On the Use of Surface Plasmon Resonance-Based Biosensors for Advanced Bioprocess Monitoring
Abstract
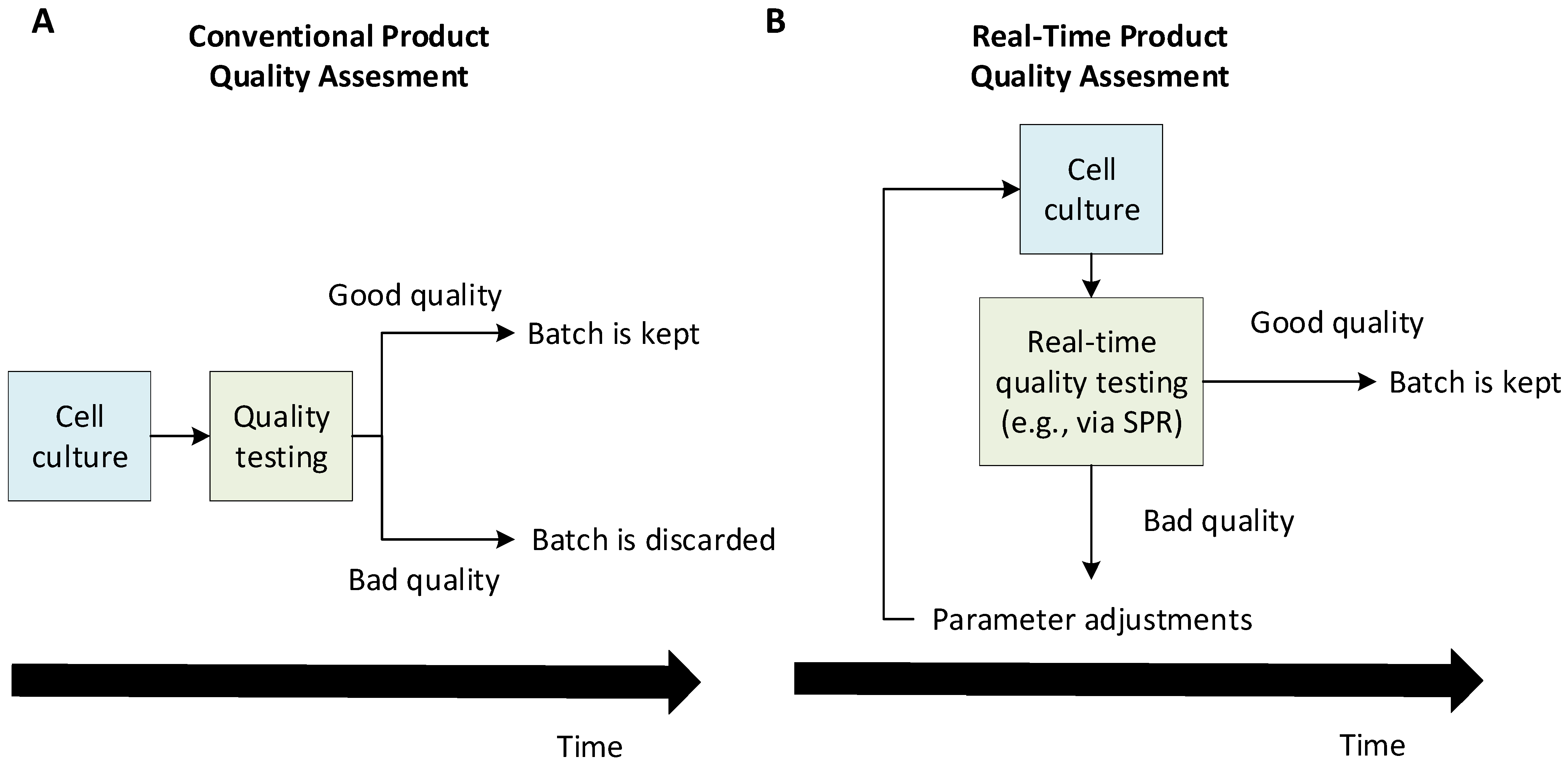
:1. Introduction
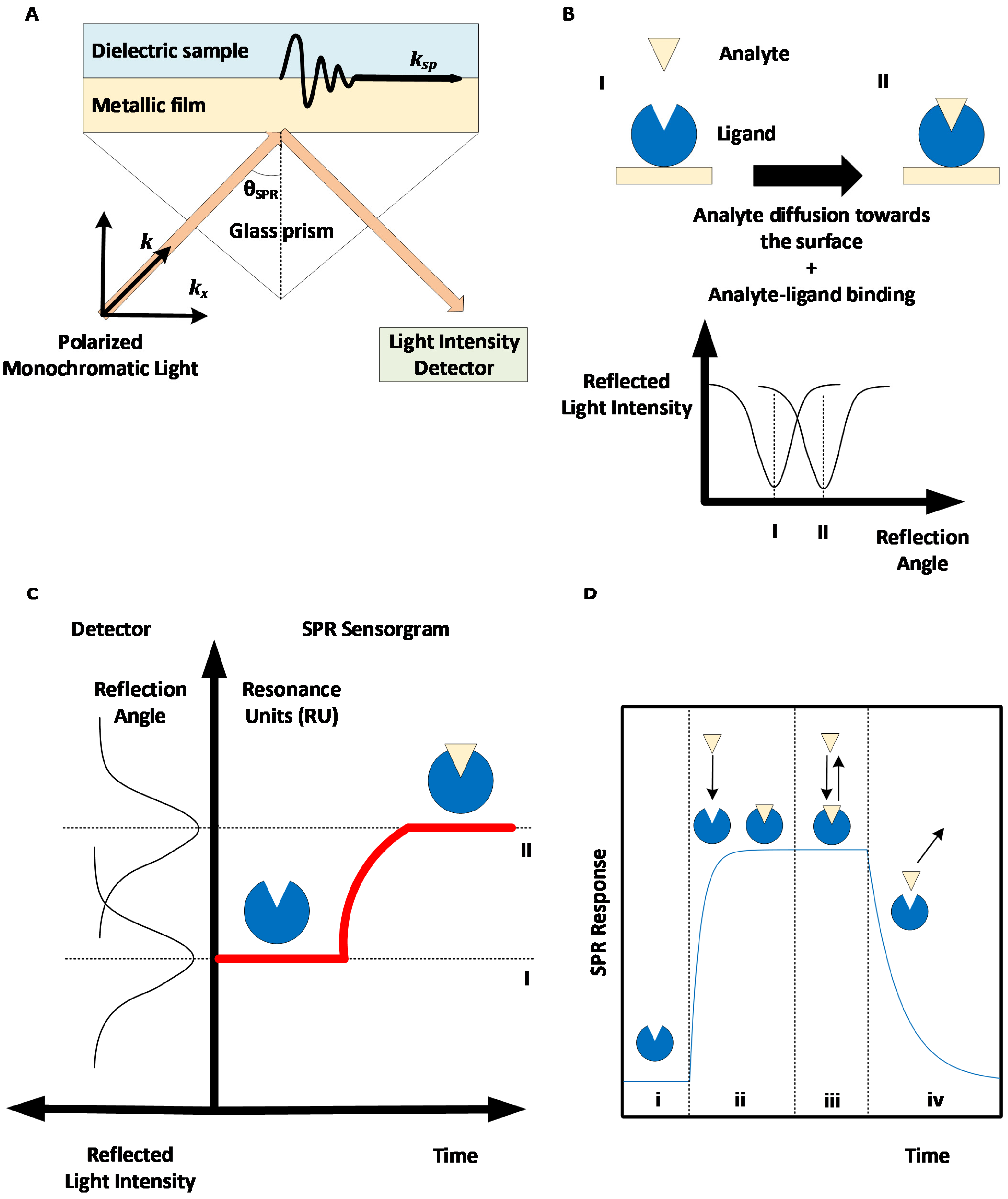
2. Surface Plasmon Resonance: Basic Principles and Methods
| Manufacturer | SPR Method | Detection | Reference |
|---|---|---|---|
| Affinité Instruments | SPR | Wavelength | [76] |
| Biacore (Cytiva) | SPR | Angle | [77] |
| Bionavis | Multi Parametric SPR (MP-SPR) | Angle | [78] |
| Biosensing Instrument | SPR | Angle | [79] |
| Carterra | SPR Imaging (SPRi) | Angle | [80] |
| Reichert Technologies | SPR | Angle | [81] |
| Sierra Sensors (Bruker) | SPR Imaging (SPRi) | Angle | [82] |
2.1. SPR to Measure Kinetics and Affinity
2.2. SPR to Measure Concentrations
3. SPR Applications to Biotherapeutics Production Monitoring
3.1. SPR for the Early Development of Biotherapeutics
3.2. SPR for the Quantification of Biotherapeutics
3.3. SPR for the Safety and Quality Assessment of Biotherapeutics
3.4. Sequential SPR Assays
4. SPR Applications to Vaccine Production Monitoring
4.1. SPR for Quantification of Vaccines Preparations
4.2. SPR for Quantification of Vaccines during Production
5. SPR Applications in Bacteria and Contaminant Monitoring
6. Limitations of SPR Biosensing in the Context of Bioprocess Monitoring
7. Recent Developments in SPR Instruments
SPR Studies with Live Cells
8. Recent Developments in SPR Data Analysis
9. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef]
- Gherghescu, I.; Delgado-Charro, M.B. The Biosimilar Landscape: An Overview of Regulatory Approvals by the EMA and FDA. Pharmaceutics 2021, 13, 48. [Google Scholar] [CrossRef]
- U. S. Food and Drug Administration. Guidance for Industry: Q8(R2) Pharmaceutical Development; U. S. Food and Drug Administration: Rockville, MD, USA, 2009. [Google Scholar]
- Yu, L.X.; Amidon, G.; Khan, M.A.; Hoag, S.W.; Polli, J.; Raju, G.K.; Woodcock, J. Understanding pharmaceutical quality by design. AAPS J 2014, 16, 771–783. [Google Scholar] [CrossRef] [Green Version]
- Grangeia, H.B.; Silva, C.; Simões, S.P.; Reis, M.S. Quality by design in pharmaceutical manufacturing: A systematic review of current status, challenges and future perspectives. Eur. J. Pharm. Biopharm. 2020, 147, 19–37. [Google Scholar] [CrossRef]
- Yu, L.X.; Kopcha, M. The future of pharmaceutical quality and the path to get there. Int. J. Pharm. 2017, 528, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Sommeregger, W.; Sissolak, B.; Kandra, K.; von Stosch, M.; Mayer, M.; Striedner, G. Quality by control: Towards model predictive control of mammalian cell culture bioprocesses. Biotechnol. J. 2017, 12, 1600546. [Google Scholar] [CrossRef] [Green Version]
- Su, Q.; Ganesh, S.; Moreno, M.; Bommireddy, Y.; Gonzalez, M.; Reklaitis, G.V.; Nagy, Z.K. A perspective on Quality-by-Control (QbC) in pharmaceutical continuous manufacturing. Comput. Chem. Eng. 2019, 125, 216–231. [Google Scholar] [CrossRef]
- U. S. Food and Drug Administration. Guidance for Industry: PAT-A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance; U. S. Food and Drug Administration: Rockville, MD, USA, 2004. [Google Scholar]
- Simon, L.L.; Pataki, H.; Marosi, G.; Meemken, F.; Hungerbühler, K.; Baiker, A.; Tummala, S.; Glennon, B.; Kuentz, M.; Steele, G.; et al. Assessment of Recent Process Analytical Technology (PAT) Trends: A Multiauthor Review. Org. Process Res. Dev. 2015, 19, 3–62. [Google Scholar] [CrossRef]
- Helgers, H.; Schmidt, A.; Lohmann, L.J.; Vetter, F.L.; Juckers, A.; Jensch, C.; Mouellef, M.; Zobel-Roos, S.; Strube, J. Towards Autonomous Operation by Advanced Process Control—Process Analytical Technology for Continuous Biologics Antibody Manufacturing. Processes 2021, 9, 172. [Google Scholar] [CrossRef]
- Kretschmann, E.; Raether, H. Notizen: Radiative Decay of Non Radiative Surface Plasmons Excited by Light. Z. Nat. A 1968, 23, 2135–2136. [Google Scholar] [CrossRef]
- Kretschmann, E. Decay of non radiative surface plasmons into light on rough silver films. Comparison of experimental and theoretical results. Opt. Commun. 1972, 6, 185–187. [Google Scholar] [CrossRef]
- Otto, A. Excitation of nonradiative surface plasma waves in silver by the method of frustrated total reflection. Z. Phys. A Hadron. Nucl. 1968, 216, 398–410. [Google Scholar] [CrossRef]
- De Crescenzo, G.; Boucher, C.; Durocher, Y.; Jolicoeur, M. Kinetic Characterization by Surface Plasmon Resonance-Based Biosensors: Principle and Emerging Trends. Cell. Mol. Bioeng. 2008, 1, 204–215. [Google Scholar] [CrossRef]
- Homola, J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Couture, M.; Zhao, S.S.; Masson, J.-F. Modern surface plasmon resonance for bioanalytics and biophysics. Phys. Chem. Chem. Phys. 2013, 15, 11190–11216. [Google Scholar] [CrossRef]
- Wang, D.S.; Fan, S.K. Microfluidic Surface Plasmon Resonance Sensors: From Principles to Point-of-Care Applications. Sensors 2016, 16, 1175. [Google Scholar] [CrossRef] [Green Version]
- Prabowo, B.A.; Purwidyantri, A.; Liu, K.-C. Surface Plasmon Resonance Optical Sensor: A Review on Light Source Technology. Biosensors 2018, 8, 80. [Google Scholar] [CrossRef] [Green Version]
- Shrivastav, A.M.; Cvelbar, U.; Abdulhalim, I. A comprehensive review on plasmonic-based biosensors used in viral diagnostics. Commun. Biol. 2021, 4, 70. [Google Scholar] [CrossRef]
- Guo, X. Surface plasmon resonance based biosensor technique: A review. J. Biophotonics 2012, 5, 483–501. [Google Scholar] [CrossRef]
- Myszka, D.G. Improving biosensor analysis. J. Mol. Recognit. JMR 1999, 12, 279–284. [Google Scholar] [CrossRef]
- Önell, A.; Andersson, K. Kinetic determinations of molecular interactions using Biacore—minimum data requirements for efficient experimental design. J. Mol. Recognit. 2005, 18, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, R.; Katsamba, P.S.; Nordin, H.; Pol, E.; Myszka, D.G. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 2006, 349, 136–147. [Google Scholar] [CrossRef] [PubMed]
- De Crescenzo, G.; Woodward, L.; Srinivasan, B. Online optimization of surface plasmon resonance-based biosensor experiments for improved throughput and confidence. J. Mol. Recognit. 2008, 21, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Mehand, M.S.; De Crescenzo, G.; Srinivasan, B. Increasing throughput of surface plasmon resonance-based biosensors by multiple analyte injections. J. Mol. Recognit. JMR 2012, 25, 208–215. [Google Scholar] [CrossRef]
- Si Mehand, M.; De Crescenzo, G.; Srinivasan, B. On-line kinetic model discrimination for optimized surface plasmon resonance experiments. J. Mol. Recognit. JMR 2014, 27, 276–284. [Google Scholar] [CrossRef]
- Mehand, M.S.; Srinivasan, B.; De Crescenzo, G. Optimizing Multiple Analyte Injections in Surface Plasmon Resonance Biosensors with Analytes having Different Refractive Index Increments. Sci. Rep. 2015, 5, 15855. [Google Scholar] [CrossRef]
- Zhang, Y.; Forssén, P.; Fornstedt, T.; Gulliksson, M.; Dai, X. An adaptive regularization algorithm for recovering the rate constant distribution from biosensor data. Inverse Probl. Sci. Eng. 2018, 26, 1464–1489. [Google Scholar] [CrossRef]
- Forssén, P.; Multia, E.; Samuelsson, J.; Andersson, M.; Aastrup, T.; Altun, S.; Wallinder, D.; Wallbing, L.; Liangsupree, T.; Riekkola, M.-L.; et al. Reliable Strategy for Analysis of Complex Biosensor Data. Anal. Chem. 2018, 90, 5366–5374. [Google Scholar] [CrossRef] [Green Version]
- Gaudreault, J.; Liberelle, B.; Durocher, Y.; Henry, O.; De Crescenzo, G. Determination of the composition of heterogeneous binder solutions by surface plasmon resonance biosensing. Sci. Rep. 2021, 11, 3685. [Google Scholar] [CrossRef]
- Pol, E.; Roos, H.; Markey, F.; Elwinger, F.; Shaw, A.; Karlsson, R. Evaluation of calibration-free concentration analysis provided by Biacore™ systems. Anal. Biochem. 2016, 510, 88–97. [Google Scholar] [CrossRef]
- Karlsson, R. Biosensor binding data and its applicability to the determination of active concentration. Biophys. Rev. 2016, 8, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, R.; Roos, H.; Fägerstam, L.; Persson, B. Kinetic and Concentration Analysis Using BIA Technology. Methods 1994, 6, 99–110. [Google Scholar] [CrossRef]
- Christensen, L.L. Theoretical analysis of protein concentration determination using biosensor technology under conditions of partial mass transport limitation. Anal. Biochem. 1997, 249, 153–164. [Google Scholar] [CrossRef]
- Vaisocherová, H.; Faca, V.M.; Taylor, A.D.; Hanash, S.; Jiang, S. Comparative study of SPR and ELISA methods based on analysis of CD166/ALCAM levels in cancer and control human sera. Biosens. Bioelectron. 2009, 24, 2143–2148. [Google Scholar] [CrossRef]
- Karlsson, R.; Fridh, V.; Frostell, Å. Surrogate potency assays: Comparison of binding profiles complements dose response curves for unambiguous assessment of relative potencies. J. Pharm. Anal. 2018, 8, 138–146. [Google Scholar] [CrossRef]
- Mandenius, C.F.; Wang, R.; Aldén, A.; Bergström, G.; Thébault, S.; Lutsch, C.; Ohlson, S. Monitoring of influenza virus hemagglutinin in process samples using weak affinity ligands and surface plasmon resonance. Anal. Chim. Acta 2008, 623, 66–75. [Google Scholar] [CrossRef]
- Nilsson, C.E.; Abbas, S.; Bennemo, M.; Larsson, A.; Hämäläinen, M.D.; Frostell-Karlsson, A. A novel assay for influenza virus quantification using surface plasmon resonance. Vaccine 2010, 28, 759–766. [Google Scholar] [CrossRef]
- Khurana, S.; King, L.R.; Manischewitz, J.; Coyle, E.M.; Golding, H. Novel antibody-independent receptor-binding SPR-based assay for rapid measurement of influenza vaccine potency. Vaccine 2014, 32, 2188–2197. [Google Scholar] [CrossRef]
- Durous, L.; Julien, T.; Padey, B.; Traversier, A.; Rosa-Calatrava, M.; Blum, L.J.; Marquette, C.A.; Petiot, E. SPRi-based hemagglutinin quantitative assay for influenza vaccine production monitoring. Vaccine 2019, 37, 1614–1621. [Google Scholar] [CrossRef]
- Bruce-Staskal, P.J.; Woods, R.M.; Borisov, O.V.; Massare, M.J.; Hahn, T.J. Corrigendum to “Hemagglutinin from multiple divergent influenza A and B viruses bind to a distinct branched, sialylated poly-LacNAc glycan by surface plasmon resonance” [Vaccine 38(43) (2020) 6757–6765]. Vaccine 2021, 39, 1544–1545. [Google Scholar] [CrossRef]
- Poltronieri, P.; Mezzolla, V.; Primiceri, E.; Maruccio, G. Biosensors for the Detection of Food Pathogens. Foods 2014, 3, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Mattiasson, B.; Teeparuksapun, K.; Hedström, M. Immunochemical binding assays for detection and quantification of trace impurities in biotechnological production. Trends Biotechnol. 2010, 28, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Dudak, F.C.; Boyaci, I.H. Rapid and label-free bacteria detection by surface plasmon resonance (SPR) biosensors. Biotechnol. J. 2009, 4, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ye, Z.; Si, C.; Ying, Y. Monitoring of Escherichia coli O157:H7 in food samples using lectin based surface plasmon resonance biosensor. Food Chem. 2013, 136, 1303–1308. [Google Scholar] [CrossRef]
- Bhandari, D.; Chen, F.-C.; Bridgman, R.C. Detection of Salmonella Typhimurium in Romaine Lettuce Using a Surface Plasmon Resonance Biosensor. Biosensors 2019, 9, 94. [Google Scholar] [CrossRef] [Green Version]
- Masdor, N.A.; Altintas, Z.; Tothill, I.E. Surface Plasmon Resonance Immunosensor for the Detection of Campylobacter jejuni. Chemosensors 2017, 5, 16. [Google Scholar] [CrossRef]
- Masdor, N.A.; Altintas, Z.; Shukor, M.Y.; Tothill, I.E. Subtractive inhibition assay for the detection of Campylobacter jejuni in chicken samples using surface plasmon resonance. Sci. Rep. 2019, 9, 13642. [Google Scholar] [CrossRef] [Green Version]
- Raghu, H.V.; Kumar, N. Rapid Detection of Listeria monocytogenes in Milk by Surface Plasmon Resonance Using Wheat Germ Agglutinin. Food Anal. Methods 2020, 13, 982–991. [Google Scholar] [CrossRef]
- Forest-Nault, C.; Gaudreault, J.; Henry, O.; Durocher, Y.; De Crescenzo, G. On the Use of Surface Plasmon Resonance Biosensing to Understand IgG-FcγR Interactions. Int. J. Mol. Sci. 2021, 22, 6616. [Google Scholar] [CrossRef]
- Cambay, F.; Henry, O.; Durocher, Y.; De Crescenzo, G. Impact of N-glycosylation on Fcγ receptor/IgG interactions: Unravelling differences with an enhanced surface plasmon resonance biosensor assay based on coiled-coil interactions. mAbs 2019, 11, 435–452. [Google Scholar] [CrossRef] [Green Version]
- Cambay, F.; Forest-Nault, C.; Dumoulin, L.; Seguin, A.; Henry, O.; Durocher, Y.; De Crescenzo, G. Glycosylation of Fcγ receptors influences their interaction with various IgG1 glycoforms. Mol. Immunol. 2020, 121, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Subedi, G.P.; Barb, A.W. The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc γ receptor. mAbs 2016, 8, 1512–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorion-Thibaudeau, J.; Durocher, Y.; De Crescenzo, G. Quantification and simultaneous evaluation of the bioactivity of antibody produced in CHO cell culture-The use of the ectodomain of FcγRI and surface plasmon resonance-based biosensor. Mol. Immunol. 2017, 82, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Soriano, B.; Chen, Q. Glycan profiling of proteins using lectin binding by Surface Plasmon Resonance. Anal. Biochem. 2017, 538, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, J.; Wang, Y.; Cai, K.; Wang, Q.; Hou, X.; Guo, W.; Zhang, F. Development of biosensor-based SPR technology for biological quantification and quality control of pharmaceutical proteins. J. Pharm. Biomed. Anal. 2009, 50, 1026–1029. [Google Scholar] [CrossRef] [PubMed]
- Coty, J.B.; Noiray, M.; Vauthier, C. Assessment of Complement Activation by Nanoparticles: Development of a SPR Based Method and Comparison with Current High Throughput Methods. Pharm. Res. 2018, 35, 129. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ding, L.; Zhou, J.; Chen, S.; Chen, F.; Zhao, C.; Xu, J.; Hu, W.; Ji, J.; Xu, H.; et al. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 2021, 171, 112685. [Google Scholar] [CrossRef]
- Mauriz, E. Recent Progress in Plasmonic Biosensing Schemes for Virus Detection. Sensors 2020, 20, 4745. [Google Scholar] [CrossRef]
- Singh, P. Surface Plasmon Resonance: A Boon for Viral Diagnostics. Ref. Modul. Life Sci. 2017. [Google Scholar] [CrossRef]
- Bellassai, N.; D’Agata, R.; Jungbluth, V.; Spoto, G. Surface Plasmon Resonance for Biomarker Detection: Advances in Non-invasive Cancer Diagnosis. Front. Chem. 2019, 7, 570. [Google Scholar] [CrossRef] [Green Version]
- Souto, D.E.P.; Volpe, J.; Gonçalves, C.d.C.; Ramos, C.H.I.; Kubota, L.T. A brief review on the strategy of developing SPR-based biosensors for application to the diagnosis of neglected tropical diseases. Talanta 2019, 205, 120122. [Google Scholar] [CrossRef] [PubMed]
- Brulé, T.; Granger, G.; Bukar, N.; Deschênes-Rancourt, C.; Havard, T.; Schmitzer, A.R.; Martel, R.; Masson, J.-F. A field-deployed surface plasmon resonance (SPR) sensor for RDX quantification in environmental waters. Analyst 2017, 142, 2161–2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zschätzsch, M.; Ritter, P.; Henseleit, A.; Wiehler, K.; Malik, S.; Bley, T.; Walther, T.; Boschke, E. Monitoring bioactive and total antibody concentrations for continuous process control by surface plasmon resonance spectroscopy. Eng. Life Sci. 2019, 19, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frostell, Å.; Mattsson, A.; Eriksson, Å.; Wallby, E.; Kärnhall, J.; Illarionova, N.B.; Estmer Nilsson, C. Nine surface plasmon resonance assays for specific protein quantitation during cell culture and process development. Anal. Biochem. 2015, 477, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chavane, N.; Jacquemart, R.; Hoemann, C.D.; Jolicoeur, M.; De Crescenzo, G. At-line quantification of bioactive antibody in bioreactor by surface plasmon resonance using epitope detection. Anal. Biochem. 2008, 378, 158–165. [Google Scholar] [CrossRef]
- Jacquemart, R.; Chavane, N.; Durocher, Y.; Hoemann, C.; De Crescenzo, G.; Jolicoeur, M. At-line monitoring of bioreactor protein production by surface plasmon resonance. Biotechnol. Bioeng. 2008, 100, 184–188. [Google Scholar] [CrossRef]
- Beeg, M.; Nobili, A.; Orsini, B.; Rogai, F.; Gilardi, D.; Fiorino, G.; Danese, S.; Salmona, M.; Garattini, S.; Gobbi, M. A Surface Plasmon Resonance-based assay to measure serum concentrations of therapeutic antibodies and anti-drug antibodies. Sci. Rep. 2019, 9, 2064. [Google Scholar] [CrossRef]
- D’Agata, R.; Bellassai, N.; Jungbluth, V.; Spoto, G. Recent Advances in Antifouling Materials for Surface Plasmon Resonance Biosensing in Clinical Diagnostics and Food Safety. Polymers 2021, 13, 1929. [Google Scholar] [CrossRef]
- D’Agata, R.; Bellassai, N.; Giuffrida, M.C.; Aura, A.M.; Petri, C.; Kögler, P.; Vecchio, G.; Jonas, U.; Spoto, G. A new ultralow fouling surface for the analysis of human plasma samples with surface plasmon resonance. Talanta 2021, 221, 121483. [Google Scholar] [CrossRef]
- Schasfoort, R.B.M. Introduction to Surface Plasmon Resonance. In Handbook of Surface Plasmon Resonance, 2nd ed.; Schasfoort, R.B.M., Ed.; The Royal Society of Chemistry: London, UK, 2017; pp. 1–26. [Google Scholar]
- Deng, S.; Wang, P.; Yu, X. Phase-Sensitive Surface Plasmon Resonance Sensors: Recent Progress and Future Prospects. Sensors 2017, 17, 2819. [Google Scholar] [CrossRef] [Green Version]
- Kashif, M.; Bakar, A.A.A.; Arsad, N.; Shaari, S. Development of phase detection schemes based on surface plasmon resonance using interferometry. Sensors 2014, 14, 15914–15938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenberg, E.; Persson, B.; Roos, H.; Urbaniczky, C. Quantitative determination of surface concentration of protein with surface plasmon resonance using radiolabeled proteins. J. Colloid Interface Sci. 1991, 143, 513–526. [Google Scholar] [CrossRef]
- Affinité Instruments. Unleashing Label Free Sensing-Surface Plasmon Resonance for Rapid Testing. Available online: https://www.affiniteinstruments.com/ (accessed on 16 September 2021).
- Cytiva. Biacore SPR-Surface Plasmon Resonance Interaction Analysis. Available online: https://www.cytivalifesciences.com/en/us/solutions/protein-research/interaction-analysis-with-biacore-surface-plasmon-resonance-spr (accessed on 16 September 2021).
- BioNavis. BioNavis: Enter the world of MP-SPR and find a solution to your research needs! Available online: https://www.bionavis.com/ (accessed on 16 September 2021).
- Biosensing Instrument. Biosensing Instrument Offers Bioanalytical Tools to Accelerate Drug Discovery Research. Available online: https://biosensingusa.com/ (accessed on 16 September 2021).
- Carterra. High Throughput Antibody Screening and Characterization. Available online: https://carterra-bio.com/ (accessed on 16 September 2021).
- Reichert Technologies. Discover the Reichert SPR Difference. Available online: https://www.reichertspr.com/ (accessed on 16 September 2021).
- Bruker. Surface Plasmon Resonance for High-Throughput Analysis. Available online: https://www.bruker.com/en/products-and-solutions/surface-plasmon-resonance.html (accessed on 16 September 2021).
- Andersson, K.; Hämäläinen, M.; Malmqvist, M. Identification and Optimization of Regeneration Conditions for Affinity-Based Biosensor Assays. A Multivariate Cocktail Approach. Anal. Chem. 1999, 71, 2475–2481. [Google Scholar] [CrossRef] [PubMed]
- Goode, J.A.; Rushworth, J.V.H.; Millner, P.A. Biosensor Regeneration: A Review of Common Techniques and Outcomes. Langmuir 2015, 31, 6267–6276. [Google Scholar] [CrossRef] [PubMed]
- Svitel, J.; Balbo, A.; Mariuzza, R.A.; Gonzales, N.R.; Schuck, P. Combined Affinity and Rate Constant Distributions of Ligand Populations from Experimental Surface Binding Kinetics and Equilibria. Biophys. J. 2003, 84, 4062–4077. [Google Scholar] [CrossRef] [Green Version]
- Svitel, J.; Boukari, H.; Van Ryk, D.; Willson, R.C.; Schuck, P. Probing the Functional Heterogeneity of Surface Binding Sites by Analysis of Experimental Binding Traces and the Effect of Mass Transport Limitation. Biophys. J. 2007, 92, 1742–1758. [Google Scholar] [CrossRef] [Green Version]
- Gorshkova, I.I.; Svitel, J.; Razjouyan, F.; Schuck, P. Bayesian Analysis of Heterogeneity in the Distribution of Binding Properties of Immobilized Surface Sites. Langmuir 2008, 24, 11577–11586. [Google Scholar] [CrossRef] [Green Version]
- Khalifa, M.B.; Choulier, L.; Lortat-Jacob, H.; Altschuh, D.; Vernet, T. BIACORE Data Processing: An Evaluation of the Global Fitting Procedure. Anal. Biochem. 2001, 293, 194–203. [Google Scholar] [CrossRef]
- De Crescenzo, G.; Grothe, S.; Zwaagstra, J.; Tsang, M.; O’Connor-McCourt, M.D. Real-time monitoring of the interactions of transforming growth factor-beta (TGF-beta ) isoforms with latency-associated protein and the ectodomains of the TGF-beta type II and III receptors reveals different kinetic models and stoichiometries of binding. J. Biol. Chem. 2001, 276, 29632–29643. [Google Scholar] [CrossRef] [Green Version]
- Sprague, E.R.; Martin, W.L.; Bjorkman, P.J. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE-gI. J. Biol. Chem. 2004, 279, 14184–14193. [Google Scholar] [CrossRef] [Green Version]
- Giannetti, A.M.; Snow, P.M.; Zak, O.; Björkman, P.J. Mechanism for Multiple Ligand Recognition by the Human Transferrin Receptor. PLoS Biol. 2003, 1, e51. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.M.; Arndt, K.M.; Plückthun, A. Model and simulation of multivalent binding to fixed ligands. Anal. Biochem. 1998, 261, 149–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Crescenzo, G.; Grothe, S.; Lortie, R.; Debanne, M.T.; O’Connor-McCourt, M. Real-Time Kinetic Studies on the Interaction of Transforming Growth Factor α with the Epidermal Growth Factor Receptor Extracellular Domain Reveal a Conformational Change Model. Biochemistry 2000, 39, 9466–9476. [Google Scholar] [CrossRef] [PubMed]
- Futamura, M.; Dhanasekaran, P.; Handa, T.; Phillips, M.C.; Lund-Katz, S.; Saito, H. Two-step mechanism of binding of apolipoprotein E to heparin: Implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J. Biol. Chem. 2005, 280, 5414–5422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bio-Rad Laboratories. ProteOn XPR36 Experimental Design and Application Guide; Bio-Rad Laboratoires: Hercules, CA, USA, 2014. [Google Scholar]
- Goldstein, B.; Coombs, D.; He, X.; Pineda, A.R.; Wofsy, C. The influence of transport on the kinetics of binding to surface receptors: Application to cells and BIAcore. J. Mol. Recognit. JMR 1999, 12, 293–299. [Google Scholar] [CrossRef]
- Sjoelander, S.; Urbaniczky, C. Integrated fluid handling system for biomolecular interaction analysis. Anal. Chem. 1991, 63, 2338–2345. [Google Scholar] [CrossRef]
- Cytiva. Accurate Comparability Assessment of a Biosimilar Interferon in Process Development; Cytiva: Marlborough, MA, USA, 2020. [Google Scholar]
- Hu, T.; Wu, L.; Sun, X.; Su, P.; Yang, Y. Comparative study on quantitation of human myoglobin by both isotope dilution mass spectrometry and surface plasmon resonance based on calibration-free analysis. Anal. Bioanal. Chem. 2020, 412, 2777–2784. [Google Scholar] [CrossRef]
- Imamura, H.; Honda, S. Calibration-free concentration analysis for an analyte prone to self-association. Anal. Biochem. 2017, 516, 61–64. [Google Scholar] [CrossRef]
- Hossler, P.; Khattak, S.F.; Li, Z.J. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology 2009, 19, 936–949. [Google Scholar] [CrossRef] [Green Version]
- Canziani, G.A.; Klakamp, S.; Myszka, D.G. Kinetic screening of antibodies from crude hybridoma samples using Biacore. Anal. Biochem. 2004, 325, 301–307. [Google Scholar] [CrossRef]
- Wu, L.; He, Y.; Hu, Y.; Lu, H.; Cao, Z.; Yi, X.; Wang, J. Real-time surface plasmon resonance monitoring of site-specific phosphorylation of p53 protein and its interaction with MDM2 protein. Analyst 2019, 144, 6033–6040. [Google Scholar] [CrossRef] [PubMed]
- Cytiva. Biacore Systems in Discovery and Early-Stage Development of Biotherapeutics Antibodies; Cytiva: Marlborough, MA, USA, 2016. [Google Scholar]
- Yun, S.; Lee, S.; Park, J.P.; Choo, J.; Lee, E.K. Modification of phage display technique for improved screening of high-affinity binding peptides. J. Biotechnol. 2019, 289, 88–92. [Google Scholar] [CrossRef]
- Zhao, A.; Tohidkia, M.R.; Siegel, D.L.; Coukos, G.; Omidi, Y. Phage antibody display libraries: A powerful antibody discovery platform for immunotherapy. Crit. Rev. Biotechnol. 2016, 36, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Munke, A.; Persson, J.; Weiffert, T.; De Genst, E.; Meisl, G.; Arosio, P.; Carnerup, A.; Dobson, C.M.; Vendruscolo, M.; Knowles, T.P.J.; et al. Phage display and kinetic selection of antibodies that specifically inhibit amyloid self-replication. Proc. Natl. Acad. Sci. USA 2017, 114, 6444–6449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.; Lentino, J.; Kopp, J.; Murray, L.; Ellison, W.; Rhee, M.; Shockey, G.; Akella, L.; Erby, K.; Heyward, W.L.; et al. Immunogenicity of a two-dose investigational hepatitis B vaccine, HBsAg-1018, using a toll-like receptor 9 agonist adjuvant compared with a licensed hepatitis B vaccine in adults. Vaccine 2018, 36, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.G.; Pitts, K.E.; Steffek, M.; Mulvihill, M.M. Determination of Affinity and Residence Time of Potent Drug-Target Complexes by Label-free Biosensing. J. Med. Chem. 2018, 61, 5154–5161. [Google Scholar] [CrossRef] [PubMed]
- Frostell-Karlsson, A.; Remaeus, A.; Roos, H.; Andersson, K.; Borg, P.; Hämäläinen, M.; Karlsson, R. Biosensor analysis of the interaction between immobilized human serum albumin and drug compounds for prediction of human serum albumin binding levels. J. Med. Chem. 2000, 43, 1986–1992. [Google Scholar] [CrossRef]
- Gonzales, N.R.; Schuck, P.; Schlom, J.; Kashmiri, S.V. Surface plasmon resonance-based competition assay to assess the sera reactivity of variants of humanized antibodies. J. Immunol. Methods 2002, 268, 197–210. [Google Scholar] [CrossRef]
- Ritter, G.; Cohen, L.S.; Williams, C., Jr.; Richards, E.C.; Old, L.J.; Welt, S. Serological analysis of human anti-human antibody responses in colon cancer patients treated with repeated doses of humanized monoclonal antibody A33. Cancer Res. 2001, 61, 6851–6859. [Google Scholar]
- Wang, W.; Lu, P.; Fang, Y.; Hamuro, L.; Pittman, T.; Carr, B.; Hochman, J.; Prueksaritanont, T. Monoclonal antibodies with identical Fc sequences can bind to FcRn differentially with pharmacokinetic consequences. Drug Metab. Dispos. 2011, 39, 1469–1477. [Google Scholar] [CrossRef] [Green Version]
- Pyzik, M.; Sand, K.M.K.; Hubbard, J.J.; Andersen, J.T.; Sandlie, I.; Blumberg, R.S. The Neonatal Fc Receptor (FcRn): A Misnomer? Front. Immunol. 2019, 10, 1540. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Vernes, J.M.; Chiang, N.; Ou, Q.; Ding, J.; Adams, C.; Hong, K.; Truong, B.T.; Ng, D.; Shen, A.; et al. Identification of IgG(1) variants with increased affinity to FcγRIIIa and unaltered affinity to FcγRI and FcRn: Comparison of soluble receptor-based and cell-based binding assays. J. Immunol. Methods 2011, 365, 132–141. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Influenza Vaccine Response during the Start of a Pandemic: Report of a WHO Informal Consultation Held in Geneve, Switzerland, 29 June–1 July 2015; World Health Organization: Geneva, Switzerland, 2016; Volume 2016. [Google Scholar]
- Suenaga, E.; Mizuno, H.; Penmetcha, K.K. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens. Bioelectron. 2012, 32, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Abadian, P.N.; Yildirim, N.; Gu, A.Z.; Goluch, E.D. SPRi-based adenovirus detection using a surrogate antibody method. Biosens. Bioelectron. 2015, 74, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Roldão, A.; Mellado, M.C.; Castilho, L.R.; Carrondo, M.J.; Alves, P.M. Virus-like particles in vaccine development. Expert Rev. Vaccines 2010, 9, 1149–1176. [Google Scholar] [CrossRef] [PubMed]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Thompson, C.M.; Petiot, E.; Lennaertz, A.; Henry, O.; Kamen, A.A. Analytical technologies for influenza virus-like particle candidate vaccines: Challenges and emerging approaches. Virol. J. 2013, 10, 141. [Google Scholar] [CrossRef] [Green Version]
- Chang, J. Adenovirus Vectors: Excellent Tools for Vaccine Development. Immune Netw 2021, 21, e6. [Google Scholar] [CrossRef]
- Glick, G.D.; Toogood, P.L.; Wiley, D.C.; Skehel, J.J.; Knowles, J.R. Ligand recognition by influenza virus. The binding of bivalent sialosides. J. Biol. Chem. 1991, 266, 23660–23669. [Google Scholar] [CrossRef]
- Connor, R.J.; Kawaoka, Y.; Webster, R.G.; Paulson, J.C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 1994, 205, 17–23. [Google Scholar] [CrossRef]
- Ohlson, S.; Jungar, C.; Strandh, M.; Mandenius, C.-F. Continuous weak-affinity immunosensing. Trends Biotechnol. 2000, 18, 49–52. [Google Scholar] [CrossRef]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Bacteriophages: Biosensing tools for multi-drug resistant pathogens. Analyst 2014, 139, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Nanduri, V.; Bhunia, A.K.; Tu, S.I.; Paoli, G.C.; Brewster, J.D. SPR biosensor for the detection of L. monocytogenes using phage-displayed antibody. Biosens. Bioelectron. 2007, 23, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.H.; Ku, S. Current Technical Approaches for the Early Detection of Foodborne Pathogens: Challenges and Opportunities. Int. J. Mol. Sci. 2017, 18, 2078. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Gao, S.; Wu, J.; Hu, X. Recent Advances in Aptamer-Based Biosensors for Detection of Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 5229. [Google Scholar] [CrossRef]
- Puttharugsa, C.; Wangkam, T.; Huangkamhang, N.; Gajanandana, O.; Himananto, O.; Sutapun, B.; Amarit, R.; Somboonkaew, A.; Srikhirin, T. Development of surface plasmon resonance imaging for detection of Acidovorax avenae subsp. citrulli (Aac) using specific monoclonal antibody. Biosens. Bioelectron. 2011, 26, 2341–2346. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.D.; Ladd, J.; Homola, J.; Jiang, S. Surface Plasmon Resonance (SPR) Sensors for the Detection of Bacterial Pathogens. In Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Zourob, M., Elwary, S., Turner, A., Eds.; Springer: New York, NY, USA, 2008; pp. 83–108. [Google Scholar]
- Kastenhofer, J.; Rajamanickam, V.; Libiseller-Egger, J.; Spadiut, O. Monitoring and control of E. coli cell integrity. J. Biotechnol. 2021, 329, 1–12. [Google Scholar] [CrossRef]
- Hofer, A.; Kroll, P.; Barmettler, M.; Herwig, C. A Reliable Automated Sampling System for On-Line and Real-Time Monitoring of CHO Cultures. Processes 2020, 8, 637. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, G.; Hein, R.; Liu, N.; Luo, X.; Davis, J.J. Antifouling Strategies for Selective In Vitro and In Vivo Sensing. Chem. Rev. 2020, 120, 3852–3889. [Google Scholar] [CrossRef]
- Wang, D.; Loo, J.F.; Chen, J.; Yam, Y.; Chen, S.-C.; He, H.; Kong, S.K.; Ho, H.P. Recent Advances in Surface Plasmon Resonance Imaging Sensors. Sensors 2019, 19, 1266. [Google Scholar] [CrossRef] [Green Version]
- Bravman, T.; Bronner, V.; Nahshol, O.; Schreiber, G. The ProteOn XPR36™ Array System—High Throughput Kinetic Binding Analysis of Biomolecular Interactions. Cell. Mol. Bioeng. 2008, 1, 216. [Google Scholar] [CrossRef]
- Yang, D.; Singh, A.; Wu, H.; Kroe-Barrett, R. Determination of High-affinity Antibody-antigen Binding Kinetics Using Four Biosensor Platforms. J. Vis. Exp. 2017, 55659. [Google Scholar] [CrossRef] [Green Version]
- Kamat, V.; Rafique, A. Exploring sensitivity & throughput of a parallel flow SPRi biosensor for characterization of antibody-antigen interaction. Anal. Biochem. 2017, 525, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Carterra. Speeding Antibody Screening for Drug Development. Available online: https://www.nature.com/articles/d42473-020-00177-x (accessed on 16 September 2021).
- Zeni, L.; Perri, C.; Cennamo, N.; Arcadio, F.; D’Agostino, G.; Salmona, M.; Beeg, M.; Gobbi, M. A portable optical-fibre-based surface plasmon resonance biosensor for the detection of therapeutic antibodies in human serum. Sci. Rep. 2020, 10, 11154. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Chen, S.; Cheng, F.; Wang, H.; Peng, W. Surface Plasmon Resonance Biosensor Based on Smart Phone Platforms. Sci. Rep. 2015, 5, 12864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lertvachirapaiboon, C.; Baba, A.; Shinbo, K.; Kato, K. A smartphone-based surface plasmon resonance platform. Anal. Methods 2018, 10, 4732–4740. [Google Scholar] [CrossRef]
- Wong, C.L.; Chua, M.; Mittman, H.; Choo, L.X.; Lim, H.Q.; Olivo, M. A Phase-Intensity Surface Plasmon Resonance Biosensor for Avian Influenza A (H5N1) Detection. Sensors 2017, 17, 2363. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.H.; Ho, H.P.; Wu, S.Y.; Kong, S.K. Detecting Phase Shifts in Surface Plasmon Resonance: A Review. Adv. Opt. Technol. 2012, 2012, 471957. [Google Scholar] [CrossRef] [Green Version]
- Kuncova-Kallio, J.; Järvinen, A. Comparison of MP-SPR NaviTM Instruments to BiacoreTM in Protein Research; WP_601.809.rg; BioNavis: Tampere, Finland, 2018. [Google Scholar]
- Kari, O.K.; Rojalin, T.; Salmaso, S.; Barattin, M.; Jarva, H.; Meri, S.; Yliperttula, M.; Viitala, T.; Urtti, A. Multi-parametric surface plasmon resonance platform for studying liposome-serum interactions and protein corona formation. Drug Deliv. Transl. Res. 2017, 7, 228–240. [Google Scholar] [CrossRef]
- Viitala, T.; Granqvist, N.; Hallila, S.; Raviña, M.; Yliperttula, M. Elucidating the signal responses of multi-parametric surface plasmon resonance living cell sensing: A comparison between optical modeling and drug-MDCKII cell interaction measurements. PLoS ONE 2013, 8, e72192. [Google Scholar] [CrossRef]
- Suutari, T.; Silen, T.; Karaman, D.S.E.; Saari, H.; Desai, D.; Kerkelä, E.; Laitinen, S.; Hanzlikova, M.; Rosenholm, J.M.; Yliperttula, M.; et al. Real-Time Label-Free Monitoring of Nanoparticle Cell Uptake. Small 2016, 12, 6289–6300. [Google Scholar] [CrossRef] [PubMed]
- Koponen, A.; Kerkelä, E.; Rojalin, T.; Lázaro-Ibáñez, E.; Suutari, T.; Saari, H.O.; Siljander, P.; Yliperttula, M.; Laitinen, S.; Viitala, T. Label-free characterization and real-time monitoring of cell uptake of extracellular vesicles. Biosens. Bioelectron. 2020, 168, 112510. [Google Scholar] [CrossRef] [PubMed]
- Chabot, V.; Miron, Y.; Grandbois, M.; Charette, P.G. Long range surface plasmon resonance for increased sensitivity in living cell biosensing through greater probing depth. Sens. Actuators B Chem. 2012, 174, 94–101. [Google Scholar] [CrossRef]
- Vala, M.; Robelek, R.; Bocková, M.; Wegener, J.; Homola, J. Real-time label-free monitoring of the cellular response to osmotic stress using conventional and long-range surface plasmons. Biosens. Bioelectron. 2013, 40, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Vala, M.; Etheridge, S.; Roach, J.A.; Homola, J. Long-range surface plasmons for sensitive detection of bacterial analytes. Sens. Actuators B Chem. 2009, 139, 59–63. [Google Scholar] [CrossRef]
- Jing, J.-Y.; Wang, Q.; Zhao, W.-M.; Wang, B.-T. Long-range surface plasmon resonance and its sensing applications: A review. Opt. Lasers Eng. 2019, 112, 103–118. [Google Scholar] [CrossRef]
- Yanase, Y.; Yoshizaki, K.; Kimura, K.; Kawaguchi, T.; Hide, M.; Uno, S. Development of SPR Imaging-Impedance Sensor for Multi-Parametric Living Cell Analysis. Sensors 2019, 19, 2067. [Google Scholar] [CrossRef] [Green Version]
- Ogura, T.; Tanaka, Y.; Toyoda, H. Whole cell-based surface plasmon resonance measurement to assess binding of anti-TNF agents to transmembrane target. Anal. Biochem. 2016, 508, 73–77. [Google Scholar] [CrossRef]
- Abali, F.; Stevens, M.; Tibbe, A.G.J.; Terstappen, L.; van der Velde, P.N.; Schasfoort, R.B.M. Isolation of single cells for protein therapeutics using microwell selection and Surface Plasmon Resonance imaging. Anal. Biochem. 2017, 531, 45–47. [Google Scholar] [CrossRef]
- Schasfoort, R.B.M.; Abali, F.; Stojanovic, I.; Vidarsson, G.; Terstappen, L. Trends in SPR Cytometry: Advances in Label-Free Detection of Cell Parameters. Biosensors 2018, 8, 102. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, R.; Pol, E.; Frostell, Å. Comparison of surface plasmon resonance binding curves for characterization of protein interactions and analysis of screening data. Anal. Biochem. 2016, 502, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Tharmalingam, T.; Wu, C.H.; Callahan, S.; Goudar, T.G. A framework for real-time glycosylation monitoring (RT-GM) in mammalian cell culture. Biotechnol. Bioeng. 2015, 112, 1146–1154. [Google Scholar] [CrossRef]
- Mou, Z.-L.; Qi, X.-N.; Liu, R.-L.; Zhang, J.; Zhang, Z.-Q. Three-dimensional cell bioreactor coupled with high performance liquid chromatography–mass spectrometry for the affinity screening of bioactive components from herb medicine. J. Chromatogr. A 2012, 1243, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-Y.; Ebel, B.; Paris, C.; Chauchard, F.; Guedon, E.; Marc, A. Real-time monitoring of antibody glycosylation site occupancy by in situ Raman spectroscopy during bioreactor CHO cell cultures. Biotechnol. Prog. 2018, 34, 486–493. [Google Scholar] [CrossRef] [PubMed]

| Target Analyte | Ligand | Range (μg/mL) | SPR Instrument | Reference |
|---|---|---|---|---|
| mAb | Antibody | 0.8–50 | Biacore T200 | [66] |
| Albumin | Antibody | 2–200 | ||
| Transferrin | Antibody | 0.2–20 | ||
| IgA | Antibody | 0.4–50 | ||
| IgG | Antibody | 2–200 | ||
| IgG1 | Antibody | 2.8–90 | ||
| IgG2 | Antibody | 1.8–60 | ||
| IgG3 | Antibody | 0.4–13 | ||
| IgG4 | Antibody | 0.2–5 | ||
| Infliximab (IFX, IgG mAb) | Antigen (TNF-α) | 0.5–8 | Bio-Rad ProteOn | [69] |
| Anti-IFX antibody | Antibody (IFX) | 5–40 | ||
| Anti-GFP antibody (total) | Protein A/G | 0.03–2 | SPR 2/4 (Bruker Daltonics SPR) | [65] |
| Anti-GFP antibody (bioactive) | Antigen (GFP) | 0.03–2 | ||
| Anti-PSMA IgG mAb | Antigen (PSMA) | 1.35–30 | Biacore 3000 | [67] |
| Trastuzumab (IgG mAb) | FcγRI | 0.03–3.75 | Biacore T100 | [55] |
| Anti-TNF-α antibody | Antigen (TNF-α) | 0.02–360 | Biacore T200 | [37] |
| Target Analyte | Ligand | Range | LOQ | LOD | SPR Instrument | Reference |
|---|---|---|---|---|---|---|
| Strains of Influenza HA and Vaccine HA | Synthetic glycans with α-2,3 or α-2,6 sialic acid conformation | 0.33–30 μg/mL | Bio-Rad ProteOn | [40] | ||
| Strains of Influenza HA | Biantennary glycan with terminal α-2,6 sialic acid | 0.01–2 μg/mL | 0.01–0.02 μg/mL | Biacore T200 | [42] | |
| HA from viral serum and Vaccine HA | Recombinant HA (Inhibition assay with Anti-HA antibody) | 1–15 μg/mL | 1 μg/mL | 0.5 μg/mL | Biacore T100 | [39] |
| Adenovirus | Antibody against anti-adenovirus antibody (Inhibition assay with anti-adenovirus antibody) | 10–5000 PFU/mL | 10 PFU/mL | SPRI-Lab+ (Horiba Scientific, Edison, USA) | [118] | |
| Influenza HA | 6′-sialyllactose-conjugate with ovalbumin as carrier | 10–100 μg/mL | Biacore 2000 & Biacore X | [38] | ||
| Vaccine HA | Fetuin glycoprotein bearing both α-2,3 and α-2,6 linkages | 0.03–20 μg/mL | 0.1 μg/mL | 0.03 μg/mL | SPR-2 (Sierra Sensors) | [41] |
| Cell culture-derived whole influenza virus | ~105–107 PFU/mL | 5.3 × 105 PFU/mL | 1.8 × 105 PFU/mL |
| Target Analyte | Ligand | Assay Type | LOD (CFU/mL) | SPR Instrument | Reference |
|---|---|---|---|---|---|
| Campylobacter jejuni | Antibody | Direct | 8 × 106 | SPR-4 (Sierra Sensors) | [48] |
| Sandwich | 4 × 104 | ||||
| Campylobacter jejuni | Antibody | Subtractive inhibition | 131 | SPR-4 (Sierra Sensors) | [49] |
| Salmonella Typhimurium in romaine lettuce | Antibody | Direct | 1.9 × 106 | Reichert Dual Channel SR7500DC | [47] |
| Sandwich | 1.6 × 106 | ||||
| Sandwich with prior incubation | 4.7 × 105 | ||||
| Escherichia coli O157:H7 in ground beef and cucumber samples | Wheat germ agglutinin (lectin) | Direct | 3 × 103 | Biacore 3000 | [46] |
| Listeria monocytogenes in milk | Wheat germ agglutinin (lectin) | Direct | 104 | Biacore 3000 | [50] |
| Listeria monocytogenes | Phage-displayed single-chain antibody | Direct | 2 × 106 | Nomadics SPR3 (SPREETA3, Texas Instruments) | [127] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaudreault, J.; Forest-Nault, C.; De Crescenzo, G.; Durocher, Y.; Henry, O. On the Use of Surface Plasmon Resonance-Based Biosensors for Advanced Bioprocess Monitoring. Processes 2021, 9, 1996. https://doi.org/10.3390/pr9111996
Gaudreault J, Forest-Nault C, De Crescenzo G, Durocher Y, Henry O. On the Use of Surface Plasmon Resonance-Based Biosensors for Advanced Bioprocess Monitoring. Processes. 2021; 9(11):1996. https://doi.org/10.3390/pr9111996
Chicago/Turabian StyleGaudreault, Jimmy, Catherine Forest-Nault, Gregory De Crescenzo, Yves Durocher, and Olivier Henry. 2021. "On the Use of Surface Plasmon Resonance-Based Biosensors for Advanced Bioprocess Monitoring" Processes 9, no. 11: 1996. https://doi.org/10.3390/pr9111996
APA StyleGaudreault, J., Forest-Nault, C., De Crescenzo, G., Durocher, Y., & Henry, O. (2021). On the Use of Surface Plasmon Resonance-Based Biosensors for Advanced Bioprocess Monitoring. Processes, 9(11), 1996. https://doi.org/10.3390/pr9111996






