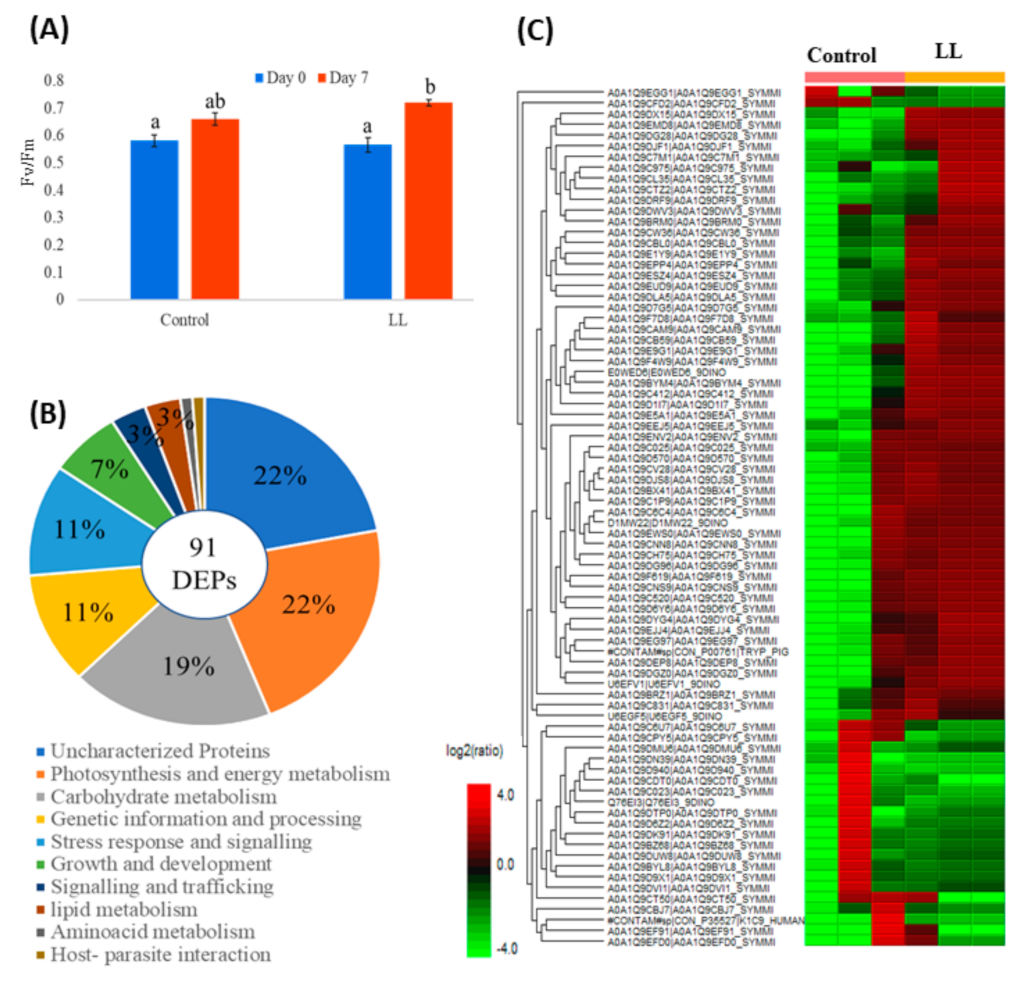
4.4. Evaluation of Single-Pot Solid-Phase-Enhanced Sample Preparation (SP3) Workflow to Reveal Proteome Regulation in Symbiodinium Tridacnidorum Under Low Light Growth
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- LaJeunesse, T.C.; Parkinson, J.E.; Gabrielson, P.W.; Jeong, H.J.; Reimer, J.D.; Voolstra, C.R.; Santos, S.R. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018, 28, 2570–2580. [Google Scholar] [CrossRef]
- Suggett, D.J.; Smith, D.J. Coral bleaching patterns are the outcome of complex biological and environmental networking. Glob. Chang. Biol. 2020, 26, 68–79. [Google Scholar] [CrossRef]
- Oakley, C.; Davy, S. Cell biology of coral bleaching. In Coral Bleaching; Springer: Cham, Switzerland, 2018; pp. 189–211. [Google Scholar]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Seneca, F.O.; Traylor-Knowles, N.; Palumbi, S.R. Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. USA 2013, 110, 1387–1392. [Google Scholar] [CrossRef]
- Fuller, Z.L.; Mocellin, V.J.L.; Morris, L.A.; Cantin, N.; Shepherd, J.; Sarre, L.; Peng, J.; Liao, Y.; Pickrell, J.; Andolfatto, P.; et al. Population genetics of the coral Acropora millepora: Toward genomic prediction of bleaching. Science 2020, 369, 268–278. [Google Scholar] [CrossRef]
- Pinzón, J.H.; Kamel, B.; Burge, C.A.; Harvell, C.D.; Medina, M.; Weil, E.; Mydlarz, L.D. Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R. Soc. Open Sci. 2015, 2, 140214. [Google Scholar] [CrossRef] [PubMed]
- Rosic, N.; Kaniewska, P.; Chan, C.K.; Ling, E.Y.; Edwards, D.; Dove, S.; Hoegh-Guldberg, O. Early transcriptional changes in the reef-building coral Acropora aspera in response to thermal and nutrient stress. BMC Genom. 2014, 15, 1052. [Google Scholar] [CrossRef] [PubMed]
- Shinzato, C.; Inoue, M.; Kusakabe, M. A snapshot of a coral “holobiont”: A transcriptome assembly of the scleractinian coral, Porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS ONE 2014, 9, e85182. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yu, L.; Zhang, H. Transcriptomic Responses to Thermal Stress and Varied Phosphorus Conditions in Fugacium kawagutii. Microorganisms 2019, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Cheng, S.; Song, B.; Zhong, Z.; Lin, X.; Li, W.; Li, L.; Zhang, Y.; Zhang, H.; Ji, Z.; et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 2015, 350, 691–694. [Google Scholar] [CrossRef]
- Bayer, T.; Aranda, M.; Sunagawa, S.; Yum, L.K.; DeSalvo, M.K.; Lindquist, E.; Coffroth, M.A.; Voolstra, C.R.; Medina, M. Symbiodinium transcriptomes: Genome insights into the dinoflagellate symbionts of reef-building corals. PLoS ONE 2012, 7, e35269. [Google Scholar] [CrossRef]
- González-Pech, R.A.; Ragan, M.A.; Chan, C.X. Signatures of adaptation and symbiosis in genomes and transcriptomes of Symbiodinium. Sci. Rep. 2017, 7, 15021. [Google Scholar] [CrossRef]
- Parkinson, J.E.; Baker, A.C.; Baums, I.B.; Davies, S.W.; Grottoli, A.G.; Kitchen, S.A.; Matz, M.V.; Miller, M.W.; Shantz, A.A.; Kenkel, C.D. Molecular tools for coral reef restoration: Beyond biomarker discovery. Conserv. Lett. 2020, 13, e12687. [Google Scholar] [CrossRef]
- Vélez-Bermúdez, I.C.; Schmidt, W. The conundrum of discordant protein and mRNA expression. Are plants special? Front. Plant Sci. 2014, 5, 619. [Google Scholar]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Mayfield, A.B. Proteomic Signatures of Corals from Thermodynamic Reefs. Microorganisms 2020, 8, 1171. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, D.A.; Yoo, M.-J.; Dixson, D.L.; Ross, C. Exposure to the Florida red tide dinoflagellate, Karenia brevis, and its associated brevetoxins induces ecophysiological and proteomic alterations in Porites astreoides. PLoS ONE 2020, 15, e0228414. [Google Scholar] [CrossRef] [PubMed]
- Warner, M.E.; Fitt, W.K.; Schmidt, G.W. Damage to photosystem II in symbiotic dinoflagellates: A determinant of coral bleaching. Proc. Natl. Acad. Sci. USA 1999, 96, 8007–8012. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Medina, R.E.; Islas-Flores, T.; Villanueva, M.A. Phosphorylation/dephosphorylation response to light stimuli of Symbiodinium proteins: Specific light-induced dephosphorylation of an HSP-like 75 kDa protein from S. microadriaticum. PeerJ. 2019, 7, e7406. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, A.B.; Hsiao, Y.Y.; Chen, H.K.; Chen, C.S. Rubisco expression in the dinoflagellate Symbiodinium sp. is influenced by both photoperiod and endosymbiotic lifestyle. Mar. Biotech. 2014, 16, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Pasaribu, B.; Weng, L.-C.; Lin, I.-P.; Camargo, E.; Tzen, J.T.; Tsai, C.-H.; Ho, S.-L.; Lin, M.-R.; Wang, L.-H.; Chen, C.-S.; et al. Morphological variability and distinct protein profiles of cultured and endosymbiotic Symbiodinium cells isolated from Exaiptasia pulchella. Sci. Rep. 2015, 5, 15353. [Google Scholar] [CrossRef] [PubMed]
- Medrano, E.; Merselis, D.G.; Bellantuono, A.J.; Rodriguez-Lanetty, M. Proteomic basis of symbiosis: A heterologous partner fails to duplicate homologous colonization in a novel cnidarian–Symbiodiniaceae mutualism. Front. Microbiol. 2019, 10, 1153. [Google Scholar] [CrossRef]
- Awad, D.; Brueck, T. Optimization of protein isolation by proteomic qualification from Cutaneotrichosporon oleaginosus. Anal. Bioanal. Chem. 2020, 412, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, S.; Stenoien, D.L.; Paša-Tolić, L. High-throughput proteomics. Annu. Rev. Anal. Chem. 2014, 7, 427–454. [Google Scholar] [CrossRef] [PubMed]
- Sielaff, M.; Kuharev, J.R.; Bohn, T.; Hahlbrock, J.; Bopp, T.; Tenzer, S.; Distler, U. Evaluation of FASP, SP3, and iST protocols for proteomic sample preparation in the low microgram range. J. Proteom. Res. 2017, 16, 4060–4072. [Google Scholar] [CrossRef]
- Batth, T.S.; Tollenaere, M.A.; Rüther, P.; Gonzalez-Franquesa, A.; Prabhakar, B.S.; Bekker-Jensen, S.; Deshmukh, A.S.; Olsen, J.V. Protein aggregation capture on microparticles enables multipurpose proteomics sample preparation. Mol. Cell Proteom. 2019, 18, 1027–1035. [Google Scholar] [CrossRef]
- Eddhif, B.; Lange, J.; Guignard, N.; Batonneau, Y.; Clarhaut, J.; Papot, S.; Geffroy-Rodier, C.; Poinot, P. Study of a novel agent for TCA precipitated proteins washing-comprehensive insights into the role of ethanol/HCl on molten globule state by multi-spectroscopic analyses. J. Proteom. 2018, 173, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J. Filter-aided sample preparation: The versatile and efficient method for proteomic analysis. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2017; pp. 15–27. [Google Scholar]
- Zougman, A.; Selby, P.J.; Banks, R.E. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics 2014, 14, 1006–1010. [Google Scholar] [CrossRef]
- Hughes, C.S.; Foehr, S.; Garfield, D.A.; Furlong, E.E.; Steinmetz, L.M.; Krijgsveld, J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. [Google Scholar] [CrossRef] [PubMed]
- Choksawangkarn, W.; Edwards, N.; Wang, Y.; Gutierrez, P.; Fenselau, C. Comparative study of workflows optimized for in-gel, in-solution, and on-filter proteolysis in the analysis of plasma membrane proteins. J. Proteom. Res. 2012, 11, 3030–3034. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, K.R.; Schroll, M.M.; Hummon, A.B. Comparison of in-solution, FASP, and S-trap based digestion methods for bottom-up proteomic studies. J. Proteom. Res. 2018, 17, 2480–2490. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.S.; Moggridge, S.; Müller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319. [Google Scholar] [CrossRef]
- Suggett, D.J.; Goyen, S.; Evenhuis, C.; Szabó, M.; Pettay, D.T.; Warner, M.E.; Ralph, P.J. Functional diversity of photobiological traits within the genus Symbiodinium appears to be governed by the interaction of cell size with cladal designation. New Phytol. 2015, 208, 370–381. [Google Scholar] [CrossRef]
- Goodman, J.K.; Zampronio, C.G.; Jones, A.M.; Hernandez-Fernaud, J.R. Updates of the In-Gel Digestion Method for Protein Analysis by Mass Spectrometry. Proteomics 2018, 18, 1800236. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Padula, M.P.; Davey, P.; Pernice, M.; Jiang, Z.; Sablok, G.; Contreras-Porcia, L.; Ralph, P.J. Proteome Analysis Reveals Extensive Light Stress-Response Reprogramming in the Seagrass Zostera muelleri (Alismatales, Zosteraceae) Metabolism. Front. Plant Sci. 2017, 7, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Stepanova, E.; Gygi, S.P.; Paulo, J.A. Filter-based protein digestion (FPD): A detergent-free and scaffold-based strategy for TMT workflows. J. Proteom. Res. 2018, 17, 1227–1234. [Google Scholar] [CrossRef]
- HaileMariam, M.; Eguez, R.V.; Singh, H.; Bekele, S.; Ameni, G.; Pieper, R.; Yu, Y. S-Trap, an ultrafast sample-preparation approach for shotgun proteomics. J. Proteom. Res. 2018, 17, 2917–2924. [Google Scholar] [CrossRef]
- Erde, J.; Loo, R.R.O.; Loo, J.A. Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J. Proteom. Res. 2014, 13, 1885–1895. [Google Scholar] [CrossRef]
- Ni, M.W.; Wang, L.; Chen, W.; Mou, H.Z.; Zhou, J.; Zheng, Z.G. Modified filter-aided sample preparation (FASP) method increases peptide and protein identifications for shotgun proteomics. Rapid Commun. Mass Spectrom. 2017, 31, 171–178. [Google Scholar] [CrossRef]
- Li, S.; Cao, X.; Wang, Y.; Zhu, Z.; Zhang, H.; Xue, S.; Tian, J. A method for microalgae proteomics analysis based on modified filter-aided sample preparation. Appl. Biochem. Biotechnol. 2017, 183, 923–930. [Google Scholar] [CrossRef]
- Yu, Y.; Xie, L.; Gunawardena, H.P.; Khatun, J.; Maier, C.; Spitzer, W.; Leerkes, M.; Giddings, M.C.; Chen, X. GOFAST: An Integrated Approach for Efficient and Comprehensive Membrane Proteome Analysis. Anal. Chem. 2012, 84, 9008–9014. [Google Scholar] [CrossRef] [PubMed]
- Hildonen, S.; Halvorsen, T.G.; Reubsaet, L. Why less is more when generating tryptic peptides in bottom-up proteomics. Proteomics 2014, 14, 2031–2041. [Google Scholar] [CrossRef]
- Hayoun, K.; Gouveia, D.D.; Grenga, L.; Pible, O.; Armengaud, J. Evaluation of sample preparation methods for fast proteotyping of microorganisms by tandem mass spectrometry. Front. Microbiol. 2019, 10, 1985. [Google Scholar] [CrossRef] [PubMed]
- Moggridge, S.; Sorensen, P.H.; Morin, G.B.; Hughes, C.S. Extending the compatibility of the SP3 paramagnetic bead processing approach for proteomics. J. Proteom. Res. 2018, 17, 1730–1740. [Google Scholar] [CrossRef]
- Dimayacyac-Esleta, B.R.T.; Tsai, C.-F.; Kitata, R.B.; Lin, P.-Y.; Choong, W.-K.; Lin, T.-D.; Wang, Y.-T.; Weng, S.-H.; Yang, P.-C.; Arco, S.D.; et al. Rapid high-pH reverse phase StageTip for sensitive small-scale membrane proteomic profiling. Anal. Chem. 2015, 87, 12016–12023. [Google Scholar] [CrossRef]
- Kuljanin, M.; Dieters-Castator, D.Z.; Hess, D.A.; Postovit, L.M.; Lajoie, G.A. Comparison of sample preparation techniques for large-scale proteomics. Proteomics 2017, 17, 1600337. [Google Scholar] [CrossRef]
- Magdeldin, S.; Yamamoto, K.; Yoshida, Y.; Xu, B.; Zhang, Y.; Fujinaka, H.; Yaoita, E.; Yates, J.R., III; Yamamoto, T. Deep proteome mapping of mouse kidney based on OFFGel prefractionation reveals remarkable protein post-translational modifications. J. Proteom. Res. 2014, 13, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Malin, K.; Mohan, G.; Rafael, H.; Fredrik, L.; Jonas, B.; Kovac, L.; Bergström Lind, S. Magnetic beads for desalting of monoclonal antibodies and antibody–drug Conjugates. Anal. Chem. 2020, 92, 9001–9007. [Google Scholar]
- Busch, A.; Hippler, M. The structure and function of eukaryotic photosystem I. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 864–877. [Google Scholar] [CrossRef]
- Bobeszko, T. Characterisation of Carbonic Anhydrase in the Symbiotic Dinoflagellate Symbiodinium; James Cook University: Townsville, Australia, 2017. [Google Scholar]
- Gierz, S.L.; Forêt, S.; Leggat, W. Transcriptomic analysis of thermally stressed Symbiodinium reveals differential expression of stress and metabolism genes. Front. Plant Sci. 2017, 8, 271. [Google Scholar] [CrossRef]
- Gierz, S.L.; Gordon, B.R.; Leggat, W. Integral light-harvesting complex expression in Symbiodinium within the coral Acropora aspera under thermal stress. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Buck, J.M.; Sherman, J.; Bártulos, C.R.; Serif, M.; Halder, M.; Henkel, J.; Falciatore, A.; Lavaud, J.; Gorbunov, M.Y.; Kroth, P.G.; et al. Lhcx proteins provide photoprotection via thermal dissipation of absorbed light in the diatom Phaeodactylum tricornutum. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xiang, T.; Nelson, W.; Rodriguez, J.; Tolleter, D.; Grossman, A.R. Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J. 2015, 82, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Trotta, A.; Suorsa, M.; Rantala, M.; Lundin, B.; Aro, E.M. Serine and threonine residues of plant STN7 kinase are differentially phosphorylated upon changing light conditions and specifically influence the activity and stability of the kinase. Plant J. 2016, 87, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Tikkanen, M.; Gollan, P.J.; Suorsa, M.; Kangasjärvi, S.; Aro, E.M. STN7 Operates in Retrograde Signaling through Controlling Redox Balance in the Electron Transfer Chain. Front. Plant Sci. 2012, 3, 277. [Google Scholar] [CrossRef]
- Puthiyaveetil, S.; Woodiwiss, T.; Knoerdel, R.; Zia, A.; Wood, M.; Hoehner, R.; Kirchhoff, H. Significance of the photosystem II core phosphatase PBCP for plant viability and protein repair in thylakoid membranes. Plant Cell Physiol. 2014, 55, 1245–1254. [Google Scholar] [CrossRef]
- Babele, P.K.; Kumar, J.; Chaturvedi, V. Proteomic De-Regulation in Cyanobacteria in Response to Abiotic Stresses. Front. Microbiol. 2019, 10, 1315. [Google Scholar] [CrossRef]
- Dizengremel, P.; Vaultier, M.N.; Le Thiec, D.; Cabané, M.; Bagard, M.; Gérant, D.; Gérard, J.; Dghim, A.A.; Richet, N.; Afif, D.; et al. Phosphoenolpyruvate is at the crossroads of leaf metabolic responses to ozone stress. New Phytol. 2012, 195, 512–517. [Google Scholar] [CrossRef]
- Hunter, W.N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007, 282, 21573–21577. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Kleczkowski, L.A. The Glycerate and Phosphorylated Pathways of Serine Synthesis in Plants: The Branches of Plant Glycolysis Linking Carbon and Nitrogen Metabolism. Front. Plant Sci. 2018, 9, 318. [Google Scholar] [CrossRef]
- Welchen, E.; García, L.; Mansilla, N.; Gonzalez, D. Coordination of plant mitochondrial biogenesis: Keeping pace with cellular requirements. Front. Plant Sci. 2014, 4, 551. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, T.D.; Hagemeyer, J.C.G.; Hoadley, K.D.; Marsh, A.G.; Warner, M.E. Partitioning of Respiration in an Animal-Algal Symbiosis: Implications for Different Aerobic Capacity between Symbiodinium spp. Front. Physiol. 2016, 7, 128. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Supasri, K.M.; Kumar, M.; Mathew, M.J.; Signal, B.; Padula, M.P.; Suggett, D.J.; Ralph, P.J. Evaluation of Filter, Paramagnetic, and STAGETips Aided Workflows for Proteome Profiling of Symbiodiniaceae Dinoflagellate. Processes 2021, 9, 983. https://doi.org/10.3390/pr9060983
Supasri KM, Kumar M, Mathew MJ, Signal B, Padula MP, Suggett DJ, Ralph PJ. Evaluation of Filter, Paramagnetic, and STAGETips Aided Workflows for Proteome Profiling of Symbiodiniaceae Dinoflagellate. Processes. 2021; 9(6):983. https://doi.org/10.3390/pr9060983
Chicago/Turabian StyleSupasri, Kanoknate M., Manoj Kumar, Mano J. Mathew, Bethany Signal, Matthew P. Padula, David J. Suggett, and Peter J. Ralph. 2021. "Evaluation of Filter, Paramagnetic, and STAGETips Aided Workflows for Proteome Profiling of Symbiodiniaceae Dinoflagellate" Processes 9, no. 6: 983. https://doi.org/10.3390/pr9060983
APA StyleSupasri, K. M., Kumar, M., Mathew, M. J., Signal, B., Padula, M. P., Suggett, D. J., & Ralph, P. J. (2021). Evaluation of Filter, Paramagnetic, and STAGETips Aided Workflows for Proteome Profiling of Symbiodiniaceae Dinoflagellate. Processes, 9(6), 983. https://doi.org/10.3390/pr9060983








