Anti-Inflammatory Effects of Abeliophyllum distichum Nakai (Cultivar Okhwang 1) Callus through Inhibition of PI3K/Akt, NF-κB, and MAPK Signaling Pathways in Lipopolysaccharide-Induced Macrophages
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material of A. distichum Nakai Cultivar OkHwang 1
2.2. Chemicals and Reagents
2.3. Callus Induction
2.4. Extraction and Fractionation
2.5. HPLC Analysis
2.6. Cell Culture
2.7. Cell Viability
2.8. NO Assay
2.9. Western Blotting Analysis
2.10. RT-PCR
2.11. Quantitative PCR Analysis
2.12. Immunofluorescence
2.13. Statistical Analysis
3. Results
3.1. Analysis of Acteoside and Isoacteoside in CAO
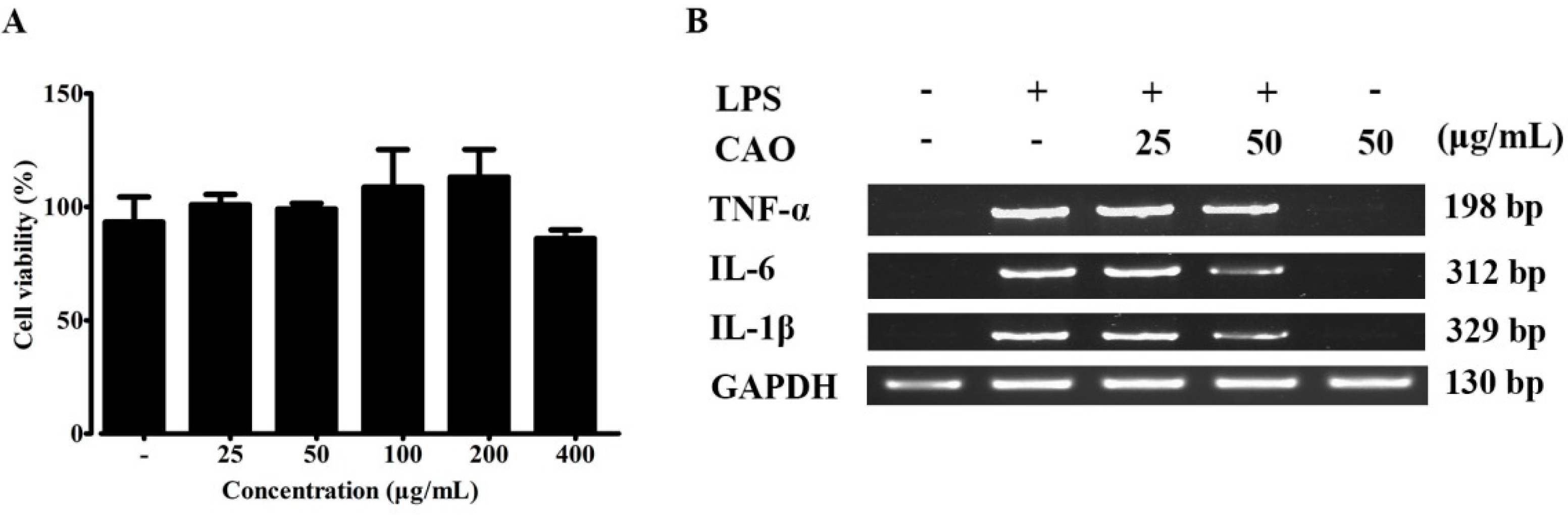
3.2. Effects of CAO on Cell Viability
3.3. Effects of CAO on Pro-Inflammatory Cytokine Secretions
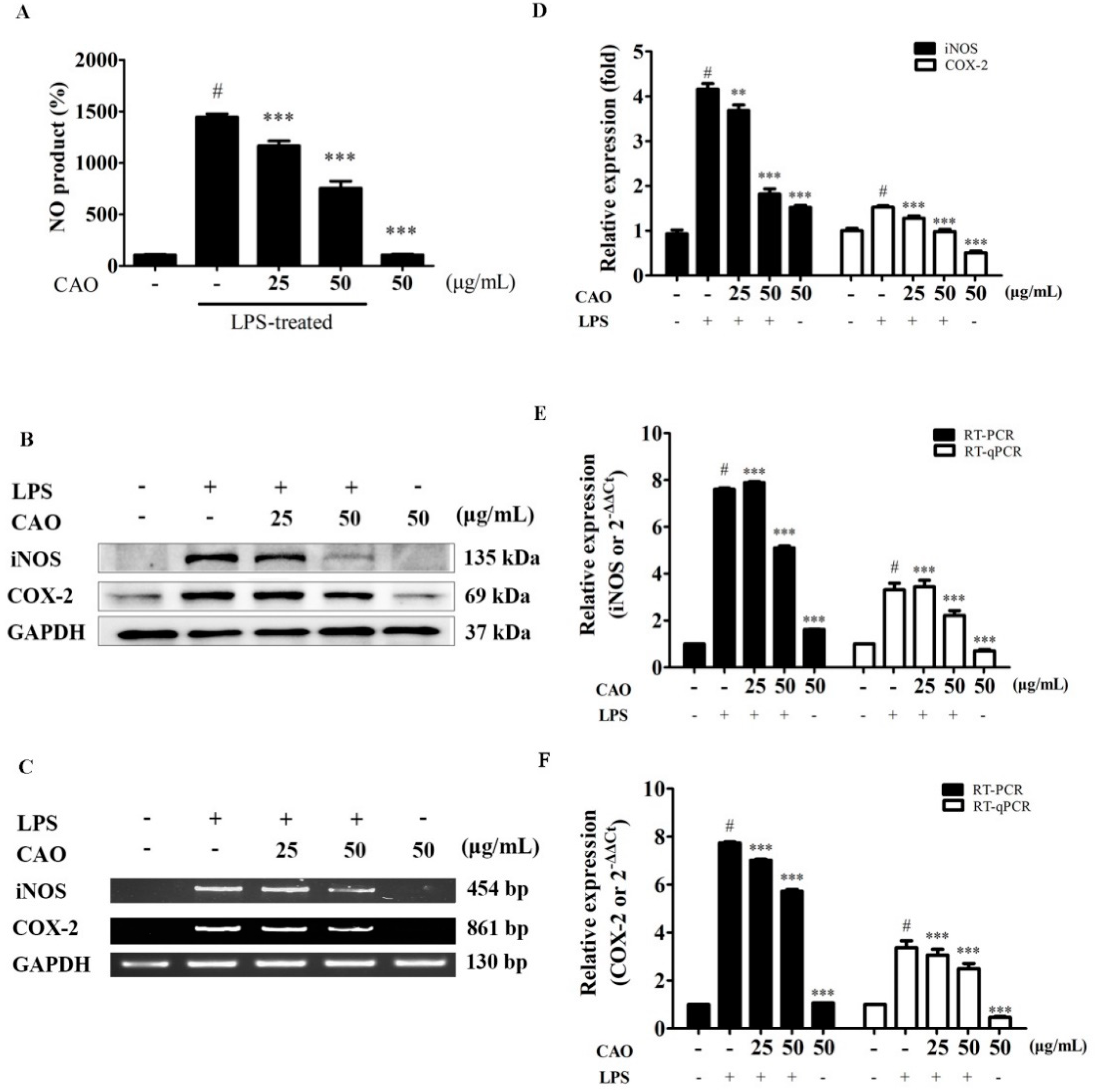
3.4. Effects of CAO on NO Production, COX-2 and iNOS Expression
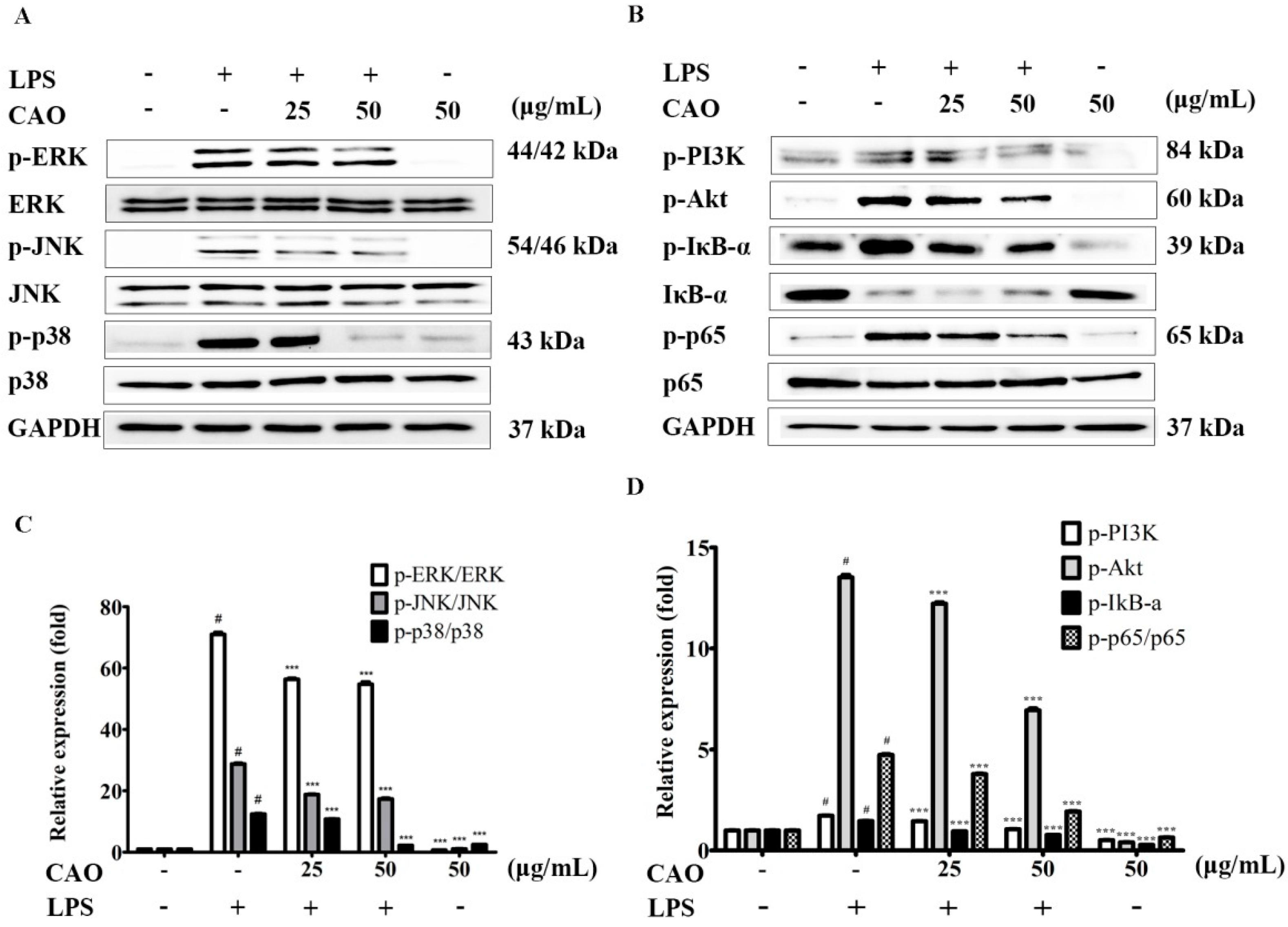
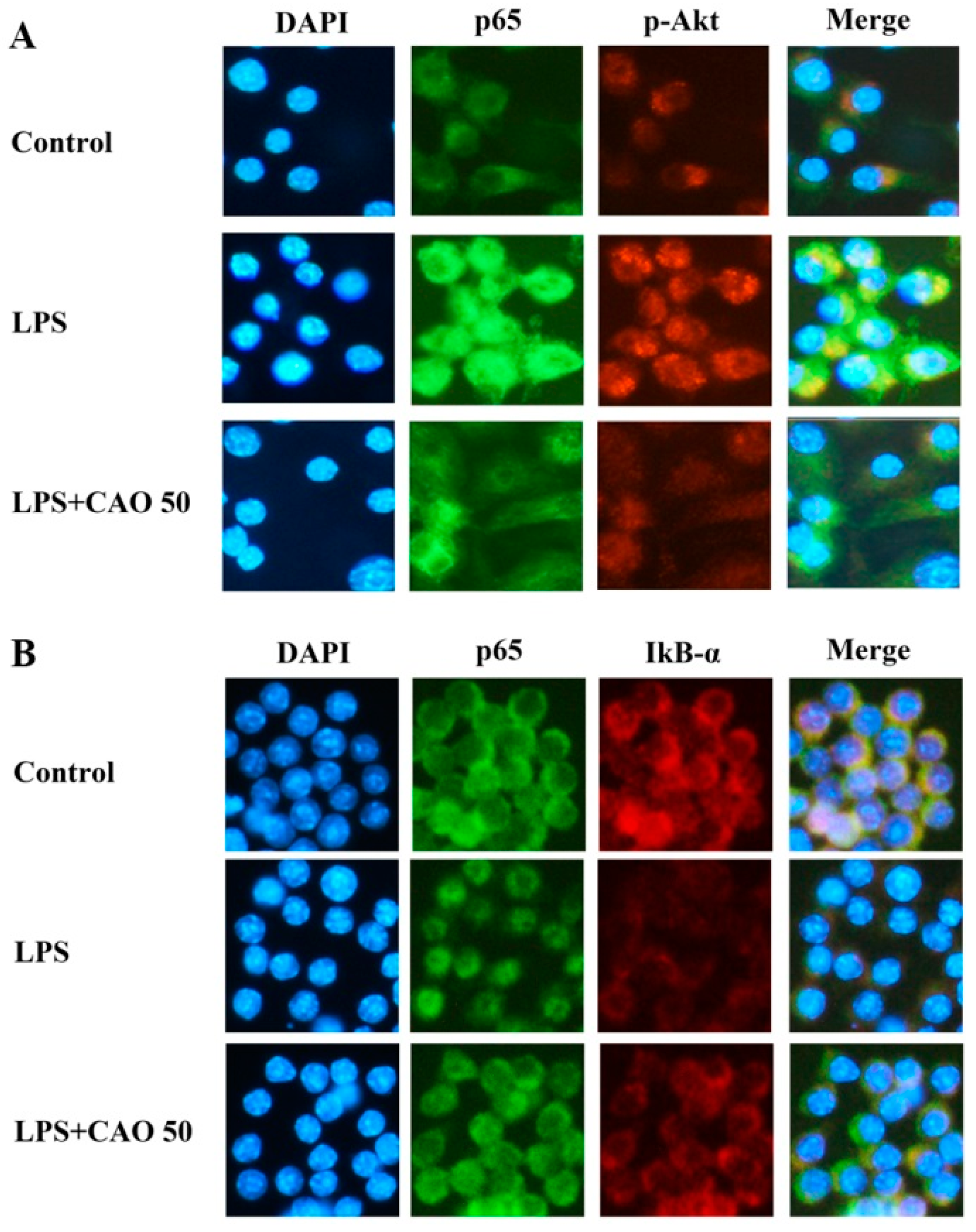
3.5. Effects of CAO on IκB/NF-κB Signaling Pathways
3.6. Effects of CAO on PI3K-Akt Activation and MAPKs Pathway
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- A current view on inflammation. Nat. Immunol. 2017, 18, 825. [CrossRef] [PubMed]
- Hattori, Y.; Hattori, S.; Kasai, K. Lipopolysaccharide activates Akt in vascular smooth muscle cells resulting in induction of inducible nitric oxide synthase through nuclear factor-kappa B activation. Eur. J. Pharmacol. 2003, 481, 153–158. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Bowie, A.G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007, 7, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Savva, A.; Roger, T. Targeting toll-like receptors: Promising therapeutic strategies for the management of sepsis-associated pathology and infectious diseases. Front. Immunol. 2013, 4, 387. [Google Scholar] [CrossRef]
- Hsu, H.Y.; Wen, M.H. Lipopolysaccharide-mediated reactive oxygen species and signal transduction in the regulation of interleukin-1 gene expression. J. Biol. Chem. 2002, 277, 22131–22139. [Google Scholar] [CrossRef] [PubMed]
- Murray, R.Z.; Stow, J.L. Cytokine secretion in macrophages: SNAREs, Rabs, and membrane trafficking. Front. Immunol. 2014, 5, 538. [Google Scholar] [CrossRef]
- Posadas, I.; Terencio, M.C.; Guillén, I.; Ferrándiz, M.L.; Coloma, J.; Payá, M.; Alcaraz, M.J. Co-regulation between cyclo-oxygenase-2 and inducible nitric oxide synthase expression in the time-course of murine inflammation. Naunyn Schmiedeberg’s Arch. Pharmacol. 2000, 361, 98–106. [Google Scholar] [CrossRef]
- Moncada, S. Nitric oxide: Physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 1991, 43, 109–142. [Google Scholar]
- Yun, H.Y.; Dawson, V.L.; Dawson, T.M. Neurobiology of nitric oxide. Crit. Rev. Neurobiol. 1996, 10, 291–316. [Google Scholar] [CrossRef]
- Bishop-Bailey, D.; Calatayud, S.; Warner, T.D.; Hla, T.; Mitchell, J.A. Prostaglandins and the regulation of tumor growth. J. Environ. Pathol. Toxicol. Oncol. 2002, 21, 9. [Google Scholar] [CrossRef]
- Ishihara, S.; Rumi, M.A.; Okuyama, T.; Kinoshita, Y. Effect of prostaglandins on the regulation of tumor growth. Curr. Med. Chem. Anticancer Agents 2004, 4, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Vane, J.R.; Botting, R.M. Mechanism of action of nonsteroidal anti-inflammatory drugs. Am. J. Med. 1998, 104, 2S–8S, discussion 21S–22S. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Kempe, S.; Kestler, H.; Lasar, A.; Wirth, T. NF-κB controls the global pro-inflammatory response in endothelial cells: Evidence for the regulation of a pro-atherogenic program. Nucleic Acids Res. 2005, 33, 5308–5319. [Google Scholar] [CrossRef]
- Karin, M.; Yamamoto, Y.; Wang, Q.M. The IKK NF-κB system: A treasure trove for drug development. Nat. Rev. Drug Discov. 2004, 3, 17–26. [Google Scholar] [CrossRef]
- Christian, F.; Smith, E.L.; Carmody, R.J. The regulation of NF-κB subunits by phosphorylation. Cells 2016, 5, 12. [Google Scholar] [CrossRef]
- Collins, P.E.; Mitxitorena, I.; Carmody, R.J. The ubiquitination of NF-κB subunits in the control of transcription. Cells 2016, 5, 23. [Google Scholar] [CrossRef]
- Kanarek, N.; London, N.; Schueler-Furman, O.; Ben-Neriah, Y. Ubiquitination and degradation of the inhibitors of NF-kappaB. Cold Spring Harb. Perspect. Biol. 2010, 2, a000166. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Baltimore, D. NF-κB: Ten years after. Cell 1996, 87, 13–20. [Google Scholar] [CrossRef]
- Hwang, P.A.; Chien, S.Y.; Chan, Y.L.; Lu, M.K.; Wu, C.H.; Kong, Z.L.; Wu, C.J. Inhibition of lipopolysaccharide (LPS)-induced inflammatory responses by Sargassum hemiphyllum sulfated polysaccharide extract in RAW 264.7 macrophage cells. J. Agric. Food Chem. 2011, 59, 2062–2068. [Google Scholar] [CrossRef]
- Kang, U.; Chang, C.S.; Kim, Y.S. Genetic structure and conservation considerations of rare endemic Abeliophyllum distichum Nakai (Oleaceae) in Korea. J. Plant Res. 2000, 113, 127–138. [Google Scholar] [CrossRef]
- Park, J.; Kim, Y.; Xi, H.; Jang, T.; Park, J.H. The complete chloroplast genome of Abeliophyllum distichum Nakai (Oleaceae), cultivar Ok Hwang 1ho: Insights of cultivar specific variations of A. distichum. Mitochondrial DNA Part B 2019, 4, 1640–1642. [Google Scholar] [CrossRef]
- Choi, J.H.; Seo, E.J.; Sung, J.; Choi, K.M.; Kim, H.; Kim, J.S.; Lee, J.; Efferth, T.; Hyun, T.K. Polyphenolic compounds, antioxidant and anti-inflammatory effects of Abeliophyllum distichum Nakai extract. J. Appl. Bot. Food Qual. 2017, 90, 266–273. [Google Scholar]
- Oh, H.; Kang, D.G.; Kwon, T.O.; Jang, K.K.; Chai, K.Y.; Yun, Y.G.; Chung, H.T.; Lee, H.S. Four glycosides from the leaves of Abeliophyllum distichum with inhibitory effects on angiotensin converting enzyme. Phytother. Res. 2003, 17, 811–813. [Google Scholar] [CrossRef]
- Xiong, Q.; Hase, K.; Tezuka, Y.; Tani, T.; Namba, T.; Kadota, S. Hepatoprotective activity of phenylethanoids from Cistanche deserticola. Planta Med. 1998, 64, 120–125. [Google Scholar] [CrossRef]
- Schlesier, K.; Harwat, M.; Böhm, V.; Bitsch, R. Assessment of antioxidant activity by using different in vitro methods. Free Radic. Res. 2002, 36, 177–187. [Google Scholar] [CrossRef]
- He, Z.D.; Lau, K.M.; Xu, H.X.; Li, P.C.; But, P.P.H. Antioxidant activity of phenylethanoid glycosides from Brandisia hancei. J. Ethnopharmacol. 2000, 71, 483–486. [Google Scholar] [CrossRef]
- Imakura, Y.; Kobayashi, S.; Mima, A. Bitter phenyl propanoid glycosides from campsis chinensis. Phytochemistry 1985, 24, 139–146. [Google Scholar] [CrossRef]
- Kitagawa, S.; Nishibe, S.; Benecke, R.; Thieme, H. Phenolic compounds from Forsythia leaves. II. Chem. Pharm. Bull. 1988, 36, 3667–3670. [Google Scholar] [CrossRef]
- Yang, X.; Guo, F.; Peng, Q.; Liu, Y.; Yang, B. Suppression of in vitro and in vivo human ovarian cancer growth by isoacteoside is mediated via sub-G1 cell cycle arrest, ROS generation, and modulation of AKT/PI3K/m-TOR signalling pathway. J. BUON 2019, 24, 285–290. [Google Scholar]
- Giri, A.; Narasu, M.L. Production of podophyllotoxin from Podophyllum hexandrum: A potential natural product for clinically useful anticancer drugs. Cytotechnology 2000, 34, 17–26. [Google Scholar] [CrossRef]
- Rao, S.R.; Ravishankar, G. Plant cell cultures: Chemical factories of secondary metabolites. Biotechnol. Adv. 2002, 20, 101–153. [Google Scholar] [PubMed]
- Hellwig, S.; Drossard, J.; Twyman, R.M.; Fischer, R. Plant cell cultures for the production of recombinant proteins. Nat. Biotechnol. 2004, 22, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Khanpour-Ardestani, N.; Sharifi, M.; Behmanesh, M. Establishment of callus and cell suspension culture of Scrophularia striata Boiss.: An in vitro approach for acteoside production. Cytotechnology 2015, 67, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Jang, T.W.; Park, J.H. Antioxidant activity and inhibitory effects on oxidative DNA damage of callus from Abeliophyllum distichum Nakai. Kor. J. Plant Res. 2018, 31, 228–236. [Google Scholar]
- Koh, D.; Seo, B.; Lee, C. Studies on the in vitro induction of callus from another culture of Abeliophyllum distichum. J. Chonbuk Nat. Univ. 1989, 31, 153–159. [Google Scholar]
- Jensen, S.R.; Franzyk, H.; Wallander, E. Chemotaxonomy of the Oleaceae: Iridoids as taxonomic markers. Phytochemistry 2002, 60, 213–231. [Google Scholar] [CrossRef]
- Kim, N.Y.; Lee, H.Y. Effect of antioxidant and skin whitening of ethanol extracts from ultrasonic pretreated Abeliophyllum distichum Nakai. Kor. J. Med. Crop. Sci. 2015, 23, 155–160. [Google Scholar] [CrossRef]
- Kim, H.W.; Lee, H.L.; Lee, D.K.; Kim, K.J. Complete plastid genome sequences of Abeliophyllum distichum Nakai (Oleaceae), a Korea endemic genus. Mitochondrial DNA Part B 2016, 1, 596–598. [Google Scholar] [CrossRef]
- Min, J.; Kim, Y.; Xi, H.; Jang, T.; Kim, G.; Park, J.; Park, J.H. The complete chloroplast genome of a new candidate cultivar, Sang Jae, of Abeliophyllum distichum Nakai (Oleaceae): Initial step of A. distichum intraspecies variations atlas. Mitochondrial DNA Part B 2019, 4, 3716–3718. [Google Scholar] [CrossRef]
- Park, J.; Min, J.; Kim, Y.; Xi, H.; Kwon, W.; Jang, T.; Kim, G.; Park, J.H. The complete chloroplast genome of a new candidate cultivar, Dae Ryun, of Abeliophyllum distichum Nakai (Oleaceae). Mitochondrial DNA Part B 2019, 4, 3713–3715. [Google Scholar] [CrossRef]
- Park, G.H.; Park, J.H.; Eo, H.J.; Song, H.M.; Lee, M.H.; Lee, J.R.; Jeong, J.B. Anti-inflammatory effect of the extracts from Abeliophyllum distichum Nakai in LPS-stimulated RAW264. 7 cells. Kor. J. Plant Res. 2014, 27, 209–214. [Google Scholar] [CrossRef]
- Haque, M.A.; Jantan, I.; Harikrishnan, H. Zerumbone suppresses the activation of inflammatory mediators in LPS-stimulated U937 macrophages through MyD88-dependent NF-κB/MAPK/PI3K-Akt signaling pathways. Int. Immunopharmacol. 2018, 55, 312–322. [Google Scholar] [CrossRef]
- Haque, M.A.; Jantan, I.; Harikrishnan, H.; Wahab, S.M.A. Magnoflorine enhances LPS-activated pro-inflammatory responses via MyD88-dependent pathways in U937 macrophages. Planta Med. 2018, 84, 1255–1264. [Google Scholar] [CrossRef]
- Roh, K.B.; Kim, H.; Shin, S.; Kim, Y.S.; Lee, J.A.; Kim, M.O.; Jung, E.; Lee, J.; Park, D. Anti-inflammatory effects of Zea mays L. husk extracts. BMC Complement. Altern. Med. 2016, 16, 1–8. [Google Scholar] [CrossRef]
- Green, S.J.; Mellouk, S.; Hoffman, S.L.; Meltzer, M.S.; Nacy, C.A. Cellular mechanisms of nonspecific immunity to intracellular infection: Cytokine-induced synthesis of toxic nitrogen oxides from L-arginine by macrophages and hepatocytes. Immunol. Lett. 1990, 25, 15–19. [Google Scholar] [CrossRef]
- Green, S.; Nacy, C.; Schreiber, R.; Granger, D.; Crawford, R.; Meltzer, M.; Fortier, A. Neutralization of gamma interferon and tumor necrosis factor alpha blocks in vivo synthesis of nitrogen oxides from L-arginine and protection against Francisella tularensis infection in Mycobacterium bovis BCG-treated mice. Infect. Immun. 1993, 61, 689–698. [Google Scholar] [CrossRef]
- Kamijo, R.; Gerecitano, J.; Shapiro, D.; Green, S.J.; Aguet, M.; Le, J.; Vilcek, J. Generation of nitric oxide and clearance of interferon-gamma after BCG infection are impaired in mice that lack the interferon-gamma receptor. J. Inflamm. 1995, 46, 23–31. [Google Scholar]
- Green, S.J.; Scheller, L.F.; Marletta, M.A.; Seguin, M.C.; Klotz, F.W.; Slayter, M.; Nelson, B.J.; Nacy, C.A. Nitric oxide: Cytokine-regulation of nitric oxide in host resistance to intracellular pathogens. Immunol. Lett. 1994, 43, 87–94. [Google Scholar] [CrossRef]
- Mohamed, S.I.A.; Jantan, I.; Haque, M.A. Naturally occurring immunomodulators with antitumor activity: An insight on their mechanisms of action. Int. Immunopharmacol. 2017, 50, 291–304. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat. Rev. Immunol. 2007, 7, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.-E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar]
- Nishida, E.; Gotoh, Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem. Sci. 1993, 18, 128–131. [Google Scholar] [CrossRef]
- Kirkwood, K.L.; Rossa, C., Jr. The potential of p38 MAPK inhibitors to modulate periodontal infections. Curr. Drug Metab. 2009, 10, 55–67. [Google Scholar] [CrossRef]
- Kaminska, B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy—From molecular mechanisms to therapeutic benefits. Biochim. Biophys. Acta Proteins Proteom. 2005, 1754, 253–262. [Google Scholar] [CrossRef]
- Kim, J.W.; Kim, C. Inhibition of LPS-induced NO production by taurine chloramine in macrophages is mediated though Ras-ERK-NF-κB. Biochem. Pharmacol. 2005, 70, 1352–1360. [Google Scholar] [CrossRef]
- Gilmore, T.D. Introduction to NF-κ B: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef] [PubMed]
- Brasier, A.R. The NF-κB regulatory network. Cardiovasc. Toxicol. 2006, 6, 111–130. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.J.; Chun, K.S.; Cha, H.H.; Han, S.S.; Keum, Y.S.; Park, K.K.; Lee, S.S. Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: Down-regulation of COX-2 and iNOS through suppression of NF-κB activation. Mutat. Res. 2001, 480, 243–268. [Google Scholar] [CrossRef]
- Makarov, S.S. NF-κB as a therapeutic target in chronic inflammation: Recent advances. Mol. Med. Today 2000, 6, 441–448. [Google Scholar] [CrossRef]
- Laird, M.H.; Rhee, S.H.; Perkins, D.J.; Medvedev, A.E.; Piao, W.; Fenton, M.J.; Vogel, S.N. TLR4/MyD88/PI3K interactions regulate TLR4 signaling. J. Leukoc. Biol. 2009, 85, 966–977. [Google Scholar] [CrossRef]


| A, RT-PCR | ||
| Gene | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
| iNOS | AATGGCAACATCAGGTCGGC CATCACT | GCTGTGTGTCACAGAAGTCTC GAACTC |
| COX-2 | GGAGAGACTATCAAGATAGT | ATGGTCAGTAGACTTTTACA |
| TNF-α | CACACTCAGATCATCTTCTC | TTGAAGAGAACCTGGGAGTA |
| IL-6 | ATTACACATGTTCTCTGGGA | TTTTACCTCTTGGTTGAAGA |
| IL-1β | CAGGATGAGGACATGAGCAC | CTCTGCAGACTCAAACTCCA |
| GAPDH | AACTTTGGCATTGTGGAAGG | ATGCAGGGATGATGTTCTGG |
| B, RT-qPCR | ||
| Gene | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
| iNOS | TGGTGGTGACAAGCACATTT | AAGGCCAAACACAGCATACC |
| COX-2 | AGAAGGAAATGGCTG CAGAA | CCCCAAAGATAGCATCTGGA |
| GAPDH | CCTCCAAGGAGTAAGAAACC | CTAGGCCCCTCCTGTTATTA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, T.-W.; Park, J.-H. Anti-Inflammatory Effects of Abeliophyllum distichum Nakai (Cultivar Okhwang 1) Callus through Inhibition of PI3K/Akt, NF-κB, and MAPK Signaling Pathways in Lipopolysaccharide-Induced Macrophages. Processes 2021, 9, 1071. https://doi.org/10.3390/pr9061071
Jang T-W, Park J-H. Anti-Inflammatory Effects of Abeliophyllum distichum Nakai (Cultivar Okhwang 1) Callus through Inhibition of PI3K/Akt, NF-κB, and MAPK Signaling Pathways in Lipopolysaccharide-Induced Macrophages. Processes. 2021; 9(6):1071. https://doi.org/10.3390/pr9061071
Chicago/Turabian StyleJang, Tae-Won, and Jae-Ho Park. 2021. "Anti-Inflammatory Effects of Abeliophyllum distichum Nakai (Cultivar Okhwang 1) Callus through Inhibition of PI3K/Akt, NF-κB, and MAPK Signaling Pathways in Lipopolysaccharide-Induced Macrophages" Processes 9, no. 6: 1071. https://doi.org/10.3390/pr9061071
APA StyleJang, T.-W., & Park, J.-H. (2021). Anti-Inflammatory Effects of Abeliophyllum distichum Nakai (Cultivar Okhwang 1) Callus through Inhibition of PI3K/Akt, NF-κB, and MAPK Signaling Pathways in Lipopolysaccharide-Induced Macrophages. Processes, 9(6), 1071. https://doi.org/10.3390/pr9061071






