Biodegradation of a Complex Phenolic Industrial Stream by Bacterial Strains Isolated from Industrial Wastewaters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phenolic Industrial Stream
2.2. Massive Sequencing, Isolation, Growth, and Identification of Bacterial Strains
2.3. Bacterial Growth in Standard and PS-Supplemented Culture Media
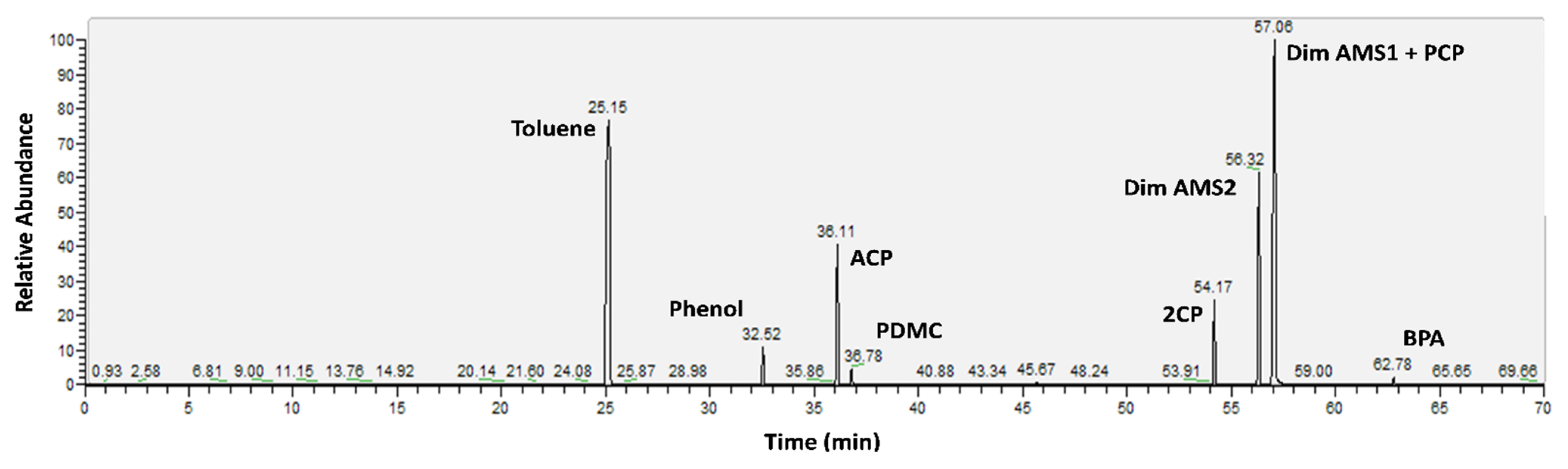
2.4. Extraction and Determination of Phenolic Compounds
2.5. Sample Preparation for Untargeted Metabolomic Analysis
2.6. Untargeted Metabolomics Analysis Using a Combined Analytical Multiplatform Based on GC-MS and UPLC-QTOF-MS
2.6.1. Metabolomic Analysis by GC-MS
2.6.2. Metabolomic Analysis by UPLC-QTOF-MS
2.7. Statistics
3. Results and Discussion
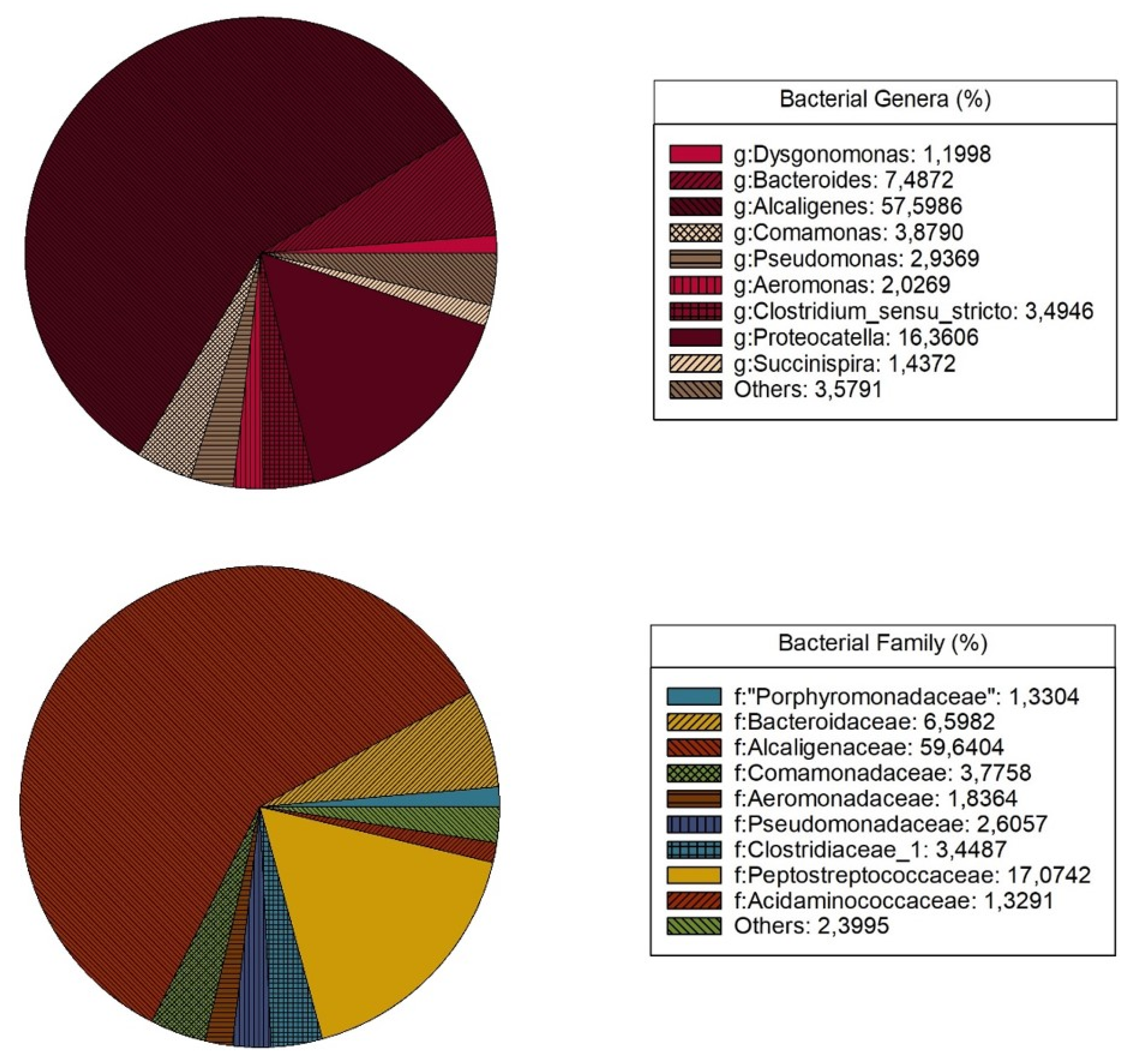
3.1. Identification of Microorganisms with Potential for Degrading Phenolic Compounds
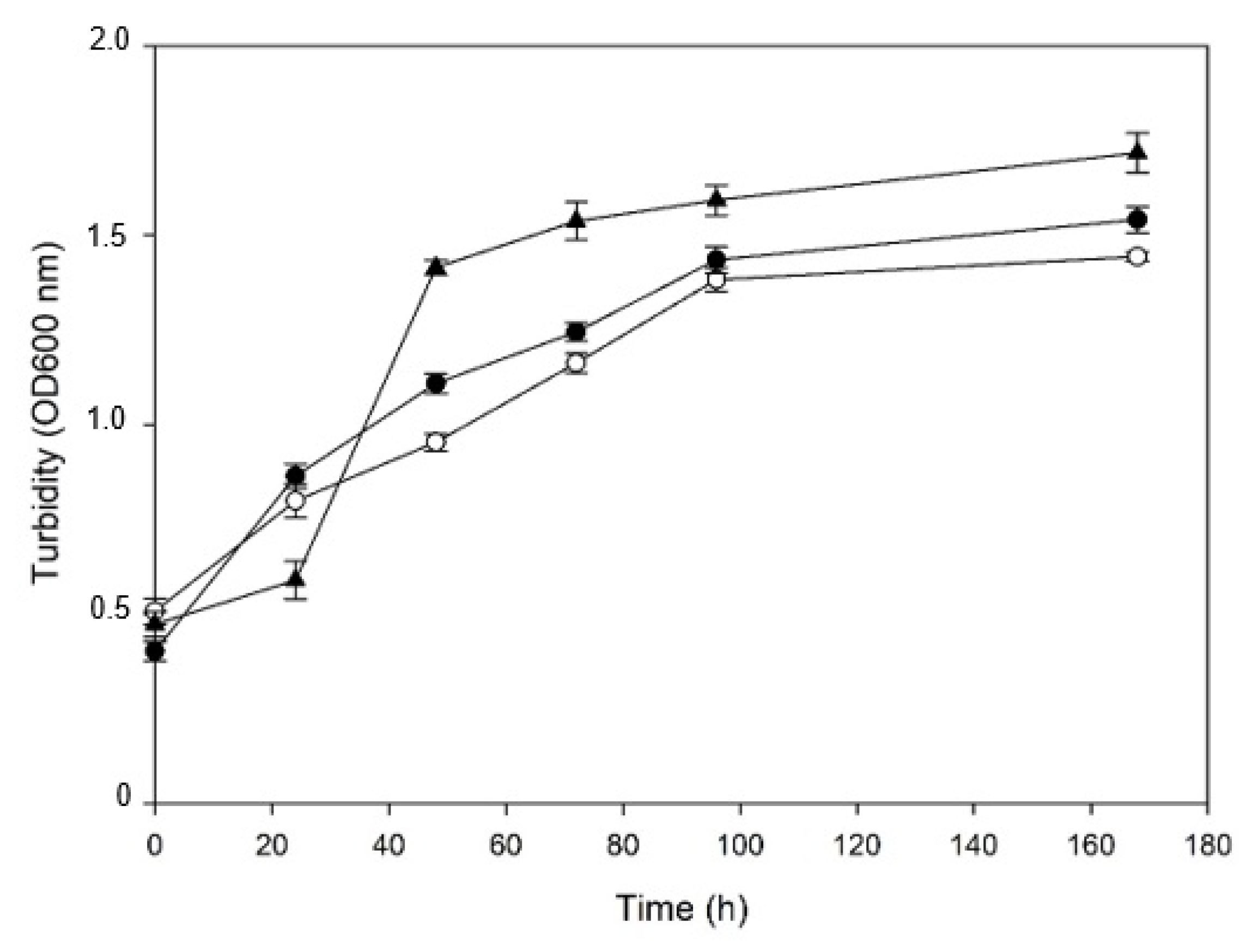
3.2. Construction of a Bacterial Consortium and Tolerance to the PS
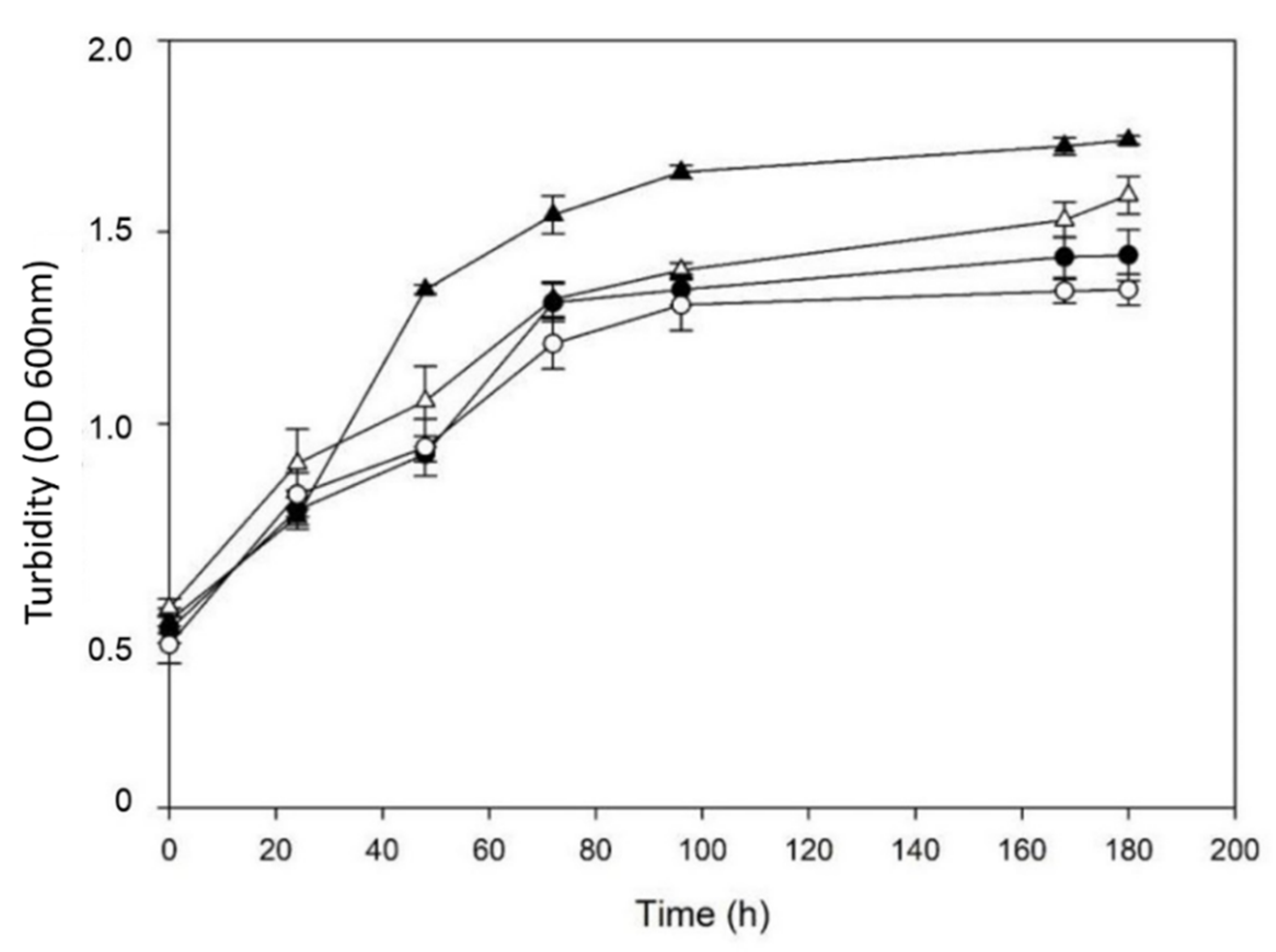
3.3. Towards Process Conditions: Assessment of a Close-to-Process Culture Medium
3.4. Performance of Semicontinuous Cultures of the Bacterial Consortium for Biodegradation of a Complex Mix of Phenolic Compounds (PS)
- (a)
- Molecules generated by degradation: This group comprises the first (methyl β-phenylglycidate) and the second (linalyl benzoate) molecules (Table 5). Methyl β-phenylglycidate and linalyl benzoate originated from biological and/or chemical degradation of phenolic compounds originally present in the industrial effluent. The chemical structure of these molecules suggests that they might originate from the mentioned processes. This hypothesis finds scientific support in the literature and is discussed below.
- (b)
- Molecules of biological synthesis: This group comprises several molecules, including geranylgeraniol, hexadecanoic acid, glycerol, and benzoic acid, among others. These originate from cell metabolism and are further released into the medium or partially extracted from the cells by any of the phenolic compounds present in the medium.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Busca, G.; Berardinelli, S.; Resini, C.; Arrighi, L. Technologies for the removal of phenol from fluid streams: A short review of recent developments. J. Hazard. Mater. 2008, 160, 265–288. [Google Scholar] [CrossRef]
- Pal, P.; Kumar, R. Treatment of coke wastewater: A critical review for developing sustainable management strategies. Separ. Purif. Rev. 2014, 43, 89–123. [Google Scholar] [CrossRef]
- Cetinkaya, A.Y.; Ozdemir, O.K. Phenol removal from synthetic solution using low pressure membranes coated with graphene oxide and carbon. Chem. Pap. 2018, 72, 327–335. [Google Scholar] [CrossRef]
- Michałowicz, J.; Duda, W. Phenols sources and toxicity. J. Environ. Stud. 2007, 16, 347–362. [Google Scholar]
- Dayana-Priyadharshini, S.; Bakthavatsalam, A.K. A comparative study on growth and degradation behavior of C. pyrenoidosa on synthetic phenol and phenolic wastewater of a coal gasification plant. J. Environ. Chem. Eng. 2019, 7, 103079. [Google Scholar] [CrossRef]
- Gao, M.; Diao, M.H.; Yuan, S.; Wang, Y.K.; Xu, H.; Wang, X.H. Effects of phenol on physicochemical properties and treatment performances of partial nitrifying granules in sequencing batch reactors. Biotechnol. Rep. 2017, 13, 13–18. [Google Scholar] [CrossRef]
- Pop, C.E.; Draga, S.; Măciucă, R.; Niță, R.; Crăciun, N.; Wolff, R. Bisphenol A Effects in Aqueous Environment on Lemna minor. Processes 2021, 9, 1512. [Google Scholar] [CrossRef]
- Rucká, L.; Nešvera, J.; Pátek, M. Biodegradation of phenol and its derivatives by engineered bacteria: Current knowledge and perspectives. World J. Microbiol. Biotechnol. 2017, 33, 174. [Google Scholar] [CrossRef] [PubMed]
- Tomei, M.C.; Mosca-Angelucci, D.; Clagnan, E.; Brusettil, L. Anaerobic biodegradation of phenol in wastewater treatment: Achievements and limits. Appl. Microbiol. Biotechnol. 2021, 105, 2195–2224. [Google Scholar] [CrossRef]
- Kaczorek, E.; Smułek, W. Special Issue “Study of Biodegradation and Bioremediation”. Processes 2021, 9, 1130. [Google Scholar] [CrossRef]
- Barman, S.R.; Banerjee, P.; Mukhopadhayay, A.; Das, P. Biodegradation of acenapthene and naphthalene by Pseudomonas mendocina: Process optimization, and toxicity evaluation. J. Environ. Chem. Eng. 2017, 5, 4803–4812. [Google Scholar] [CrossRef]
- Barik, M.; Das, C.P.; Verma, A.K.; Sahoo, S.; Sahoo, N.K. Metabolic profiling of phenol biodegradation by an indigenous Rhodococcus pyridinivorans strain PDB9T N-1 isolated from paper pulp wastewater. Int. Biodeter. Biodegrad. 2021, 158, 105168. [Google Scholar] [CrossRef]
- Kong, L.; Gao, Y.; Zhou, Q.; Zhao, X.; Sun, Z. Biochar accelerates PAHs biodegradation in petroleum-polluted soil by biostimulation strategy. J. Hazard. Mater. 2018, 343, 276–284. [Google Scholar] [CrossRef]
- Liang, X.; Guo, C.; Liao, C.; Liu, S.; Wick, L.Y.; Peng, D.; Yi, X.; Lu, G.; Yin, H.; Lin, Z.; et al. Drivers and applications of integrated clean-up technologies for surfactant-enhanced remediation of environments contaminated with polycyclic aromatic hydrocarbons (PAHs). Environ. Pollut. 2017, 225, 129–140. [Google Scholar] [CrossRef]
- Nybo, S.E.; Lamberts, J.T. Integrated use of LC/MS/MS and LC/Q-TOF/MS targeted metabolomics with automated label-free microscopy for quantification of purine metabolites in cultured mammalian cells. Purinergic Signal. 2019, 15, 17–25. [Google Scholar] [CrossRef]
- Begley, P.; Francis-McIntyre, S.; Dunn, W.B.; Broadhurst, D.I.; Halsall, A.; Tseng, A.; Knowles, J.; Consortium, H.; Goodacre, R.; Kell, D.B. Development and Performance of a Gas Chromatography−Time-of-Flight Mass Spectrometry Analysis for Large-Scale Nontargeted Metabolomic Studies of Human Serum. Anal. Chem. 2009, 81, 7038–7046. [Google Scholar] [CrossRef]
- Jagiello, Z.; López-García, A.; Aguirre, J.I.; Dylewski, L.Z. Distance to landfill and human activities affects the debris incorporation into the white stork nests in urbanized landscape in central Spain. Environ. Sci. Pollut. Res. 2020, 27, 30893–30898. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, A.; Rupérez, F.J.; Mendes, T.O.; Pérez-Rial, S.; Girón-Martínez, A.; Terrón-Expósito, R.; Díaz-Gil, J.J.; Peces-Barba, G.; Barbas, C.; García, A. A metabolomic approach shows sphingosine 1-phosphate and lysophospholipids as mediators of the therapeutic effect of liver growth factor in emphysema. J. Pharm. Biomed. Anal. 2017, 139, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Annadurai, G.; Juang, R.S.; Lee, D.J. Microbiological degradation of phenol using mixed liquors of Pseudomonas putida and activated sludge. Waste Manag. 2002, 22, 703–710. [Google Scholar] [CrossRef]
- Lin, Y.H.; Gu, Y.J. Biodegradation Kinetic Studies of Phenol and p-Cresol in a Batch and Continuous Stirred-Tank Bioreactor with Pseudomonas putida ATCC 17484 Cells. Processes 2021, 9, 133. [Google Scholar] [CrossRef]
- Jiang, Y.; Wen, J.; Bai, J.; Jia, X.; Hu, Z. Biodegradation of phenol at high initial concentration by Alcaligenes faecalis. J. Hazard. Mater. 2007, 147, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Nair, C.I.; Jayachandran, K.; Shashidhar, S. Biodegradation of phenol. Afr. J. Biotechnol. 2008, 7, 4951–4958. [Google Scholar] [CrossRef]
- Sun, W.; Sun, X.; Cupples, A.M. Identification of Desulfosporosinus as toluene-assimilating microorganisms from a methanogenic consortium. Int. Biodeterior. Biodegradation 2014, 88, 13–19. [Google Scholar] [CrossRef]
- Hermon, L.; Denonfoux, J.; Hellal, J.; Joulian, C.; Ferreira, S.; Vuilleumier, S.; Imfeld, G. Dichloromethane biodegradation in multi-contaminated groundwater: Insights from biomolecular and compound-specific isotope analyses. Water Res. 2018, 142, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Chaillan, F. Identification and biodegradation potential of tropical aerobic hydrocarbon-degrading microorganisms. Res. Microbiol. 2004, 155, 587–595. [Google Scholar] [CrossRef]
- Haddadi, A.; Shavandi, M. Biodegradation of phenol in hypersaline conditions by Halomonas sp. strain PH2-2 isolated from saline soil. Int. Biodeterior. Biodegradation 2013, 85, 29–34. [Google Scholar] [CrossRef]
- Tena-Garitaonaindia, M.; Llamas, I.; Toral, L.; Sampedro, I. Chemotaxis of halophilic bacterium Halomonas anticariensis FP35 towards the environmental pollutants phenol and naphthalene. Sci. Total Environ. 2019, 669, 631–636. [Google Scholar] [CrossRef]
- Kaczorek, E.; Urbanowicz, M.; Olszanowski, A. The influence of surfactants on cell surface properties of Aeromonas hydrophila during diesel oil biodegradation. Colloid Surf. B 2010, 81, 363–368. [Google Scholar] [CrossRef]
- Kumar, M.; Jaiswal, S.; Sodhi, K.K.; Shree, P.; Singh, D.K.; Agrawal, P.K.; Shukla, P. Antibiotics bioremediation: Perspectives on its ecotoxicity and resistance. Environ. Int. 2019, 124, 448–461. [Google Scholar] [CrossRef]
- Field, J.A.; Sierra-Alvarez, R. Microbial degradation of chlorinated benzenes. Biodegradation 2008, 19, 463–480. [Google Scholar] [CrossRef]
- Peng, Y.H.; Shih, Y.H. Microbial degradation of some halogenated compounds: Biochemical and molecular features. In Biodegradation of Hazardous and Special Products; Chamy, R., Ed.; InTechOpen: Valparaiso, Chile, 2013; pp. 51–69. [Google Scholar]
- Bera, S.; Roy, S.A.; Mohanty, K. Biodegradation of phenol by a native mixed bacterial culture isolated from crude oil contaminated site. Int. Biodeterior. Biodegrad. 2017, 121, 107–113. [Google Scholar] [CrossRef]
- Szőköl, J.; Rucká, L.; Šimčíková, M.; Halada, P.; Nešvera, J.; Pátek, M. Induction and carbon catabolite repression of phenol degradation genes in Rhodococcus erythropolis and Rhodococcus jostii. Appl. Microbiol. Biotechnol. 2014, 98, 8267–8279. [Google Scholar] [CrossRef] [PubMed]
- Basha, K.; Rajendran, A.; Thangavelu, V. Recent advances in the biodegradation of phenol: A review. Asian J. Exp. Biol. Sci. 2010, 1, 219–234. [Google Scholar]
- Ren, Y.; Peng, L.; Zhao, G.; Wei, C. Degradation of m-cresol via the ortho cleavage pathway by Citrobacter farmeri SC01. Biochem. Eng. J. 2014, 88, 108–114. [Google Scholar] [CrossRef]
- Hage, A.; Schoemaker, H.E.; Wever, R.; Zennaro, E.; Heipieper, H.J. Determination of the toxicity of several aromatic carbonylic compounds and their reduced derivatives on Phanerochaete chrysosporium using a Pseudomonas putida test system. Biotechnol. Bioeng. 2001, 73, 69–73. [Google Scholar] [CrossRef]
- Yang, Z.H.; Zeng, R.; Chang, X.; Li, X.K.; Wang, G.H. Toxicity of aromatic ketone to yeast cell and improvement of the asymmetric reduction of aromatic ketone catalyzed by yeast cell with the introduction of resin adsorption. Food Technol. Biotechnol. 2008, 46, 322–327. [Google Scholar]
- Muhr, E.; Leicht, O.; González-Sierra, S.; Thanbichler, M.; Heider, J.A. Fluorescent Bioreporter for Acetophenone and 1-Phenylethanol derived from a Specifically Induced Catabolic Operon. Front. Microbiol. 2016, 28, 1561. [Google Scholar] [CrossRef] [Green Version]
- Havel, J.; Reineke, W. Microbial degradation of chlorinated acetophenones. Appl. Environ. Microbiol. 1993, 59, 2706–2712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamerbeek, N.M.; Olsthoorn, A.J.J.; Fraaije, M.W.; Janssen, D.B. Substrate specificity and enantioselectivity of 4- hydroxyacetophenone monooxygenase. Appl. Environ. Microbiol. 2003, 69, 419–426. [Google Scholar] [CrossRef] [Green Version]
- Janicki, T.; Krupiński, M.; Długoński, J. Degradation and toxicity reduction of the endocrine disruptors nonylphenol, 4-tert-octylphenol and 4-cumylphenol by the non-ligninolytic fungus Umbelopsis isabellina. Bioresour. Technol. 2016, 200, 223–229. [Google Scholar] [CrossRef]
- Wubbolts, M.G.; Hoven, J.; Meigert, B.; Witholt, B. Efficient production of optically active styrene epoxides in two-liquid phase cultures. Enzyme Microb. Technol. 1994, 16, 887–894. [Google Scholar] [CrossRef]
- Panke, S.; Meyer, A.; Huber, C.M.; Witholt, B.; Wubbolts, M.G. An alkane-responsive expression system for the production of fine chemicals. Appl. Environ. Microbiol. 1999, 65, 2324–2332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jussupova, D.B.; Golovleva, L.A.; Zhubanova, A.A.; Akimbekov, N. Pathways of α-methylstyrene Oxidation by P. aeruginosa DS-26. Eurasian Chem. Technol. J. 2013, 15, 337–344. [Google Scholar] [CrossRef]
- Chan, S.I.; Yu, S.S. Controlled oxidation of hydrocarbons by the membrane-bound methane monooxygenase: The case for a tricopper cluster. Acc. Chem. Res. 2008, 41, 969–979. [Google Scholar] [CrossRef]
- Moreno, R.; Rojo, F. Enzymes for Aerobic Degradation of Alkanes in Bacteria. In Aerobic Utilization of Hydrocarbons, Oils and Lipids. Handbook of Hydrocarbon and Lipid Microbiology; Rojo, F., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–25. [Google Scholar] [CrossRef]
- Archelas, A.; Furstoss, R. Synthesis of enantiopure epoxides through biocatalytic approaches. Annu. Rev. Microbiol. 1997, 51, 491–525. [Google Scholar] [CrossRef]
- Cropley, R.L.; Williams, F.J.; Urquhart, A.J.; Vaughan, O.P.H.; Tikhov, M.S.; Lambert, R.M. Efficient Epoxidation of a Terminal Alkene Containing Allylic Hydrogen Atoms: Trans-Methylstyrene on Cu{111}. J. Am. Chem. Soc. 2005, 127, 6069–6076. [Google Scholar] [CrossRef]
- Garin, D.L.; Gamber, M.; Rowe, B.J. Epoxidation of Alpha-Methylstyrene and its Lewis Acid Rearrangement to 2-Phenylpropanal. J. Chem. Educ. 1996, 73, 555. [Google Scholar] [CrossRef]
- León, R.; Vila, M.; Hernánz, D.; Vílchez, C. Production of phytoene by herbicide-treated microalgae Dunaliella bardawil in two-phase systems. Biotechnol. Bioeng. 2005, 92, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Ram, S.; Mitra, M.; Shah, F.; Tirkey, S.R.; Mishra, S. Bacteria as an alternate biofactory for carotenoid production: A review of its applications, opportunities and challenges. J. Funct. Foods 2020, 67, 103867. [Google Scholar] [CrossRef]
- Fukaya, Y.; Takemura, M.; Koyanagi, T.; Maoka, T.; Shindo, K.; Misawa, N. Structural and functional analysis of the carotenoid biosynthesis genes of a Pseudomonas strain isolated from the excrement of Autumn Darter. Biosci. Biotechnol. Biochem. 2018, 82, 1043–1052. [Google Scholar] [CrossRef] [Green Version]


| Phenol | ACP | PMDC | 2CP | DimAMS1 | PCP + Heavies * | |
|---|---|---|---|---|---|---|
| Structure | 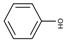 | 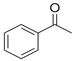 | 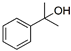 | 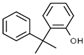 |  | 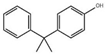 |
| Concentration (%, w/v) | 5.5 | 18 | - | 5.8 | 20.7 | 50 |
| Solubility in water (g/L, 20 °C) | 83 | 5.5 | Insoluble | <0.01 | 0.12 | 0.017–0.019 |
| Microorganism Genus | Biodegradation Capability | Reference |
|---|---|---|
| Desulfosporosinus | Toluene degradation | [23] |
| Hyphomicrobium | Dicloromethane and dimethylsulfoxide degradation | [24] |
| Flavobacterium | Phenol degradation | [25] |
| Halomonas | Phenol degradation Nafthalene degradation | [26,27] [27] |
| Aeromonas | Potential for bioremediation of industrial effluents | [28] |
| Pseudomonas | Aromatic compounds degradation | [11] |
| Alcaligenes | Phenol and phenol derivates degradation | [29] |
| Comamonas | Tetrabromobisphenol degradation 4-chlorophenol degradation | [30] [31] |
| Morphological Features | Bacterial Species | Culture Medium |
|---|---|---|
| White colonies with halo around | Pseudomonas putida | TSB/LB |
| Colonies with no uniform edge | Aeromonas hydrophila | LB |
| Yellowish colonies of circular shape | Alcaligenes faecalis | LB |
| White colonies of circular shape | Aeromonas salmonicida | TSB/LB |
| Mushroom-shape white colonies | Pseudomonas parafulva | TSB |
| Orange colonies of circular shape | Exiguobacterium aurantiacum | LB |
| PS | 710 ppm | 1070 ppm | 1420 ppm | 1780 ppm |
|---|---|---|---|---|
| DGR (%) | DGR (%) | DGR (%) | DGR (%) | |
| Phenol | 100 | 98.2 | 98.1 | 97.7 |
| ACP | 99.8 | 80.8 | 95.9 | 95.3 |
| PMDC | - | 9.6 | - | 23.2 |
| 2CP | 45.2 | 79.5 | 73 | 60.8 |
| DiAMS1 | 91.5 | 91.5 | 90.8 | 92.8 |
| PCP | - | 57.8 | 47.4 | 36.4 |
| Metabolite | Retention Time (min) | Formula | Molecular Ion (m/z) | Genesis |
|---|---|---|---|---|
| 2-Methoxynaphthalene | 5.168 | C11H10O | 159.08 | Chemical synthesis |
| Methyl β-phenylglycidate | - | C12H14O3 | - | Possibly from DimAMS degradation |
| Linalyl benzoate | 6.341 | C17H22O2 | 259.16 | Possibly from DimAMS degradation |
| 10-Eicosane | 25.639 | C20H42 | 298.34 | Chemical synthesis |
| Stearamide | 32.084 | C22H45NO2 | 284.29 | Chemical synthesis |
| Cinitapride | 26.818 | C21H30N4O4 | 403.23 | Chemical synthesis |
| Doxazosin | 30.077 | C23H25N5O5 | 469.21 | Chemical synthesis |
| Geranylgeraniol | - | C20H34O | - | Possibly from isoprenoid-derivatives degradation |
| Rifamycin | 33.439 | C39H49NO14 | 720.30 | Water contaminant |
| Acetamide | 4.32 | C2H5NO | 59.07 | Chemical origin |
| Ganoderenic acid A | 29.626 | C30H44O7 | 537.28 | Biological origin |
| Hexadecanoic acid | 10.78 | C16H32O2 | 256.42 | Fatty acid in membrane lipids |
| Urea | 4.65 | CH4N2O | 60.06 | Possible bacterial origin |
| Glycerol | 92.09 | C3H8O3 | 4.83 | TAG hydrolysis, biological origin |
| Benzoic | 7.18 | C7H6O2 | 122.12 | β-oxidation of fatty acids in bacterial cells |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartolomé, A.; Rodríguez-Moro, G.; Fuentes, J.-L.; Lopes, M.; Frontela, J.; Lázaro, J.; Cuaresma, M.; Gómez-Ariza, J.-L.; García-Barrera, T.; Vílchez, C. Biodegradation of a Complex Phenolic Industrial Stream by Bacterial Strains Isolated from Industrial Wastewaters. Processes 2021, 9, 1964. https://doi.org/10.3390/pr9111964
Bartolomé A, Rodríguez-Moro G, Fuentes J-L, Lopes M, Frontela J, Lázaro J, Cuaresma M, Gómez-Ariza J-L, García-Barrera T, Vílchez C. Biodegradation of a Complex Phenolic Industrial Stream by Bacterial Strains Isolated from Industrial Wastewaters. Processes. 2021; 9(11):1964. https://doi.org/10.3390/pr9111964
Chicago/Turabian StyleBartolomé, Alejandra, Gema Rodríguez-Moro, Juan-Luis Fuentes, Mariana Lopes, Juana Frontela, Jesús Lázaro, María Cuaresma, José-Luis Gómez-Ariza, Tamara García-Barrera, and Carlos Vílchez. 2021. "Biodegradation of a Complex Phenolic Industrial Stream by Bacterial Strains Isolated from Industrial Wastewaters" Processes 9, no. 11: 1964. https://doi.org/10.3390/pr9111964
APA StyleBartolomé, A., Rodríguez-Moro, G., Fuentes, J.-L., Lopes, M., Frontela, J., Lázaro, J., Cuaresma, M., Gómez-Ariza, J.-L., García-Barrera, T., & Vílchez, C. (2021). Biodegradation of a Complex Phenolic Industrial Stream by Bacterial Strains Isolated from Industrial Wastewaters. Processes, 9(11), 1964. https://doi.org/10.3390/pr9111964









