Advanced 3D Cell Culture Techniques in Micro-Bioreactors, Part II: Systems and Applications
Abstract
1. Introduction
- “3D cell culture” AND “microbioreactor”
- (bioreactor OR microbioreactor OR micro-bioreactor) AND (“three-dimensional cell culture” OR “3D cell culture” OR “3-D cell culture”)
- (microbioreactor OR micro-bioreactor) AND “tissue engineering”
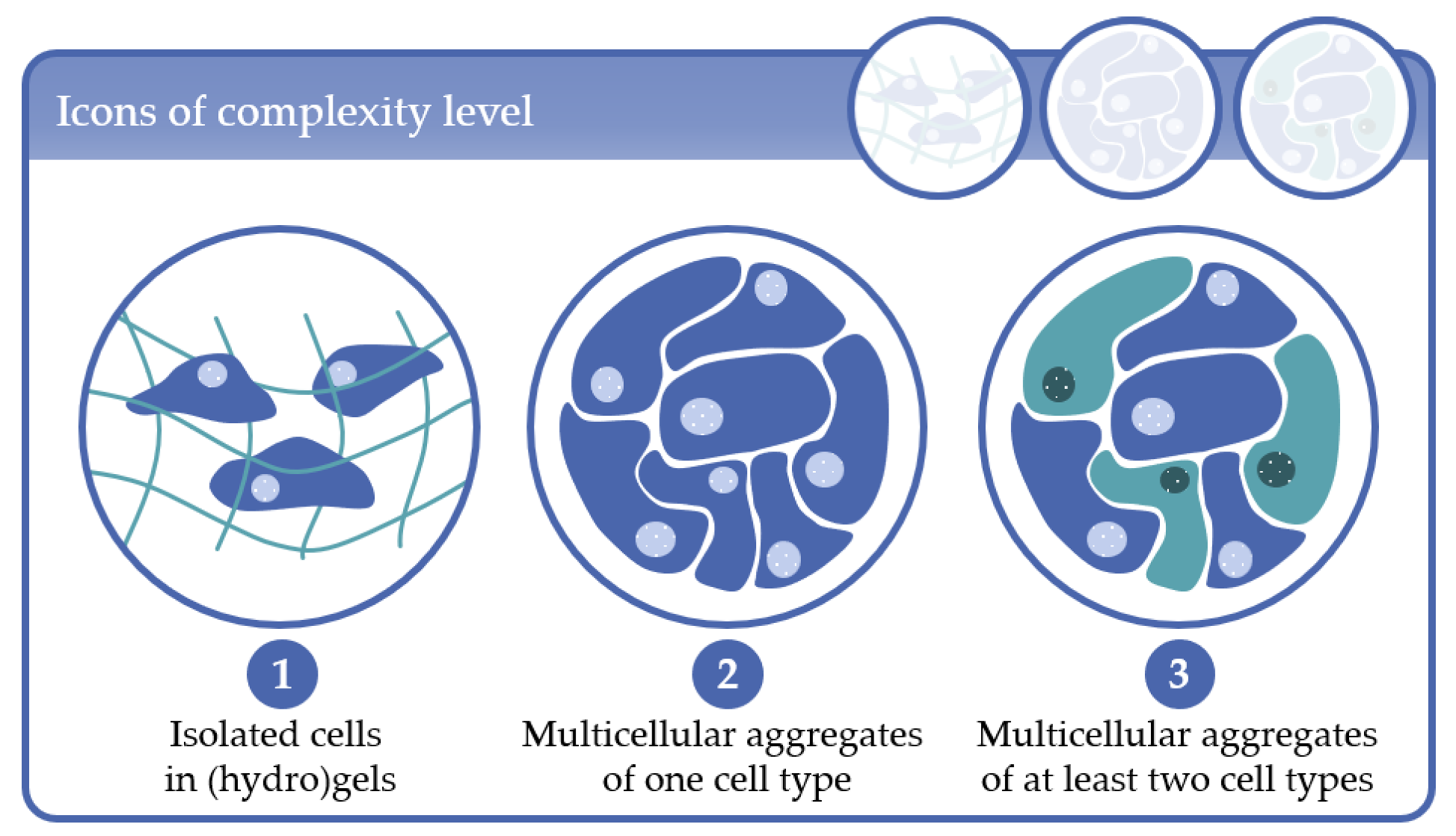
- Complexity level 1: cells immobilized in (hydro)gels as mono-culture (one cell type) or co-culture (at least two cell types);
- Complexity level 2: multicellular aggregates consisting of one cell type in 3D scaffolds or in scaffold-free cultures;
- Complexity level 3: multicellular aggregates consisting of at least two cell types in 3D scaffold-based or in scaffold-free cultures.
2. Complexity Level 1: Isolated Cells in (Hydro)Gels
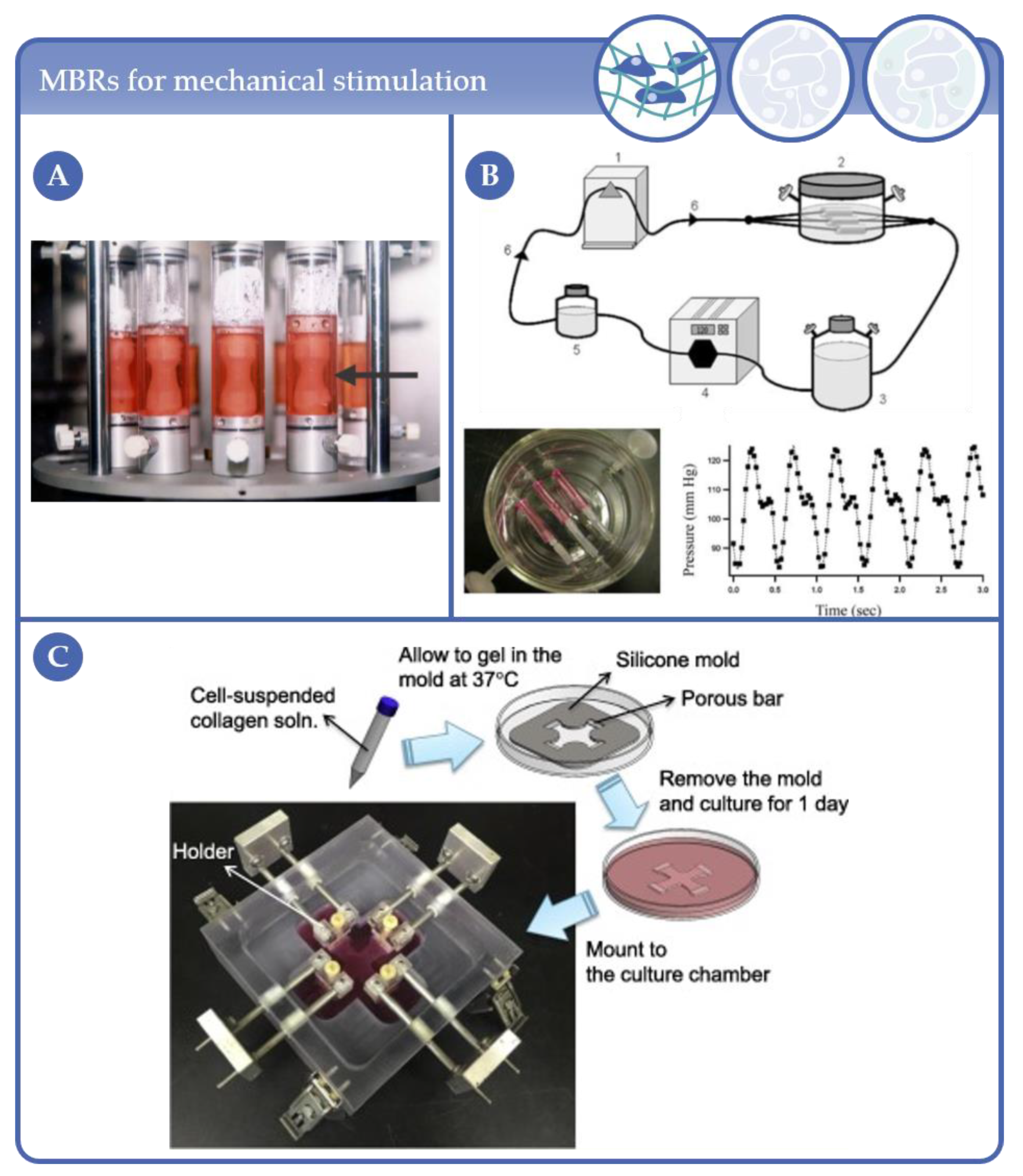
2.1. Early MBR Designs in Complexity Level 1
2.2. Latest MBR Developments in Complexity Level 1
3. Complexity Level 2: Multicellular Aggregates of One Cell Type
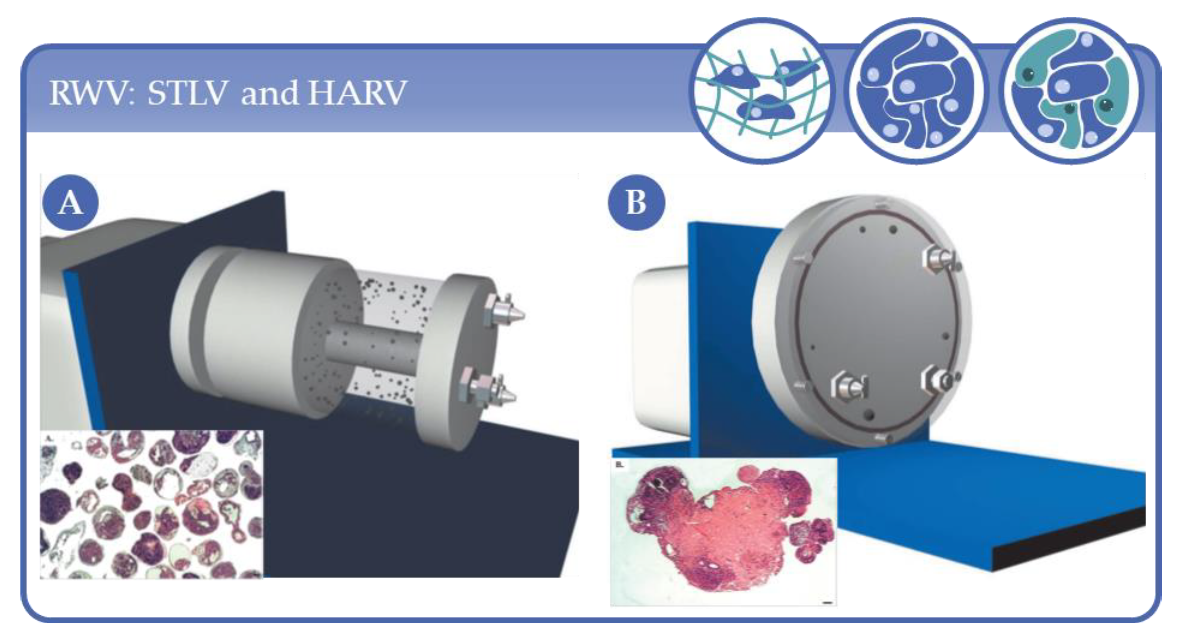
3.1. Applications Based on Rotary Wall Vessel Systems
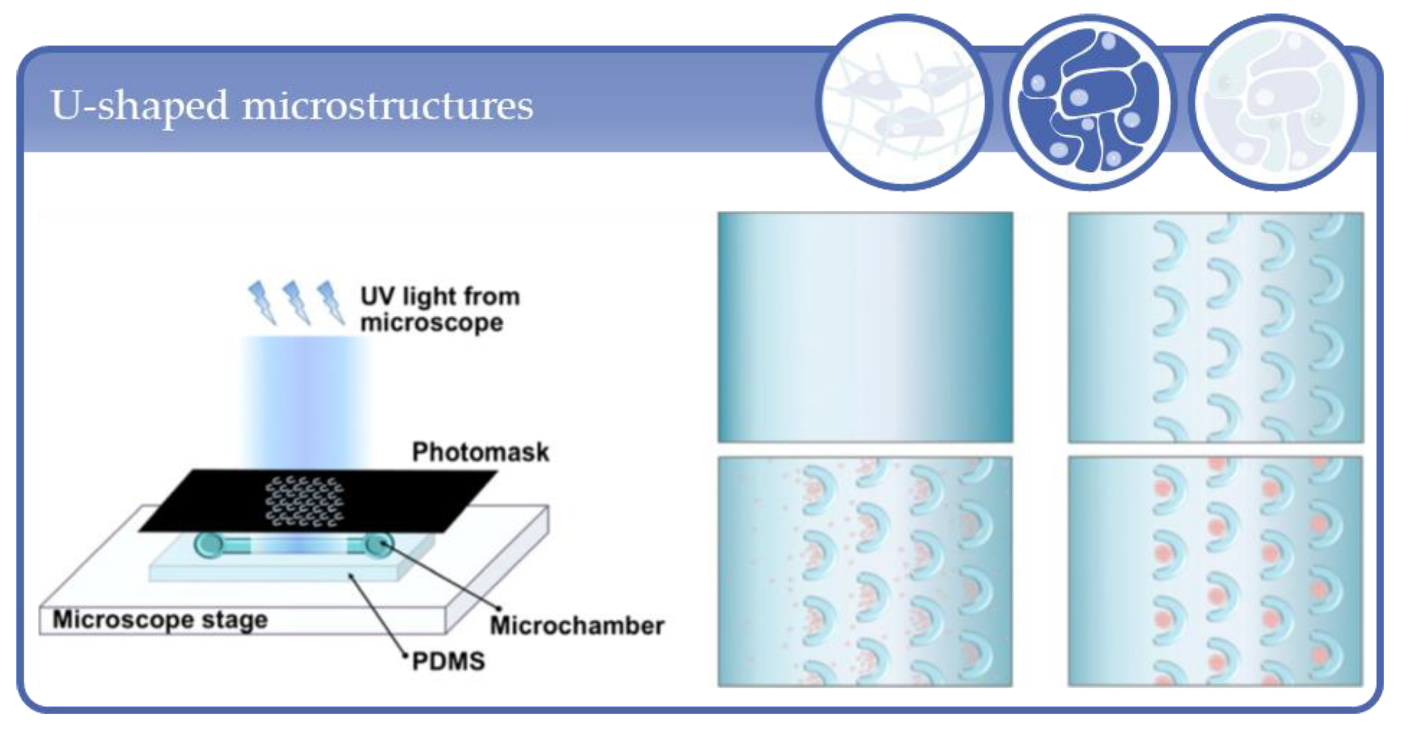
3.2. Applications Based on Microcavity/Microwell Arrangements
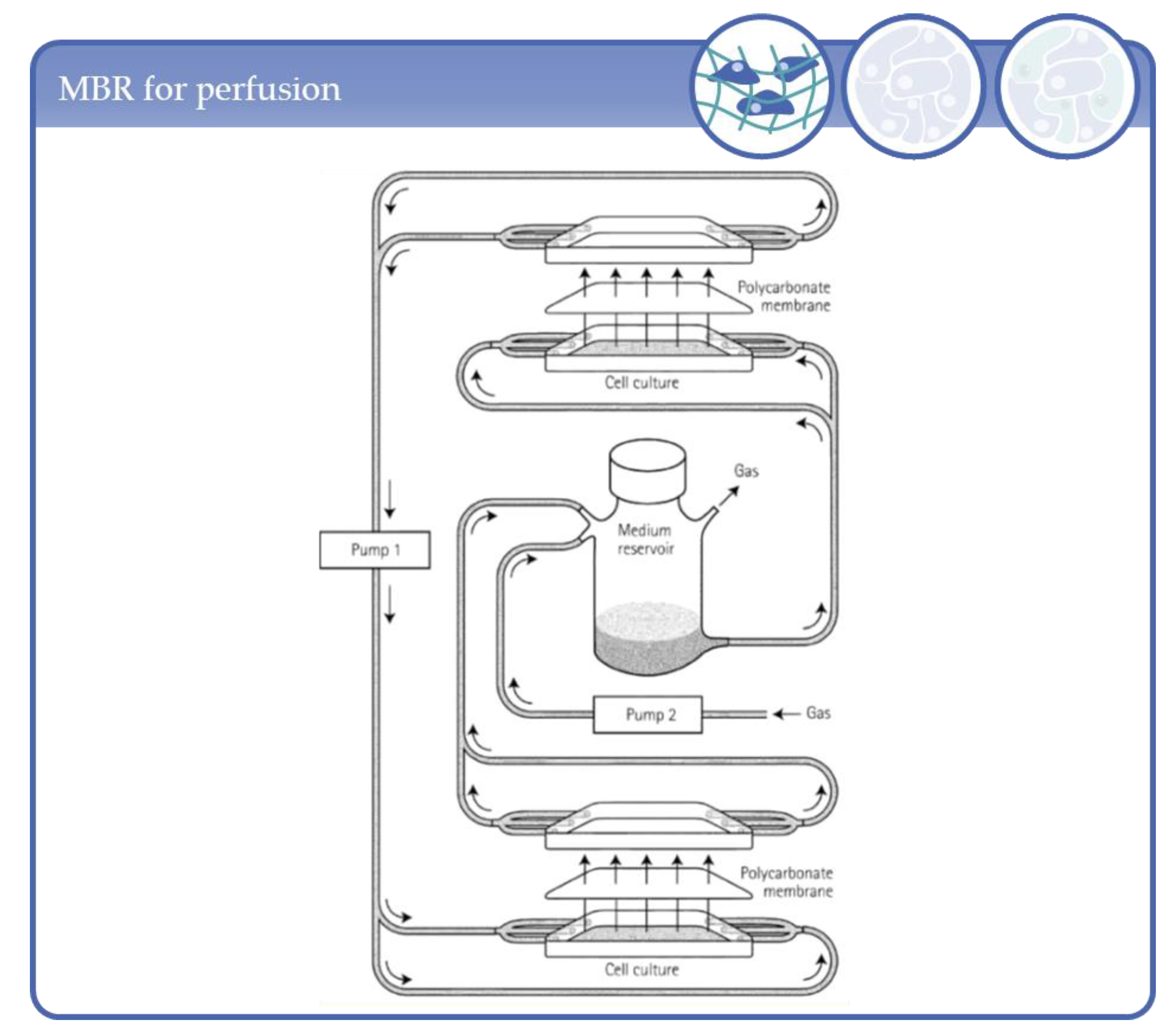
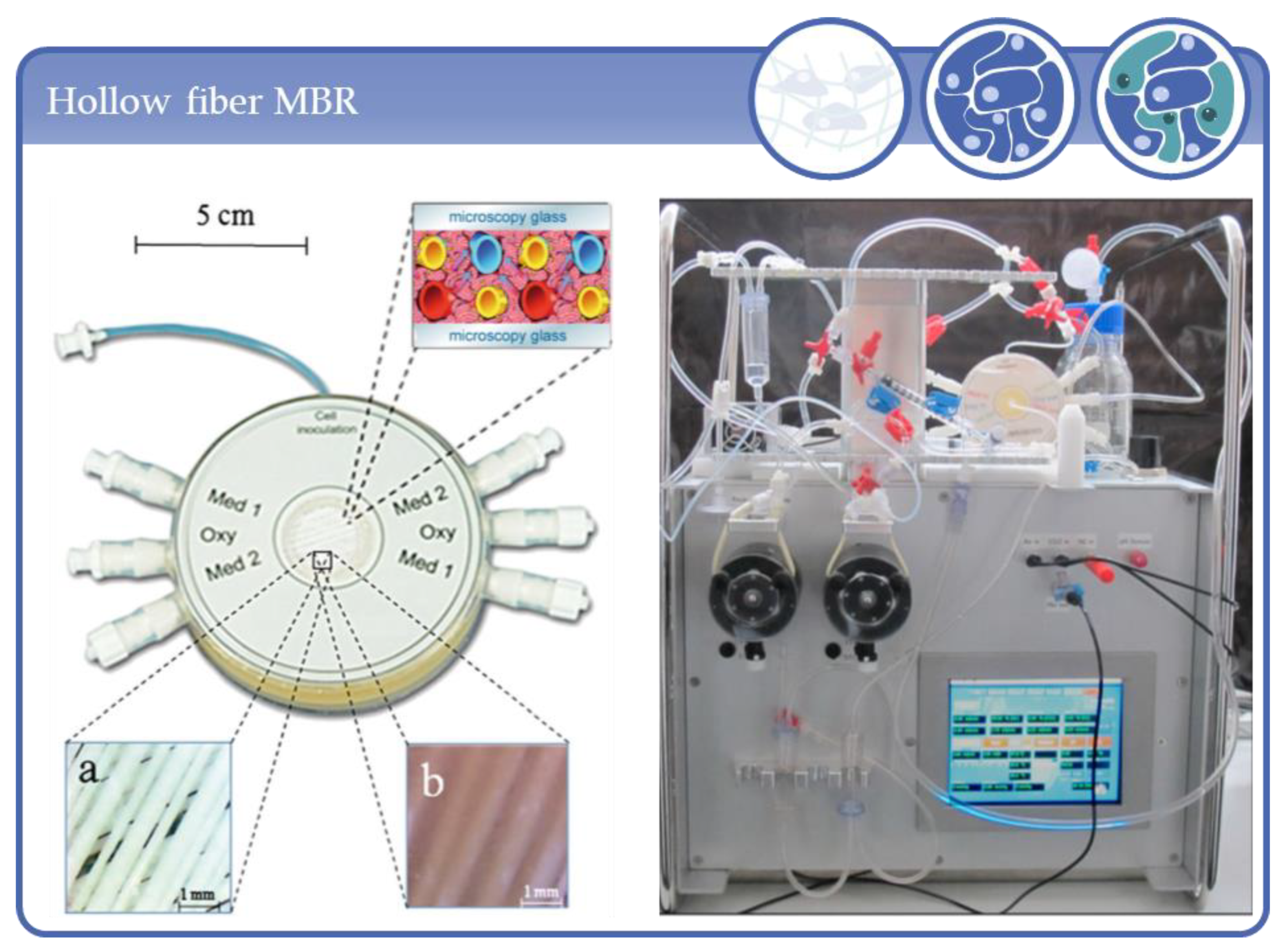
3.3. Applications Based on Hollow Fiber Systems
3.4. Stirred Micro-Bioreactors
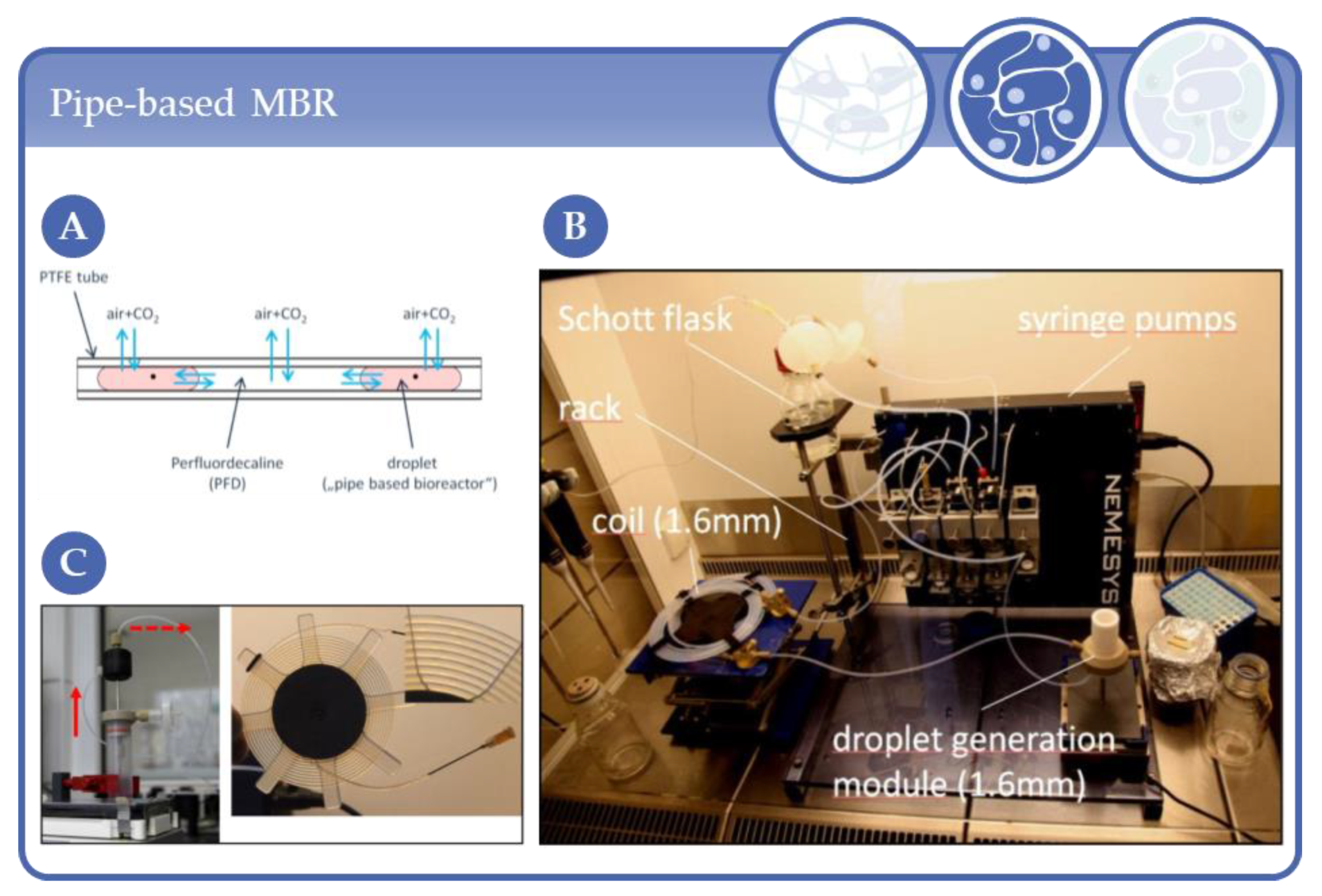
3.5. Perfusion of Scaffolds in Tube-Like Systems
3.6. Applications in Microscope Slide/Cover Slip Format Micro-Bioreactors
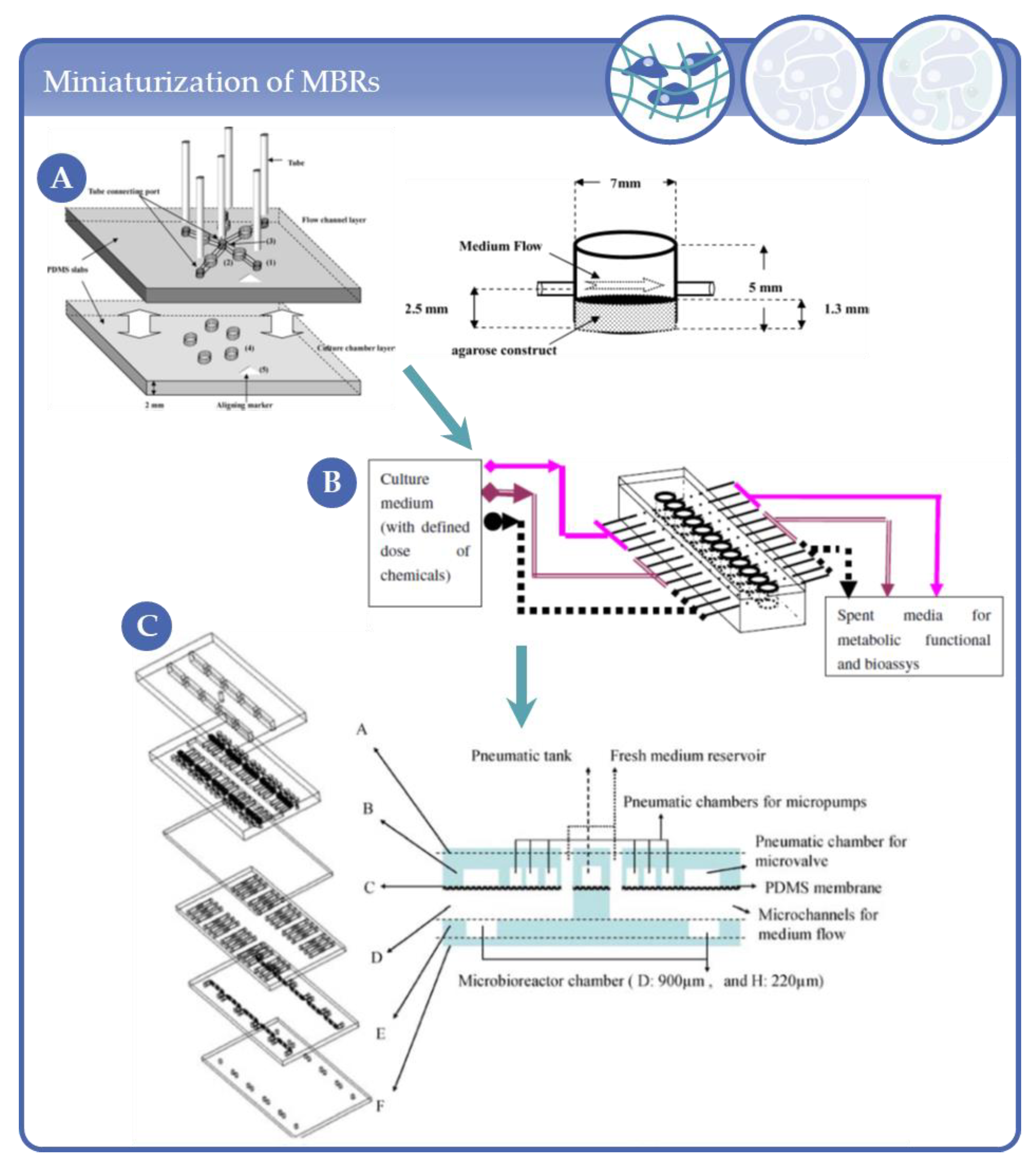
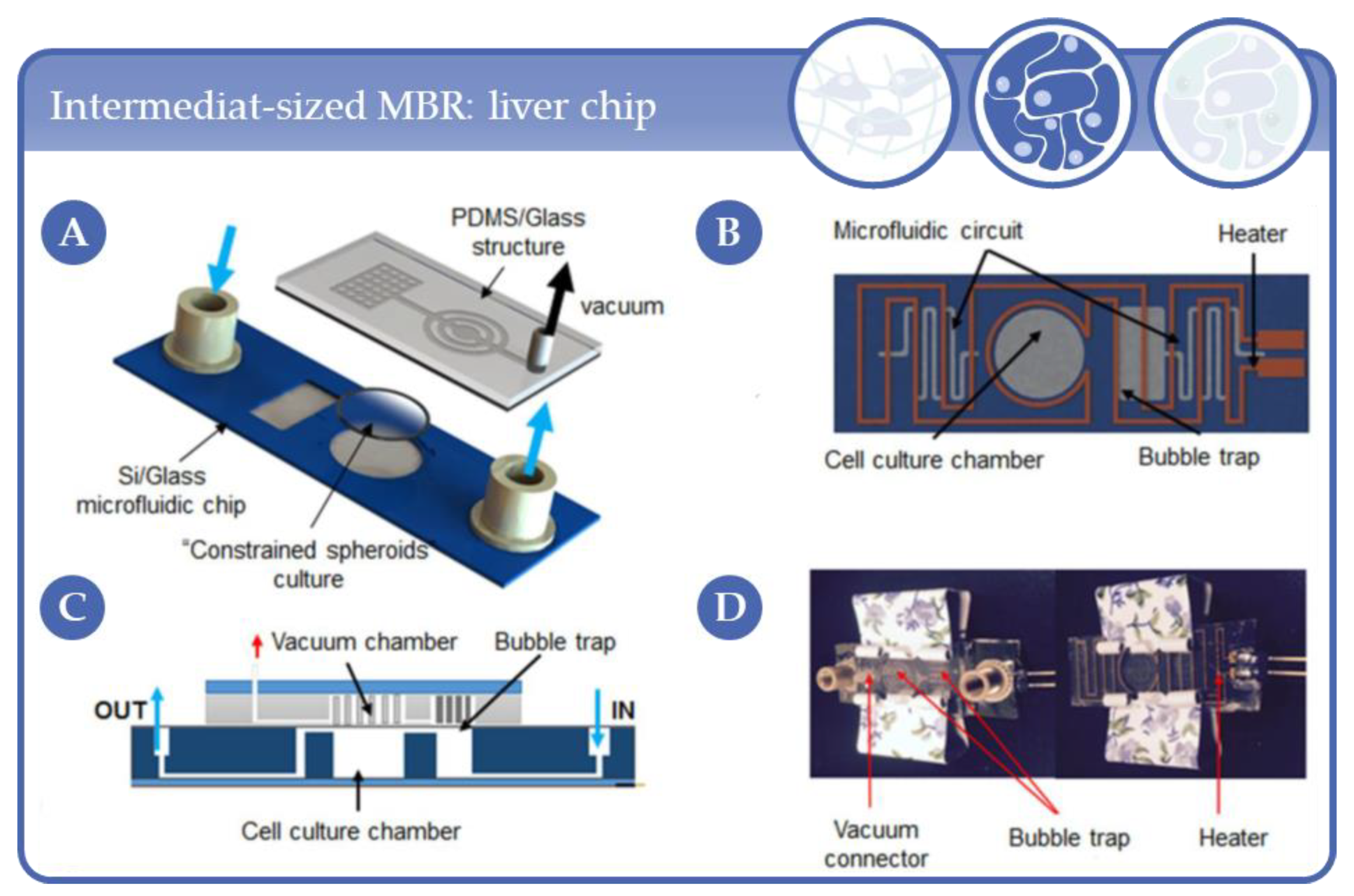
3.7. Applications in Intermediat-Sized, Chip-Like Micro-Bioreactors
3.8. Other Formats
4. Complexity Level 3: Multicellular Aggregates of at Least Two Cell Types
4.1. Stirred Sytems for Generation and Cultivation of Complexity Level 3 Spheroids
4.2. Rotating Wall Vessels
4.3. Fluidic Micro-Bioreactors: Microfluidic Chips for Complexity Level 3 Applications
4.4. Microfluidic Multiwell Plates and Other Formats for HTS Applications
4.5. Scaffold-Based Fluidic Micro-Bioreactors
4.6. Decellularized Tissues in Fluidic Micro-Bioreactors
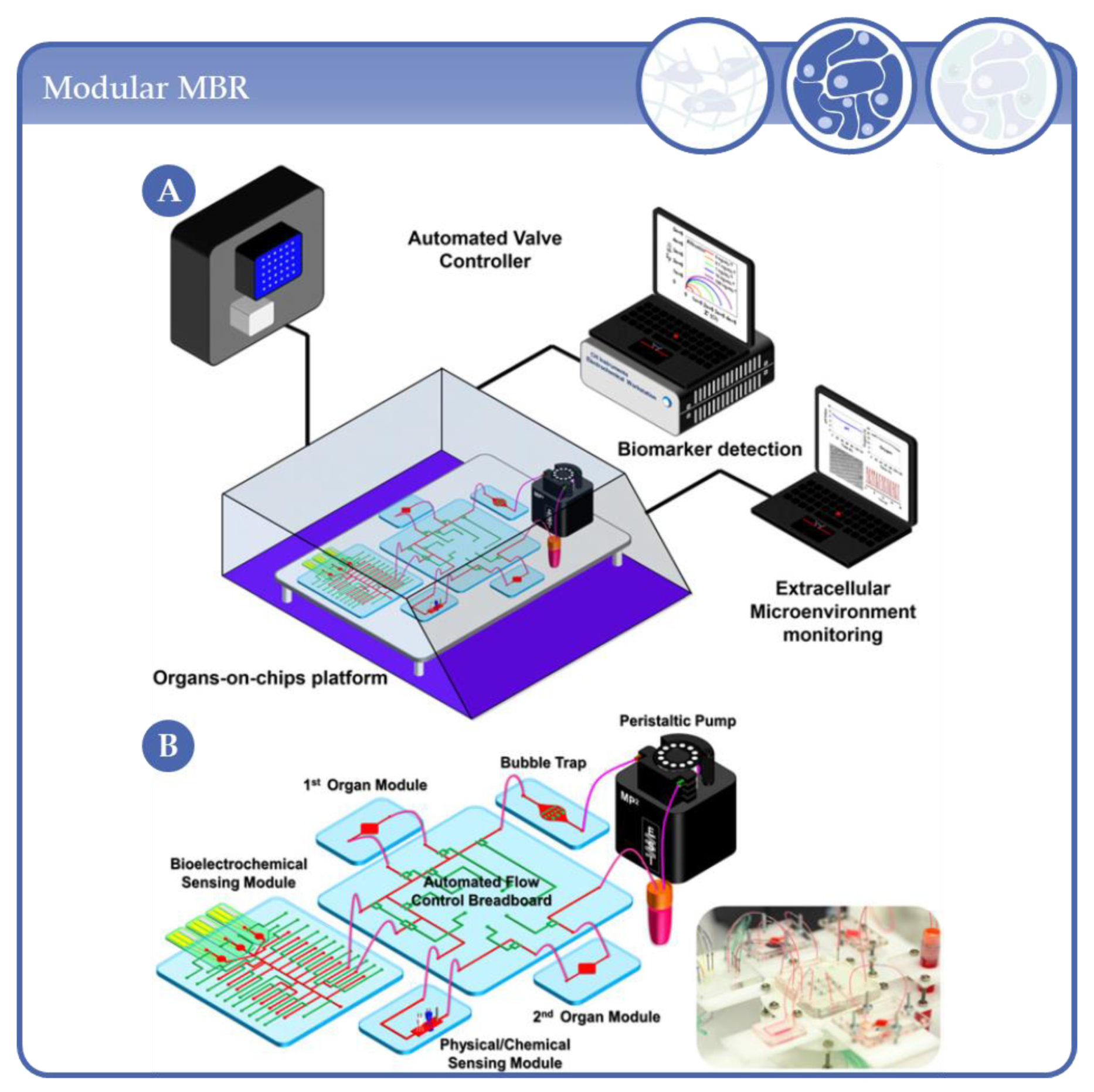
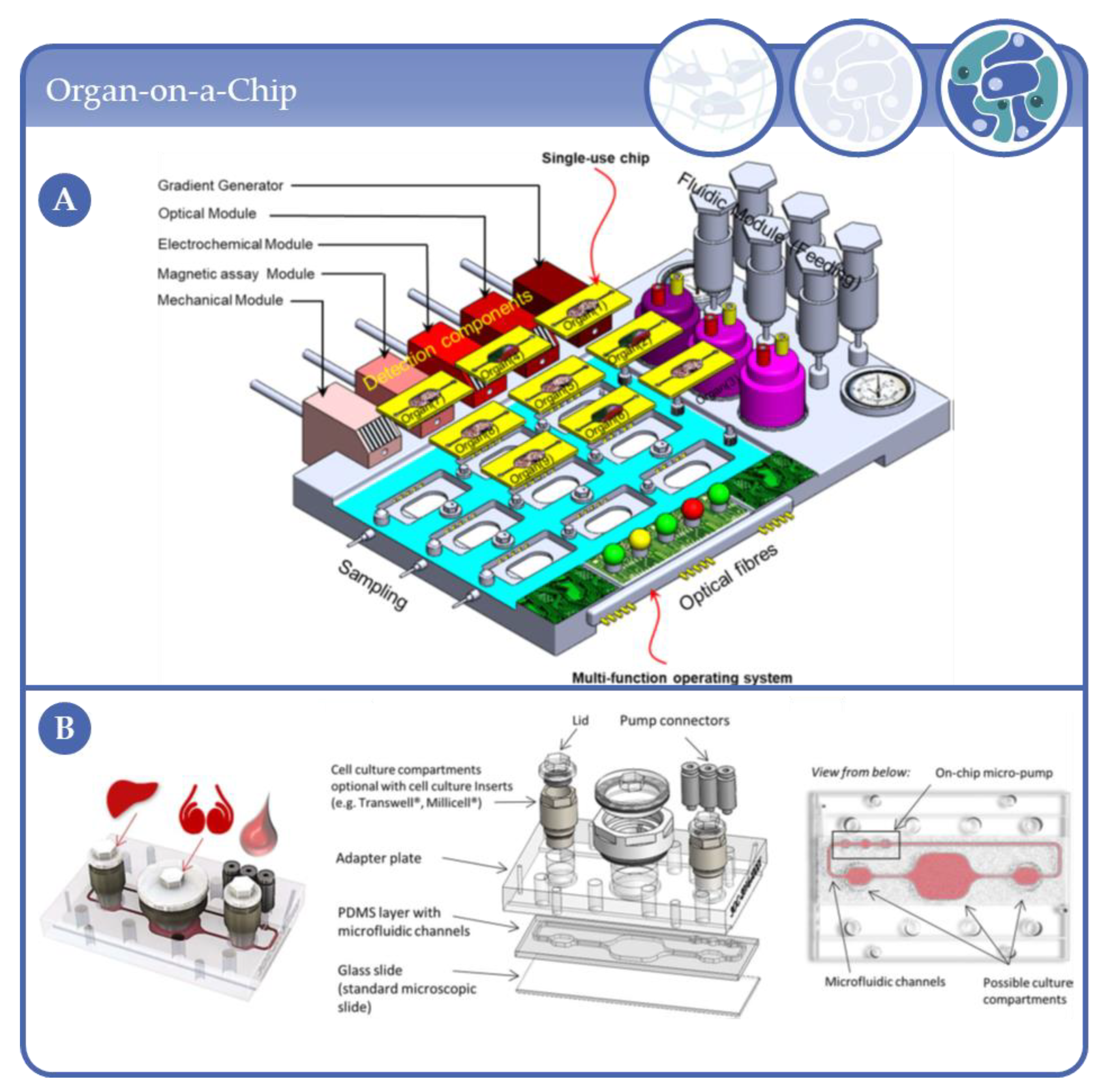
4.7. From Microfluidic Bioreactors of Complexity Level 3 to Organ-on-Chip Systems
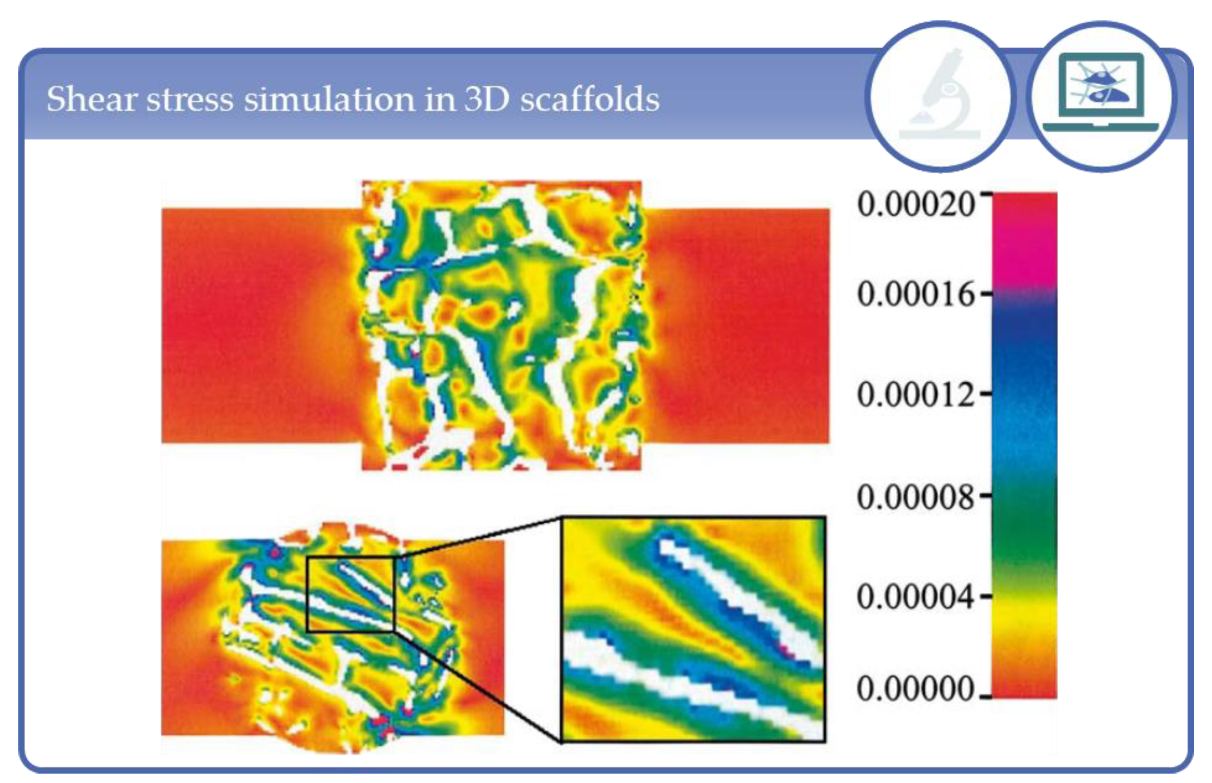
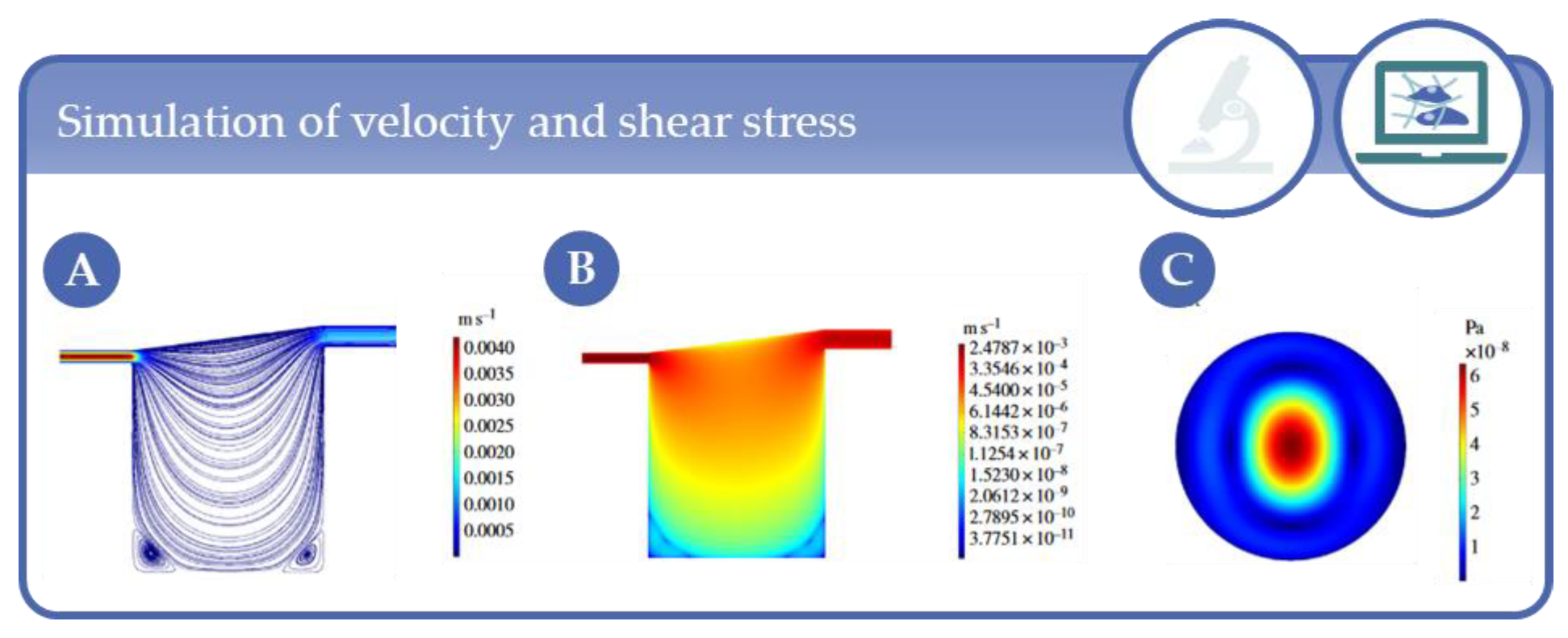
5. Simulation Studies
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Vacanti, J.P.; Langer, R. Tissue engineering: The design and fabrication of living replacement devices for surgical reconstruction and transplantation. Lancet 1999, 354, S32–S34. [Google Scholar] [CrossRef]
- Vunjak-Novakovic, G.; Freed, L.E.; Biron, R.J.; Langer, R. Effects of mixing on the composition and morphology of tissue-engineered cartilage. AIChE J. 1996, 42, 850–860. [Google Scholar] [CrossRef]
- Carrier, R.L.; Papadaki, M.; Rupnick, M.; Schoen, F.J.; Bursac, N.; Langer, R.; Freed, L.E.; Vunjak-Novakovic, G. Cardiac tissue engineering: Cell seeding, cultivation parameters, and tissue construct char-acterization. Biotechnol. Bioeng. 1999, 64, 580–589. [Google Scholar] [CrossRef]
- Niklason, L.E.; Langer, R.S. Advances in tissue engineering of blood vessels and other tissues. Transpl. Immunol. 1997, 5, 303–306. [Google Scholar] [CrossRef]
- Niklason, L.E.; Gao, J.; Abbott, W.M.; Hirschi, K.K.; Houser, S.; Marini, R.; Langer, R. Functional Arteries Grown in Vitro. Science 1999, 284, 489–493. [Google Scholar] [CrossRef]
- Altman, G.H.; Horan, R.L.; Martin, I.; Farhadi, J.; Stark, P.R.H.; Volloch, V.; Richmond, J.C.; Vunjak-Novakovic, G.; Kaplan, D.L. Cell differentiation by mechanical stress. FASEB J. 2001, 16, 1–13. [Google Scholar] [CrossRef]
- Powell, C.A.; Smiley, B.L.; Mills, J.; VanDenburgh, H.H. Mechanical stimulation improves tissue-engineered human skeletal muscle. Am. J. Physiol. Physiol. 2002, 283, C1557–C1565. [Google Scholar] [CrossRef]
- Hahn, M.S.; McHale, M.K.; Wang, E.; Schmedlen, R.H.; West, J.L. Physiologic Pulsatile Flow Bioreactor Conditioning of Poly(ethylene glycol)-based Tissue Engineered Vascular Grafts. Ann. Biomed. Eng. 2006, 35, 190–200. [Google Scholar] [CrossRef]
- Wilkes, R.P.; McNulty, A.K.; Feeley, T.D.; Schmidt, M.A.; Kieswetter, K. Bioreactor for Application of Subatmospheric Pressure to Three-Dimensional Cell Culture. Tissue Eng. 2007, 13, 3003–3010. [Google Scholar] [CrossRef]
- Moretti, M.; Freed, L.E.; Padera, R.; Laganà, K.; Boschetti, F.; Raimondi, M.T. An integrated experimental–computational approach for the study of engineered cartilage constructs subjected to combined regimens of hydrostatic pressure and interstitial perfusion. Biomed Mater. Eng. 2008, 18, 273–278. [Google Scholar] [CrossRef]
- Lee, P.-Y.; Liu, Y.-C.; Wang, M.-X.; Hu, J.-J. Fibroblast-seeded collagen gels in response to dynamic equibiaxial mechanical stimuli: A biomechanical study. J. Biomech. 2018, 78, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Langer, R.; Vacanti, J.P. Tissue engineering. Science 1993, 260, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Langer, R.S.; Vacanti, J.P. Tissue Engineering: The Challenges Ahead. Sci. Am. 1999, 280, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, A.; Dumlu, G.; Walles, H.; Maringka, M.; Ringes-Lichtenberg, S.; Ruhparwar, A.; Mertsching, H.; Haverich, A. A multifunctional bioreactor for three-dimensional cell (co)-culture. Biomaterials 2005, 26, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.-S.; Cho, J.; Tay, F.; Heng, J.Y.Y.; Ho, R.; Kazarian, S.G.; Williams, D.R.; Boccaccini, A.R.; Polak, J.M.; Mantalaris, A. The use of murine embryonic stem cells, alginate encapsulation, and rotary microgravity bioreactor in bone tissue engineering. Biomaterials 2009, 30, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Briegleb, W. The clinostat-a tool for analyzing the influence of acceleration on solid-liquid systems. In Proceedings of Workshop on space biology, Cologne, Germany, 9–11 March 1983; ESA-SP-206; European Space Agency: Paris, France, 1983. [Google Scholar]
- Wu, M.H.; Urban, J.P.G.; Cui, Z.; Cui, Z. Development of PDMS microbioreactor with well-defined and homogenous culture environment for chondrocyte 3-D culture. Biomed. Microdevices 2006, 8, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Xu, X.; Trainor, N.; Triffitt, J.; Urban, J.; Tirlapur, U. Application of multiple parallel perfused microbioreactors and three-dimensional stem cell culture for toxicity testing. Toxicol. Vitr. 2007, 21, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-H.; Huang, S.-B.; Cui, Z.; Cui, Z.; Sinha, A. A high throughput perfusion-based microbioreactor platform integrated with pneumatic micropumps for three-dimensional cell culture. Biomed. Microdevices 2007, 10, 309–319. [Google Scholar] [CrossRef]
- Ling, Y.; Rubin, J.; Deng, Y.; Huang, C.; Demirci, U.; Karp, J.M.; Khademhosseini, A. A cell-laden microfluidic hydrogel. Lab Chip 2007, 7, 756–762. [Google Scholar] [CrossRef]
- Chang, R.; Nam, J.; Holtorf, H.; Emami, K.; Jeevarajan, A.; Wu, H.; Sun, W. Direct cell writing of 3D tissue micro-organs for drug metabolism study. J. Biotechnol. 2008, 136, S144–S145. [Google Scholar] [CrossRef]
- Schätti, O.; Grad, S.; Goldhahn, J.; Salzmann, G.; Li, Z.; Alini, M.; Stoddart, M.J. A combination of shear and dynamic compression leads to mechanically induced chondrogenesis of human mesenchymal stem cells. Eur. Cells Mater. 2011, 22, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, M.A.; Grad, S.; Kaup, T.; Hänni, M.; Schneider, E.; Gogolewski, S.; Alini, M. Tribology Approach to the Engineering and Study of Articular Cartilage. Tissue Eng. 2004, 10, 1436–1445. [Google Scholar] [CrossRef] [PubMed]
- Cochis, A.; Grad, S.; Stoddart, M.J.; Farè, S.; Altomare, L.; Azzimonti, B.; Alini, M.; Rimondini, L. Bioreactor mechanically guided 3D mesenchymal stem cell chondrogenesis using a biocompatible novel thermo-reversible methylcellulose-based hydrogel. Sci. Rep. 2017, 7, srep45018. [Google Scholar] [CrossRef] [PubMed]
- Gharravi, A.M.; Orazizadeh, M.; Ansari-Asl, K.; Banoni, S.; Izadi, S.; Hashemitabar, M. Design and Fabrication of Anatomical Bioreactor Systems Containing Alginate Scaffolds for Cartilage Tissue Engineering. Avicenna J. Med Biotechnol. 2012, 4, 65–74. [Google Scholar]
- Correia, C.; Pereira, A.L.; Duarte, A.R.; Frias, A.M.; Pedro, A.J.; Oliveira, J.T.; Sousa, R.A.; Reis, R.L. Dynamic Culturing of Cartilage Tissue: The Significance of Hydrostatic Pressure. Tissue Eng. Part A 2012, 18, 1979–1991. [Google Scholar] [CrossRef]
- Santoro, R.; Olivares, A.L.; Brans, G.; Wirz, D.; Longinotti, C.; Lacroix, D.; Martin, I.; Wendt, D. Bioreactor based engineering of large-scale human cartilage grafts for joint resurfacing. Biomaterials 2010, 31, 8946–8952. [Google Scholar] [CrossRef]
- Jaeger, A.A.; Das, C.K.; Morgan, N.Y.; Pursley, R.H.; McQueen, P.G.; Hall, M.D.; Pohida, T.J.; Gottesman, M.M. Microfabricated polymeric vessel mimetics for 3-D cancer cell culture. Biomaterials 2013, 34, 8301–8313. [Google Scholar] [CrossRef]
- Sriram, R.; Van Criekinge, M.; Hansen, A.F.; Wang, Z.J.; Vigneron, D.B.; Wilson, D.M.; Keshari, K.R.; Kurhanewicz, J. Real-time measurement of hyperpolarized lactate production and efflux as a biomarker of tumor aggressiveness in an MR compatible 3D cell culture bioreactor. NMR Biomed. 2015, 28, 1141–1149. [Google Scholar] [CrossRef]
- Wu, M.-H.; Kuo, C.-Y. Application of high throughput perfusion micro 3-D cell culture platform for the precise study of cellular responses to extracellular conditions -effect of serum concentrations on the physiology of articular chondrocytes. Biomed. Microdevices 2010, 13, 131–141. [Google Scholar] [CrossRef]
- Huang, S.-B.; Wu, M.-H.; Wang, S.-S.; Sinha, A. Microfluidic cell culture chip with multiplexed medium delivery and efficient cell/scaffold loading mechanisms for high-throughput perfusion 3-dimensional cell culture-based assays. Biomed. Microdevices 2011, 13, 415–430. [Google Scholar] [CrossRef]
- Huang, S.-B.; Wang, S.-S.; Hsieh, C.-H.; Lin, Y.C.; Lai, C.-S.; Wu, M.-H. An integrated microfluidic cell culture system for high-throughput perfusion three-dimensional cell culture-based assays: Effect of cell culture model on the results of chemosensitivity assays. Lab Chip 2012, 13, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Kreiner, M.; Edrada-Ebel, R.; Cui, Z.; Van Der Walle, C.F.; Mardon, H.J. Perfusion culture enhanced human endometrial stromal cell growth in alginate-multivalent integrin α5β1 ligand scaffolds. J. Biomed. Mater. Res. Part A 2011, 99, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-H.; Chen, Y.-D.; Huang, S.-F.; Wang, H.-M.; Wu, M.-H. The Effect of Primary Cancer Cell Culture Models on the Results of Drug Chemosensitivity Assays: The Application of Perfusion Microbioreactor System as Cell Culture Vessel. BioMed Res. Int. 2015, 2015, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Moraes, C.; Wang, G.; Sun, Y.; A Simmons, C. A microfabricated platform for high-throughput unconfined compression of micropatterned biomaterial arrays. Biomaterials 2010, 31, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Pagano, G.; Ventre, M.; Iannone, M.; Greco, F.; Maffettone, P.L.; Netti, P.A. Optimizing design and fabrication of microfluidic devices for cell cultures: An effective approach to control cell microenvironment in three dimensions. Biomicrofluidics 2014, 8, 046503. [Google Scholar] [CrossRef]
- Goldman, S.M.; Barabino, G.A. Cultivation of agarose-based microfluidic hydrogel promotes the development of large, full-thickness, tissue-engineered articular cartilage constructs. J. Tissue Eng. Regen. Med. 2014, 11, 572–581. [Google Scholar] [CrossRef]
- Vecchiatini, R.; Penolazzi, L.; Lambertini, E.; Angelozzi, M.; Morganti, C.; Mazzitelli, S.; Trombelli, L.; Nastruzzi, C.; Piva, R. Effect of dynamic three-dimensional culture on osteogenic potential of human periodontal ligament-derived mesenchymal stem cells entrapped in alginate microbeads. J. Periodontal Res. 2014, 50, 544–553. [Google Scholar] [CrossRef]
- Rödling, L.; Volz, E.M.; Raic, A.; Brändle, K.; Franzreb, M.; Lee-Thedieck, C. Magnetic Macroporous Hydrogels as a Novel Approach for Perfused Stem Cell Culture in 3D Scaffolds via Contactless Motion Control. Adv. Heal. Mater. 2018, 7, e1701403. [Google Scholar] [CrossRef]
- Holtfreter, J. A study of the mechanics of gastrulation. J. Exp. Zool. 1944, 95, 171–212. [Google Scholar] [CrossRef]
- Inch, W.R.; A McCredie, J.; Sutherland, R.M. Growth of nodular carcinomas in rodents compared with multi-cell spheroids in tissue culture. Growth 1970, 34, 271–282. [Google Scholar]
- Sutherland, R.M.; A McCredie, J.; Inch, W.R. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J. Natl. Cancer Inst. 1971, 46, 113–120. [Google Scholar] [PubMed]
- Schwarz, R.P.; Wolf, D.A. Rotating bio-reactor cell culture apparatus. U.S. Patent No 4,988,623, 1991. [Google Scholar]
- Schwarz, R.P.; Wolf, D.A.; Trinh, T.T. Horizontally rotated cell culture system with a coaxial tubular oxygenator. U.S. Patent No 5,026,650, 1991. [Google Scholar]
- Gerecht-Nir, S.; Cohen, S.; Itskovitz-Eldor, J. Bioreactor cultivation enhances the efficiency of human embryoid body (hEB) formation and differentiation. Biotechnol. Bioeng. 2004, 86, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Aucamp, J.; Calitz, C.; Bronkhorst, A.J.; Wrzesinski, K.; Hamman, S.; Gouws, C.; Pretorius, P.J. Cell-free DNA in a three-dimensional spheroid cell culture model: A preliminary study. Int. J. Biochem. Cell Biol. 2017, 89, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Bramley, J.C.; Drummond, C.G.; Lennemann, N.J.; Good, C.A.; Kim, K.S.; Coyne, C.B. A Three-Dimensional Cell Culture System To Model RNA Virus Infections at the Blood-Brain Barrier. mSphere 2017, 2, e00206-17. [Google Scholar] [CrossRef]
- Drummond, C.G.; Nickerson, C.A.; Coyne, C.B. A Three-Dimensional Cell Culture Model To Study Enterovirus Infection of Polarized Intestinal Epithelial Cells. mSphere 2015, 1. [Google Scholar] [CrossRef]
- Sainz, B.; TenCate, V.; Uprichard, S.L. Three-dimensional Huh7 cell culture system for the study of Hepatitis C virus infection. Virol. J. 2009, 6, 103. [Google Scholar] [CrossRef]
- Papafragkou, E.; Hewitt, J.; Park, G.W.; Greening, G.; Vinjé, J. Challenges of Culturing Human Norovirus in Three-Dimensional Organoid Intestinal Cell Culture Models. PLoS ONE 2013, 8, e63485. [Google Scholar] [CrossRef]
- Carterson, A.J.; Zu Bentrup, K.H.; Ott, C.M.; Clarke, M.S.; Pierson, D.L.; Vanderburg, C.R.; Buchanan, K.L.; Nickerson, C.A.; Schurr, M.J. A549 Lung Epithelial Cells Grown as Three-Dimensional Aggregates: Alternative Tissue Culture Model for Pseudomonas aeruginosa Pathogenesis. Infect. Immun. 2005, 73, 1129–1140. [Google Scholar] [CrossRef]
- Crabbé, A.; Liu, Y.; Matthijs, N.; Rigole, P.; De La Fuente-Nùñez, C.; Davis, R.; Ledesma, M.A.; Sarker, S.; Van Houdt, R.; Hancock, R.E.W.; et al. Antimicrobial efficacy against Pseudomonas aeruginosa biofilm formation in a three-dimensional lung epithelial model and the influence of fetal bovine serum. Sci. Rep. 2017, 7, srep43321. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, C.A.; Goodwin, T.J.; Terlonge, J.; Ott, C.M.; Buchanan, K.L.; Uicker, W.C.; Emami, K.; Leblanc, C.L.; Ramamurthy, R.; Clarke, M.S.; et al. Three-Dimensional Tissue Assemblies: Novel Models for the Study of Salmonella enterica Serovar Typhimurium Pathogenesis. Infect. Immun. 2001, 69, 7106–7120. [Google Scholar] [CrossRef] [PubMed]
- Zu Bentrup, K.H.; Ramamurthy, R.; Ott, C.M.; Emami, K.; Nelman-Gonzalez, M.; Wilson, J.W.; Richter, E.G.; Goodwin, T.J.; Alexander, J.S.; Pierson, D.L.; et al. Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microbes Infect. 2006, 8, 1813–1825. [Google Scholar] [CrossRef] [PubMed]
- Radtke, A.L.; Wilson, J.W.; Sarker, S.; Nickerson, C.A. Analysis of Interactions of Salmonella Type Three Secretion Mutants with 3-D Intestinal Epithelial Cells. PLoS ONE 2010, 5, e15750. [Google Scholar] [CrossRef] [PubMed]
- De Weirdt, R.; Crabbé, A.; Roos, S.; Vollenweider, S.; Lacroix, C.; Van Pijkeren, J.P.; Britton, R.A.; Sarker, S.; Van De Wiele, T.; Nickerson, C.A. Glycerol Supplementation Enhances L. reuteri’s Protective Effect against S. Typhimurium Colonization in a 3-D Model of Colonic Epithelium. PLoS ONE 2012, 7, e37116. [Google Scholar] [CrossRef] [PubMed]
- Smith, Y.C.; Grande, K.K.; Rasmussen, S.B.; O’Brien, A.D. Novel Three-Dimensional Organoid Model for Evaluation of the Interaction of Uropathogenic Escherichia coli with Terminally Differentiated Human Urothelial Cells. Infect. Immun. 2006, 74, 750–757. [Google Scholar] [CrossRef]
- Carvalho, H.M.; Teel, L.D.; Goping, G.; O’Brien, A.D. A three-dimensional tissue culture model for the study of attach and efface lesion formation by enteropathogenic and enterohaemorrhagic Escherichia coli. Cell. Microbiol. 2005, 7, 1771–1781. [Google Scholar] [CrossRef]
- Warren, C.A.; Destura, R.V.; Sevilleja, J.E.A.D.; Barroso, L.F.; Carvalho, H.; Barrett, L.J.; O’Brien, A.D.; Guerrant, R.L. Detection of Epithelial-Cell Injury, and Quantification of Infection, in the HCT-8 Organoid Model of Cryptosporidiosis. J. Infect. Dis. 2008, 198, 143–149. [Google Scholar] [CrossRef]
- Łaniewski, P.; Gomez, A.; Hire, G.; So, M.; Herbst-Kralovetz, M.M. Human Three-Dimensional Endometrial Epithelial Cell Model To Study Host Interactions with Vaginal Bacteria and Neisseria gonorrhoeae. Infect. Immun. 2017, 85, e01049-16. [Google Scholar] [CrossRef]
- David, J.; Sayer, N.M.; Sarkar-Tyson, M. The use of a three-dimensional cell culture model to investigate host–pathogen interactions of Francisella tularensis in human lung epithelial cells. Microbes Infect. 2014, 16, 735–745. [Google Scholar] [CrossRef]
- Chang, T.T.; Hughes-Fulford, M. Molecular mechanisms underlying the enhanced functions of three-dimensional hepatocyte aggregates. Biomaterials 2014, 35, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Detamore, M.S.; Athanasiou, K.A. Use of a Rotating Bioreactor toward Tissue Engineering the Temporomandibular Joint Disc. Tissue Eng. 2005, 11, 1188–1197. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, M.; Bursac, N.; Langer, R.; Merok, J.; Vunjak-Novakovic, G.; Freed, L.E. Tissue engineering of functional cardiac muscle: Molecular, structural, and electrophysiological studies. Am. J. Physiol. Circ. Physiol. 2001, 280, H168–H178. [Google Scholar] [CrossRef] [PubMed]
- Rungarunlert, S.; Klincumhom, N.; Bock, I.; Nemes, C.; Techakumphu, M.; Pirity, M.K.; Dinnyes, A. Enhanced cardiac differentiation of mouse embryonic stem cells by use of the slow-turning, lateral vessel (STLV) bioreactor. Biotechnol. Lett. 2011, 33, 1565–1573. [Google Scholar] [CrossRef]
- Botchwey, E.A.; Pollack, S.R.; Levine, E.M.; Laurencin, C.T. Bone tissue engineering in a rotating bioreactor using a microcarrier matrix system. J. Biomed. Mater. Res. 2001, 55, 242–253. [Google Scholar] [CrossRef]
- Goldstein, A.S. Effect of convection on osteoblastic cell growth and function in biodegradable polymer foam scaffolds. Biomaterials 2001, 22, 1279–1288. [Google Scholar] [CrossRef]
- Marlovits, S.; Tichy, B.; Truppe, M.; Gruber, D.; Vécsei, V. Chondrogenesis of Aged Human Articular Cartilage in a Scaffold-Free Bioreactor. Tissue Eng. 2003, 9, 1215–1226. [Google Scholar] [CrossRef]
- Montani, C.; Steimberg, N.; Boniotti, J.; Biasiotto, G.; Zanella, I.; Diafera, G.; Biunno, I.; Caimi, L.; Mazzoleni, G.; Di Lorenzo, D. Fibroblasts maintained in 3 dimensions show a better differentiation state and higher sensitivity to estrogens. Toxicol. Appl. Pharmacol. 2014, 280, 421–433. [Google Scholar] [CrossRef]
- Samuelson, L.; Gerber, D.A. Improved Function and Growth of Pancreatic Cells in a Three-Dimensional Bioreactor Environment. Tissue Eng. Part C Methods 2013, 19, 39–47. [Google Scholar] [CrossRef]
- Valmikinathan, C.M.; Hoffman, J.; Yu, X. Impact of scaffold micro and macro architecture on Schwann cell proliferation under dynamic conditions in a rotating wall vessel bioreactor. Mater. Sci. Eng. C 2011, 31, 22–29. [Google Scholar] [CrossRef]
- Hjelm, B.E.; Berta, A.N.; Nickerson, C.A.; Arntzen, C.J.; Herbst-Kralovetz, M.M. Development and Characterization of a Three-Dimensional Organotypic Human Vaginal Epithelial Cell Model1. Biol. Reprod. 2010, 82, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Lamarca, H.; Ott, C.; Zu Bentrup, K.H.; Leblanc, C.; Pierson, D.; Nelson, A.; Scandurro, A.; Whitley, G.S.; Nickerson, C.; Morris, C. Three-dimensional growth of extravillous cytotrophoblasts promotes differentiation and invasion. Placenta 2005, 26, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Sarker, S.F.; Crabbé, A.; Nickerson, C.A.; Prestwich, G.D. The generation of 3-D tissue models based on hyaluronan hydrogel-coated microcarriers within a rotating wall vessel bioreactor. Biomaterials 2010, 31, 8426–8435. [Google Scholar] [CrossRef] [PubMed]
- Smit, T.; Calitz, C.; Willers, C.; Svitina, H.; Hamman, J.; Fey, S.J.; Gouws, C.; Wrzesinski, K. Characterization of an Alginate Encapsulated LS180 Spheroid Model for Anti-colorectal Cancer Compound Screening. ACS Med. Chem. Lett. 2020, 11, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Wrzesinski, K.; Fey, S.J. Metabolic Reprogramming and the Recovery of Physiological Functionality in 3D Cultures in Micro-Bioreactors. Bioengineering 2018, 5, 22. [Google Scholar] [CrossRef]
- Yamashita, T.; Takayama, K.; Sakurai, F.; Mizuguchi, H. Billion-scale production of hepatocyte-like cells from human induced pluripotent stem cells. Biochem. Biophys. Res. Commun. 2018, 496, 1269–1275. [Google Scholar] [CrossRef]
- Cortiella, J.; Niles, J.; Cantu, A.; Brettler, A.; Pham, A.; Vargas, G.; Winston, S.; Wang, J.; Walls, S.; Nichols, J.E. Influence of Acellular Natural Lung Matrix on Murine Embryonic Stem Cell Differentiation and Tissue Formation. Tissue Eng. Part A 2010, 16, 2565–2580. [Google Scholar] [CrossRef]
- Lei, X.-H.; Ning, L.-N.; Cao, Y.-J.; Liu, S.; Zhang, S.-B.; Qiu, Z.-F.; Hu, H.-M.; Zhang, H.-S.; Liu, S.; Duan, E. NASA-Approved Rotary Bioreactor Enhances Proliferation of Human Epidermal Stem Cells and Supports Formation of 3D Epidermis-Like Structure. PLoS ONE 2011, 6, e26603. [Google Scholar] [CrossRef]
- Li, W.-J.; Tuan, R.S. Fabrication and Application of Nanofibrous Scaffolds in Tissue Engineering. Curr. Protoc. Cell Biol. 2009, 42, 25.2.1–25.2.12. [Google Scholar] [CrossRef]
- Quail, D.F.; Maciel, T.J.; Rogers, K.; Postovit, L.-M. A Unique 3D In Vitro Cellular Invasion Assay. J. Biomol. Screen. 2012, 17, 1088–1095. [Google Scholar] [CrossRef][Green Version]
- Giselbrecht, S.; Truckenmüller, R. Formkörper, Verfahren zu seiner Herstellung und Verwendung. DE Patent WO2006/007948 A1, 30 June 2005. [Google Scholar]
- Gottwald, E.; Giselbrecht, S.; Augspurger, C.; Lahni, B.; Dambrowsky, N.; Truckenmüller, R.; Piotter, V.; Gietzelt, T.; Wendt, O.; Pfleging, W.; et al. A chip-based platform for the in vitro generation of tissues in three-dimensional organization. Lab Chip 2007, 7, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Gottwald, E.; Kleintschek, T.; Giselbrecht, S.; Truckenmüller, R.; Altmann, B.; Worgull, M.; Döpfert, J.; Schad, L.; Heilmann, M. Characterization of a chip-based bioreactor for three-dimensional cell cultivation via Magnetic Resonance Imaging. Zeitschrift Medizinische Physik 2013, 23, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Hoesl, M.A.U.; Kleimaier, D.; Hu, R.; Malzacher, M.; Nies, C.; Gottwald, E.; Schad, L.R. 23Na Triple-quantum signal of in vitro human liver cells, liposomes, and nanoparticles: Cell viability assessment vs. separation of intra- and extracellular signal. J. Magn. Reson. Imaging 2019, 50, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Kleimaier, D.; Goerke, S.; Nies, C.; Zaiss, M.; Kunz, P.; Bachert, P.; Ladd, M.E.; Gottwald, E.; Schad, L.R. The cellular heat shock response monitored by chemical exchange saturation transfer MRI. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Neubauer, A.; Nies, C.; Schepkin, V.D.; Hu, R.; Malzacher, M.; Chacón-Caldera, J.; Thiele, D.; Gottwald, E.; Schad, L.R. Tracking protein function with sodium multi quantum spectroscopy in a 3D-tissue culture based on microcavity arrays. Sci. Rep. 2017, 7, 3943. [Google Scholar] [CrossRef]
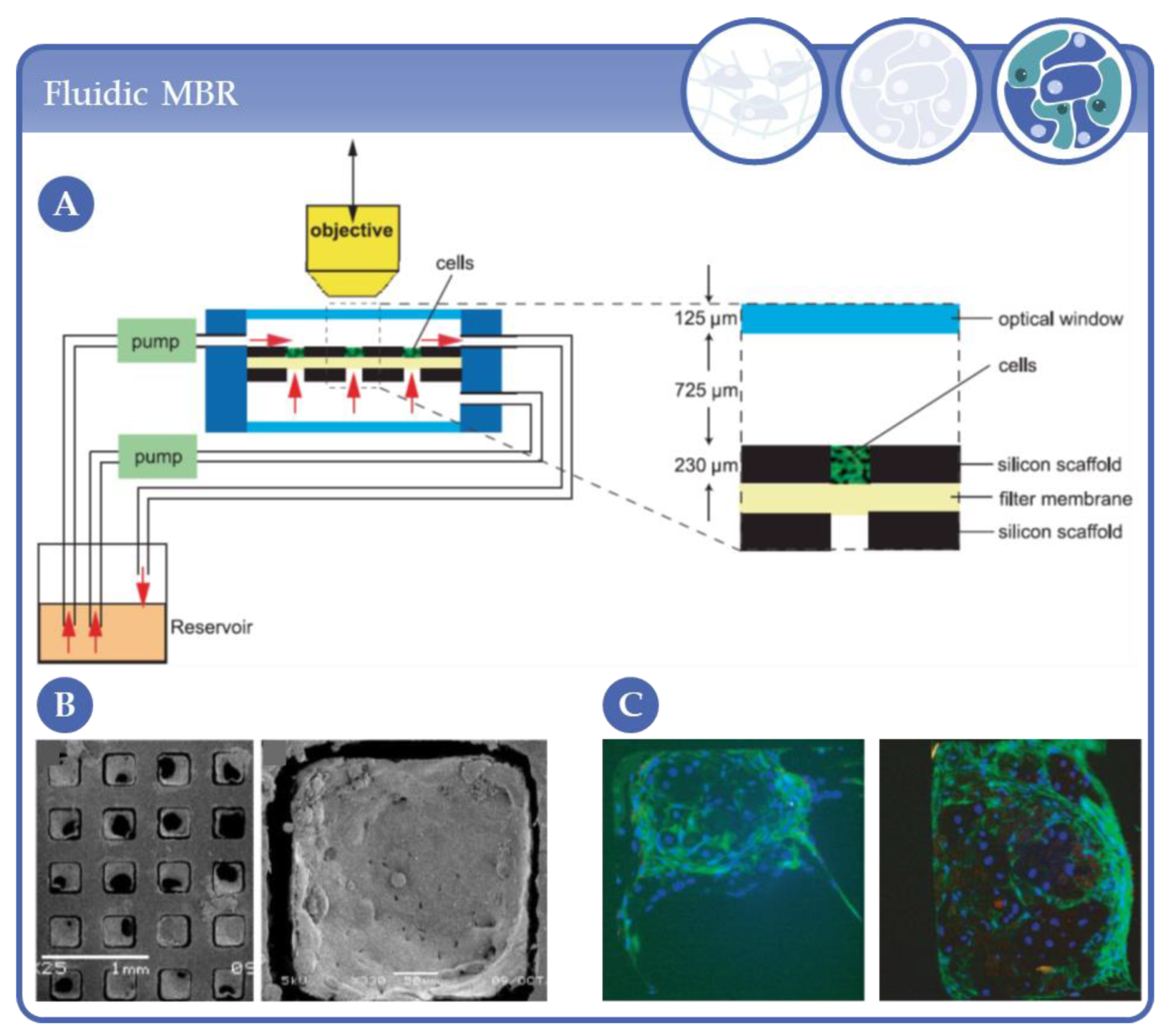
- Powers, M.J.; Domansky, K.; Kaazempur-Mofrad, M.R.; Kalezi, A.; Capitano, A.; Upadhyaya, A.; Kurzawski, P.; Wack, K.E.; Stolz, D.B.; Kamm, R.; et al. A microfabricated array bioreactor for perfused 3D liver culture. Biotechnol. Bioeng. 2002, 78, 257–269. [Google Scholar] [CrossRef]
- Sivaraman, A.; Leach, J.K.; Townsend, S.; Iida, T.; Hogan, B.J.; Stolz, D.B.; Fry, R.; Samson, L.D.; Tannenbaum, S.R.; Griffith, L.G. A Microscale In Vitro Physiological Model of the Liver: Predictive Screens for Drug Metabolism and Enzyme Induction. Curr. Drug Metab. 2005, 6, 569–591. [Google Scholar] [CrossRef]
- Powers, M.J.; Janigian, D.M.; Wack, K.E.; Baker, C.S.; Stolz, D.B.; Griffith, L.G. Functional Behavior of Primary Rat Liver Cells in a Three-Dimensional Perfused Microarray Bioreactor. Tissue Eng. 2002, 8, 499–513. [Google Scholar] [CrossRef]
- Yates, C.; Shepard, C.R.; Papworth, G.; Dash, A.; Stolz, D.B.; Tannenbaum, S.; Griffith, L.; Wells, A. Novel Three-Dimensional Organotypic Liver Bioreactor to Directly Visualize Early Events in Metastatic Progression. Adv. Cancer Res. 2007, 97, 225–246. [Google Scholar] [CrossRef]
- Weise, F.; Fernekorn, U.; Hampl, J.; Klett, M.; Schober, A. Analysis and comparison of oxygen consumption of HepG2 cells in a monolayer and three-dimensional high density cell culture by use of a matrigrid®. Biotechnol. Bioeng. 2013, 110, 2504–2512. [Google Scholar] [CrossRef]
- Giselbrecht, S.; Gietzelt, T.; Gottwald, E.; Trautmann, C.; Truckenmüller, R.; Weibezahn, K.F.; Welle, A. 3D tissue culture substrates produced by microthermoforming of pre-processed polymer films. Biomed. Microdevices 2006, 8, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Bingel, C.; Koeneke, E.; Ridinger, J.; Bittmann, A.; Sill, M.; Peterziel, H.; Wrobel, J.K.; Rettig, I.; Milde, T.; Fernekorn, U.; et al. Three-dimensional tumor cell growth stimulates autophagic flux and recapitulates chemotherapy resistance. Cell Death Dis. 2017, 8, e3013. [Google Scholar] [CrossRef] [PubMed]
- Knazek, R.A.; Gullino, P.M.; Kohler, P.O.; Dedrick, R.L. Cell Culture on Artificial Capillaries: An Approach to Tissue Growth in vitro. Science 1972, 178, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, S.L.; Shatford, R.A.; Cerra, F.B.; Hu, W.-S. Bilirubin Conjugation in a Three Compartment Hollow Fiber Bioreactor. In Proceedings of the Twelfth Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Philadelphia, PA, USA, 1–4 November 1990; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2005; pp. 443–444. [Google Scholar]
- Pless, G.; Steffen, I.; Zeilinger, K.; Sauer, I.M.; Katenz, E.; Kehr, D.C.; Roth, S.; Mieder, T.; Schwartlander, R.; Müller, C.; et al. Evaluation of Primary Human Liver Cells in Bioreactor Cultures for Extracorporeal Liver Support on the Basis of Urea Production. Artif. Organs 2006, 30, 686–694. [Google Scholar] [CrossRef]
- Gerlach, J.C.; Encke, J.; Hole, O.; Müller, C.; Courtney, J.M.; Neuhaus, P. Hepatocyte culture between three dimensionally arranged biomatrix-coated independent artifi-cial capillary systems and sinusoidal endothelial cell co-culture compartments. Int. J. Artif. Organs. 1994, 17, 301–306. [Google Scholar] [CrossRef]
- Gerlach, J.; Schnoy, N.; Smith, M.D.; Neuhaus, P. Hepatocyte Culture between Woven Capillary Networks: A Microscopy Study. Artif. Organs 1994, 18, 226–230. [Google Scholar] [CrossRef]
- Ring, A.; Gerlach, J.; Peters, G.; Pazin, B.J.; Minervini, C.F.; Turner, M.E.; Thompson, R.L.; Triolo, F.; Gridelli, B.; Miki, T. Hepatic Maturation of Human Fetal Hepatocytes in Four-Compartment Three-Dimensional Perfusion Culture. Tissue Eng. Part C Methods 2010, 16, 835–845. [Google Scholar] [CrossRef]
- Zeilinger, K.; Schreiter, T.; Darnell, M.; Söderdahl, T.; Lübberstedt, M.; Dillner, B.; Knobeloch, D.; Nüssler, A.K.; Gerlach, J.C.; Andersson, T.B. Scaling Down of a Clinical Three-Dimensional Perfusion Multicompartment Hollow Fiber Liver Bioreactor Developed for Extracorporeal Liver Support to an Analytical Scale Device Useful for Hepatic Pharmacological In Vitro Studies. Tissue Eng. Part C Methods 2011, 17, 549–556. [Google Scholar] [CrossRef]
- Ulvestad, M.; Darnell, M.; Molden, E.; Ellis, E.; Åsberg, A.; Andersson, T.B. Evaluation of Organic Anion-Transporting Polypeptide 1B1 and CYP3A4 Activities in Primary Human Hepatocytes and HepaRG Cells Cultured in a Dynamic Three-Dimensional Bioreactor System. J. Pharmacol. Exp. Ther. 2012, 343, 145–156. [Google Scholar] [CrossRef]
- Gerlach, J.C.; Encke, J.; Hole, O.; Müller, C.; Ryan, C.J.; Neuhaus, P. Bioreactor for a larger scale hepatocyte in vitro perfusion. Transplantation 1994, 58, 984–988. [Google Scholar] [CrossRef] [PubMed]
- Lübberstedt, M.; Müller-Vieira, U.; Biemel, K.M.; Darnell, M.; A Hoffmann, S.; Knöspel, F.; Wönne, E.C.; Knobeloch, D.; Nussler, A.K.; Gerlach, J.C.; et al. Serum-free culture of primary human hepatocytes in a miniaturized hollow-fibre membrane bioreactor for pharmacologicalin vitrostudies. J. Tissue Eng. Regen. Med. 2012, 9, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- De Bartolo, L.; Salerno, S.; Curcio, E.; Piscioneri, A.; Rende, M.; Morelli, S.; Tasselli, F.; Bader, A.; Drioli, E. Human hepatocyte functions in a crossed hollow fiber membrane bioreactor. Biomaterials 2009, 30, 2531–2543. [Google Scholar] [CrossRef] [PubMed]
- Schmelzer, E.; Triolo, F.; Turner, M.E.; Thompson, R.L.; Zeilinger, K.; Reid, L.M.; Gridelli, B.; Gerlach, J.C. Three-Dimensional Perfusion Bioreactor Culture Supports Differentiation of Human Fetal Liver Cells. Tissue Eng. Part A 2010, 16, 2007–2016. [Google Scholar] [CrossRef]
- Darnell, M.; Ulvestad, M.; Ellis, E.; Weidolf, L.; Andersson, T.B. In Vitro Evaluation of Major In Vivo Drug Metabolic Pathways Using Primary Human Hepatocytes and HepaRG Cells in Suspension and a Dynamic Three-Dimensional Bioreactor System. J. Pharmacol. Exp. Ther. 2012, 343, 134–144. [Google Scholar] [CrossRef]
- Hoekstra, R.; Nibourg, G.A.A.; Van Der Hoeven, T.V.; Plomer, G.; Seppen, J.; Ackermans, M.T.; Camus, S.; Kulik, W.; Van Gulik, T.M.; Elferink, R.P.O.; et al. Phase 1 and Phase 2 Drug Metabolism and Bile Acid Production of HepaRG Cells in a Bioartificial Liver in Absence of Dimethyl Sulfoxide. Drug Metab. Dispos. 2013, 41, 562–567. [Google Scholar] [CrossRef]
- Flendrig, L.M.; Velde, A.A.T.; Chamuleau, R.A. Semipermeable Hollow Fiber Membranes in Hepatocyte Bioreactors: A Prerequisite for a Successful Bioartificial Liver? Artif. Organs 2008, 21, 1177–1181. [Google Scholar] [CrossRef]
- Tapia, F.; Vogel, T.; Genzel, Y.; Behrendt, I.; Hirschel, M.; Gangemi, J.D.; Reichl, U. Production of high-titer human influenza A virus with adherent and suspension MDCK cells cultured in a single-use hollow fiber bioreactor. Vaccine 2014, 32, 1003–1011. [Google Scholar] [CrossRef]
- Rodday, B.; Hirschhaeuser, F.; Walenta, S.; Mueller-Klieser, W. Semiautomatic Growth Analysis of Multicellular Tumor Spheroids. J. Biomol. Screen. 2011, 16, 1119–1124. [Google Scholar] [CrossRef]
- Tostões, R.M.; Leite, S.B.; Serra, M.; Jensen, J.; Björquist, P.; Carrondo, M.J.T.; Brito, C.; Alves, P.M. Human liver cell spheroids in extended perfusion bioreactor culture for repeated-dose drug testing. Hepatology 2012, 55, 1227–1236. [Google Scholar] [CrossRef]
- Egger, D.; Fischer, M.; Clementi, A.; Ribitsch, V.; Hansmann, J.; Kasper, C. Development and Characterization of a Parallelizable Perfusion Bioreactor for 3D Cell Culture. Bioengeering 2017, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Egger, D.; Spitz, S.; Fischer, M.; Handschuh, S.; Glosmann, M.; Friemert, B.; Egerbacher, M.; Kasper, C. Application of a Parallelizable Perfusion Bioreactor for Physiologic 3D Cell Culture. Cells Tissues Organs 2017, 203, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Birru, B.; Mekala, N.K.; Rao, S. Improved osteogenic differentiation of umbilical cord blood MSCs using custom made perfusion bioreactor. Biomed. J. 2018, 41, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.; Schwarz, S.; Meier-Staude, R.; Sudhop, S.; Clausen-Schaumann, H.; Schieker, M.; Huber, R. A Perfusion Bioreactor System for Cell Seeding and Oxygen-Controlled Cultivation of Three-Dimensional Cell Cultures. Tissue Eng. Part C Methods 2018, 24, 585–595. [Google Scholar] [CrossRef]
- Spitkovsky, D.; Lemke, K.; Förster, T.; Römer, R.; Wiedemeier, S.; Hescheler, J.; Sachinidis, A.; Gastrock, G. Generation of Cardiomyocytes in Pipe-Based Microbioreactor Under Segmented Flow. Cell. Physiol. Biochem. 2016, 38, 1883–1896. [Google Scholar] [CrossRef]
- Vetsch, J.R.; Betts, D.C.; Muller, R.; Hofmann, S. Flow velocity-driven differentiation of human mesenchymal stromal cells in silk fibroin scaffolds: A combined experimental and computational approach. PLoS ONE 2017, 12, e0180781. [Google Scholar] [CrossRef]
- Sart, S.; Tomasi, R.F.-X.; Amselem, G.; Baroud, C.N. Multiscale cytometry and regulation of 3D cell cultures on a chip. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Toh, Y.-C.; Zhang, C.; Zhang, J.; Khong, Y.M.; Chang, S.; Samper, V.D.; Van Noort, D.; Hutmacher, D.W.; Yu, H. A novel 3D mammalian cell perfusion-culture system in microfluidic channels. Lab Chip 2007, 7, 302–309. [Google Scholar] [CrossRef]
- Ong, S.-M.; Zhang, C.; Toh, Y.-C.; Kim, S.H.; Foo, H.L.; Tan, C.H.; Van Noort, D.; Park, S.; Yu, H. A gel-free 3D microfluidic cell culture system. Biomaterials 2008, 29, 3237–3244. [Google Scholar] [CrossRef]
- Zhang, C.; Chia, S.-M.; Ong, S.-M.; Zhang, S.; Toh, Y.-C.; Van Noort, D.; Yu, H. The controlled presentation of TGF-β1 to hepatocytes in a 3D-microfluidic cell culture system. Biomaterials 2009, 30, 3847–3853. [Google Scholar] [CrossRef]
- Izzo, L.; Tunesi, M.; Boeri, L.; Laganà, M.; Giordano, C.; Raimondi, M.T. Influence of the static magnetic field on cell response in a miniaturized optically accessible bioreactor for 3D cell culture. Biomed. Microdevices 2019, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Laganà, M.; Raimondi, M.T. A miniaturized, optically accessible bioreactor for systematic 3D tissue engineering research. Biomed. Microdevices 2011, 14, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Tunesi, M.; Fusco, F.; Fiordaliso, F.; Corbelli, A.; Biella, G.; Raimondi, M.T. Optimization of a 3D Dynamic Culturing System for In Vitro Modeling of Frontotemporal Neurodegeneration-Relevant Pathologic Features. Front. Aging Neurosci. 2016, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Deng, R.; Tong, W.H.; Huan, L.; Way, N.C.; IslamBadhan, A.; Iliescu, C.; Yu, H. A perfusion incubator liver chip for 3D cell culture with application on chronic hepatotoxicity testing. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Fu, C.-Y.; Tseng, S.-Y.; Yang, S.-M.; Hsu, L.; Liu, C.-H.; Chang, H.-Y. A microfluidic chip with a U-shaped microstructure array for multicellular spheroid formation, culturing and analysis. Biofabrication 2014, 6, 015009. [Google Scholar] [CrossRef]
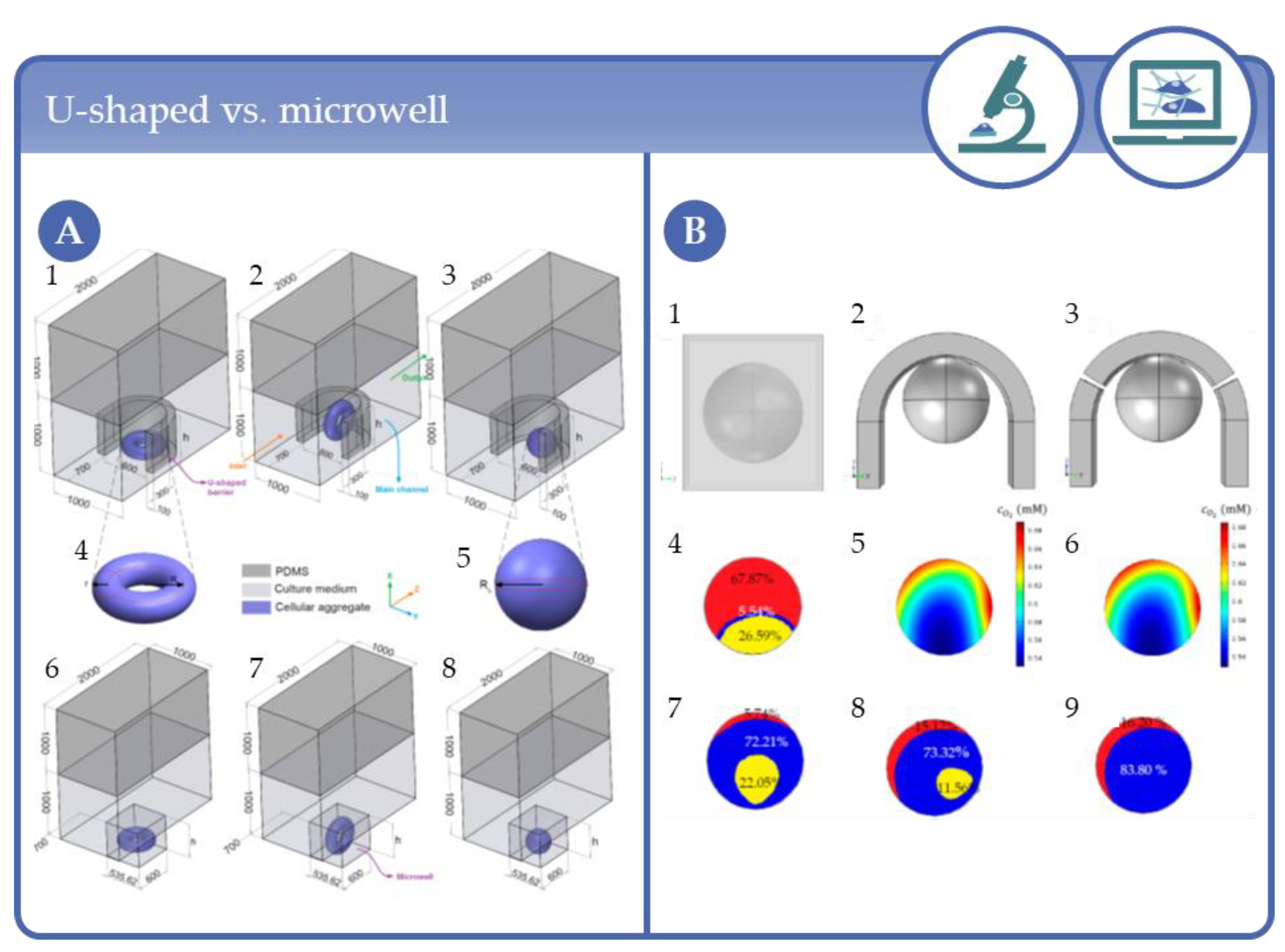
- Barisam, M.; Saidi, M.S.; Kashaninejad, N.; Vadivelu, R.K.; Nguyen, A.V. Numerical Simulation of the Behavior of Toroidal and Spheroidal Multicellular Aggregates in Microfluidic Devices with Microwell and U-Shaped Barrier. Micromachines 2017, 8, 358. [Google Scholar] [CrossRef]
- Barisam, M.; Saidi, M.S.; Kashaninejad, N.; Nguyen, A.V. Prediction of Necrotic Core and Hypoxic Zone of Multicellular Spheroids in a Microbioreactor with a U-Shaped Barrier. Micromachines 2018, 9, 94. [Google Scholar] [CrossRef]
- Lee, S.H.; Hong, S.; Song, J.; Cho, B.; Han, E.J.; Kondapavulur, S.; Kim, D.; Lee, L.P. Microphysiological Analysis Platform of Pancreatic Islet β-Cell Spheroids. Adv. Heal. Mater. 2018, 7, 1701111. [Google Scholar] [CrossRef]
- Cimetta, E.; Sirabella, D.; Yeager, K.; Davidson, K.; Simon, J.; Moon, R.T.; Vunjak-Novakovic, G. Microfluidic bioreactor for dynamic regulation of early mesodermal commitment in human pluripotent stem cells. Lab Chip 2012, 13, 355–364. [Google Scholar] [CrossRef]
- Christoffersson, J.; Bergström, G.; Schwanke, K.; Kempf, H.; Zweigerdt, R.; Mandenius, C.-F. A Microfluidic Bioreactor for Toxicity Testing of Stem Cell Derived 3D Cardiac Bodies. Adv. Struct. Saf. Stud. 2016, 159–168. [Google Scholar] [CrossRef]
- Christoffersson, J.; Mandenius, C.-F. Using a Microfluidic Device for Culture and Drug Toxicity Testing of 3D Cells. Breast Cancer 2019, 1994, 235–241. [Google Scholar] [CrossRef]
- Wen, Y.; Zhang, X.; Yang, S.-T. Microplate-reader compatible perfusion microbioreactor array for modular tissue culture and cytotoxicity assays. Biotechnol. Prog. 2010, 26, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, G.N.; Sikavitsas, V.I.; Dolder, J.V.D.; Sheffield, T.L.; Ambrose, C.G.; Jansen, J.A.; Mikos, A.G. Fluid flow increases mineralized matrix deposition in 3D perfusion culture of marrow stromal osteoblasts in a dose-dependent manner. Proc. Natl. Acad. Sci. USA 2002, 99, 12600–12605. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, G.N.; Sikavitsas, V.I.; Mikos, A.G. Technical Note: Design of a Flow Perfusion Bioreactor System for Bone Tissue-Engineering Applications. Tissue Eng. 2003, 9, 549–554. [Google Scholar] [CrossRef]
- Sikavitsas, V.I.; Bancroft, G.N.; Holtorf, H.L.; Jansen, J.A.; Mikos, A.G. Mineralized matrix deposition by marrow stromal osteoblasts in 3D perfusion culture increases with increasing fluid shear forces. Proc. Natl. Acad. Sci. USA 2003, 100, 14683–14688. [Google Scholar] [CrossRef]
- Frangos, J.A.; McIntire, L.V.; Eskin, S.G. Shear stress induced stimulation of mammalian cell metabolism. Biotechnol. Bioeng. 1988, 32, 1053–1060. [Google Scholar] [CrossRef]
- Gomes, M.E.; Sikavitsas, V.I.; Behravesh, E.; Reis, R.L.; Mikos, A.G. Effect of flow perfusion on the osteogenic differentiation of bone marrow stromal cells cultured on starch-based three-dimensional scaffolds. J. Biomed. Mater. Res. 2003, 67, 87–95. [Google Scholar] [CrossRef]
- Bartnikowski, M.; Klein, T.J.; Melchels, F.P.; Woodruff, M.A. Effects of scaffold architecture on mechanical characteristics and osteoblast response to static and perfusion bioreactor cultures. Biotechnol. Bioeng. 2014, 111, 1440–1451. [Google Scholar] [CrossRef]
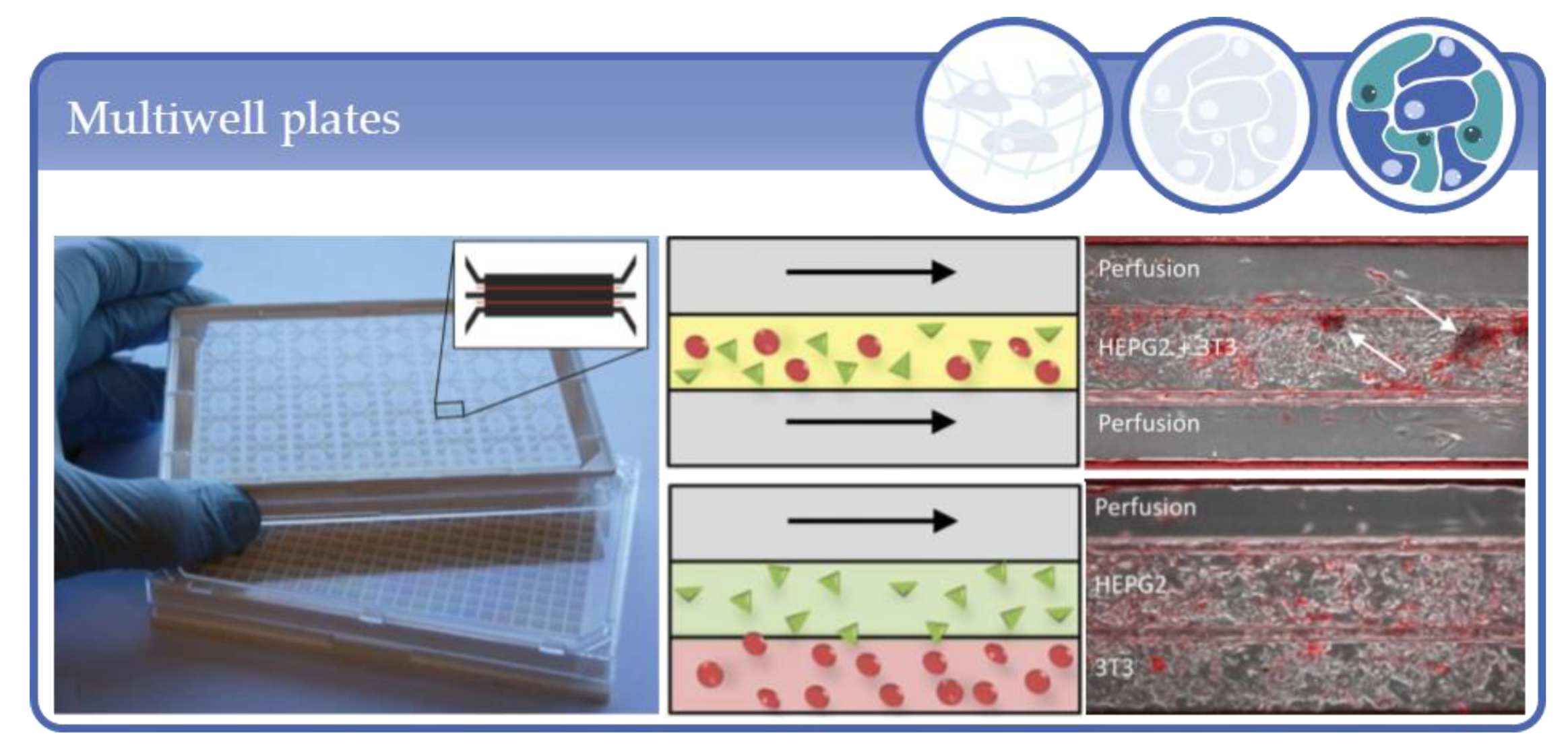
- Leclerc, E.; Sakai, Y.; Fujii, T. Perfusion culture of fetal human hepatocytes in microfluidic environments. Biochem. Eng. J. 2004, 20, 143–148. [Google Scholar] [CrossRef]
- Baudoin, R.; Griscom, L.; Prot, J.M.; Legallais, C.; Leclerc, E. Behavior of HepG2/C3A cell cultures in a microfluidic bioreactor. Biochem. Eng. J. 2011, 53, 172–181. [Google Scholar] [CrossRef]
- Baudoin, R.; Alberto, G.; Legendre, A.; Paullier, P.; Naudot, M.; Fleury, M.-J.; Jacques, S.; Griscom, L.; Leclerc, E. Investigation of expression and activity levels of primary rat hepatocyte detoxication genes under various flow rates and cell densities in microfluidic biochips. Biotechnol. Prog. 2014, 30, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Legendre, A.; Baudoin, R.; Alberto, G.; Paullier, P.; Naudot, M.; Bricks, T.; Brocheton, J.; Jacques, S.; Cotton, J.; Leclerc, E. Metabolic Characterization of Primary Rat Hepatocytes Cultivated in Parallel Microfluidic Biochips. J. Pharm. Sci. 2013, 102, 3264–3276. [Google Scholar] [CrossRef] [PubMed]
- Prot, J.M.; Aninat, C.; Griscom, L.; Razan, F.; Brochot, C.; Guillouzo, C.G.; Legallais, C.; Corlu, A.; Leclerc, E. Improvement of HepG2/C3a cell functions in a microfluidic biochip. Biotechnol. Bioeng. 2011, 108, 1704–1715. [Google Scholar] [CrossRef] [PubMed]
- Prot, J.-M.; Videau, O.; Brochot, C.; Legallais, C.; Bénech, H.; Leclerc, E. A cocktail of metabolic probes demonstrates the relevance of primary human hepatocyte cultures in a microfluidic biochip for pharmaceutical drug screening. Int. J. Pharm. 2011, 408, 67–75. [Google Scholar] [CrossRef]
- Tania, M.; Hsu, M.N.; Png, S.N.; Leo, H.L.; Toh, G.W.; Birgersson, E. Perfusion enhanced polydimethylsiloxane based scaffold cell culturing system for multi-well drug screening platform. Biotechnol. Prog. 2014, 30, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Candini, O.; Grisendi, G.; Foppiani, E.M.; Brogli, M.; Aramini, B.; Masciale, V.; Spano, C.; Petrachi, T.; Veronesi, E.; Conte, P.; et al. A Novel 3D In Vitro Platform for Pre-Clinical Investigations in Drug Testing, Gene Therapy, and Immuno-oncology. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, M.; Grayson, W.L.; Marolt, D.; Gimble, J.M.; Kregar-Velikonja, N.; Vunjak-Novakovic, G. Bone Grafts Engineered from Human Adipose-Derived Stem Cells in Perfusion Bioreactor Culture. Tissue Eng. Part A 2010, 16, 179–189. [Google Scholar] [CrossRef]
- Grayson, W.L.; Marolt, D.; Bhumiratana, S.; Fröhlich, M.; Guo, X.E.; Vunjak-Novakovic, G. Optimizing the medium perfusion rate in bone tissue engineering bioreactors. Biotechnol. Bioeng. 2010, 108, 1159–1170. [Google Scholar] [CrossRef]
- Grayson, W.L.; Bhumiratana, S.; Cannizzaro, C.; Chao, P.-H.G.; Lennon, D.P.; Caplan, A.I.; Vunjak-Novakovic, G. Effects of Initial Seeding Density and Fluid Perfusion Rate on Formation of Tissue-Engineered Bone. Tissue Eng. Part A 2008, 14, 1809–1820. [Google Scholar] [CrossRef]
- Ostrovidov, S.; Jiang, J.; Sakai, Y.; Fujii, T. Membrane-Based PDMS Microbioreactor for Perfused 3D Primary Rat Hepatocyte Cultures. Biomed. Microdevices 2004, 6, 279–287. [Google Scholar] [CrossRef]
- Ostrovidov, S.; Sakai, Y.; Fujii, T. Integration of a pump and an electrical sensor into a membrane-based PDMS microbioreactor for cell culture and drug testing. Biomed. Microdevices 2011, 13, 847–864. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.F.; Vaquette, C.; Baldwin, J.; Chhaya, M.; Gomes, M.E.; Reis, R.L.; Theodoropoulos, C.; Hutmacher, D.W. Biofabrication of customized bone grafts by combination of additive manufacturing and bioreactor knowhow. Biofabrication 2014, 6, 035006. [Google Scholar] [CrossRef] [PubMed]
- De Lora, J.A.; Velasquez, J.L.; Carroll, N.J.; Freyer, J.P.; Shreve, A.P. Centrifugal Generation of Droplet-Based 3D Cell Cultures. SLAS Technol. Transl. Life Sci. Innov. 2020, 25, 436–445. [Google Scholar] [CrossRef]
- Hongo, T.; Kajikawa, M.; Ishida, S.; Ozawa, S.; Ohno, Y.; Sawada, J.-I.; Umezawa, A.; Ishikawa, Y.; Kobayashi, T.; Honda, H. Three-dimensional high-density culture of HepG2 cells in a 5-ml radial-flow bioreactor for construction of artificial liver. J. Biosci. Bioeng. 2005, 99, 237–244. [Google Scholar] [CrossRef]
- Kensah, G.; Gruh, I.; Viering, J.; Schümann, H.; Dahlmann, J.; Meyer, H.; Skvorc, D.; Bär, A.; Akhyari, P.; Heisterkamp, A.; et al. A Novel Miniaturized Multimodal Bioreactor for Continuous In Situ Assessment of Bioartificial Cardiac Tissue During Stimulation and Maturation. Tissue Eng. Part C Methods 2011, 17, 463–473. [Google Scholar] [CrossRef]
- Scaglione, S.; Zerega, B.; Badano, R.; Benatti, U.; Fato, M.M.; Quarto, R. A three-dimensional traction/torsion bioreactor system for tissue engineering. Int. J. Artif. Organs 2010, 33, 362–369. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Shaegh, S.A.M.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef]
- Kleinhans, C.; Mohan, R.R.; Vacun, G.; Schwarz, T.L.; Haller, B.; Sun, Y.; Kahlig, A.; Kluger, P.J.; Finne-Wistrand, A.; Walles, H.; et al. A perfusion bioreactor system efficiently generates cell-loaded bone substitute materials for addressing critical size bone defects. Biotechnol. J. 2015, 10, 1727–1738. [Google Scholar] [CrossRef]
- Linti, C.; Zipfel, A.; Schenk, M.; Dauner, M.; Doser, M.; Viebahn, R.; Becker, H.; Planck, H. Cultivation of Porcine Hepatocytes in Polyurethane Nonwovens as Part of a Biohybrid Liver Support System. Int. J. Artif. Organs 2002, 25, 994–1000. [Google Scholar] [CrossRef]
- Mauney, J.R.; Sjostorm, S.; Blumberg, J.; Horan, R.; O’Leary, J.P.; Vunjak-Novakovic, G.; Volloch, V.; Kaplan, D.L. Mechanical Stimulation Promotes Osteogenic Differentiation of Human Bone Marrow Stromal Cells on 3-D Partially Demineralized Bone Scaffolds In Vitro. Calcif. Tissue Int. 2004, 74, 458–468. [Google Scholar] [CrossRef]
- Driessen-Mol, A.A.; Driessen, N.J.B.; Rutten, M.C.M.; Hoerstrup, S.P.; Bouten, C.V.C.; Baaijens, F.P.T. Tissue Engineering of Human Heart Valve Leaflets: A Novel Bioreactor for a Strain-Based Conditioning Approach. Ann. Biomed. Eng. 2005, 33, 1778–1788. [Google Scholar] [CrossRef]
- A Thompson, C.; Colon-Hernandez, P.; Pomerantseva, I.; MacNeil, B.D.; Nasseri, B.; Vacanti, J.P.; Oesterle, S.N. A Novel Pulsatile, Laminar Flow Bioreactor for the Development of Tissue-Engineered Vascular Structures. Tissue Eng. 2002, 8, 1083–1088. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Henle, P.; Roth, W.; Bock, R.; Boeuf, S.; Richter, W. Design and characterization of a new bioreactor for continuous ultra-slow uniaxial distraction of a three-dimensional scaffold-free stem cell culture. Biotechnol. Prog. 2010, 27, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Wendt, D.; Marsano, A.; Jakob, M.; Heberer, M.; Martin, I. Oscillating perfusion of cell suspensions through three-dimensional scaffolds enhances cell seeding efficiency and uniformity. Biotechnol. Bioeng. 2003, 84, 205–214. [Google Scholar] [CrossRef]
- Saini, S.; Wick, T.M. Concentric Cylinder Bioreactor for Production of Tissue Engineered Cartilage: Effect of Seeding Density and Hydrodynamic Loading on Construct Development. Biotechnol. Prog. 2003, 19, 510–521. [Google Scholar] [CrossRef]
- Wang, B.; Wang, G.; To, F.; Butler, J.R.; Claude, A.; McLaughlin, R.M.; Williams, L.N.; Curry, A.L.D.J.; Liao, J. Myocardial Scaffold-Based Cardiac Tissue Engineering: Application of Coordinated Mechanical and Electrical Stimulations. Langmuir 2013, 29, 11109–11117. [Google Scholar] [CrossRef]
- Santoro, M.; Lamhamedi-Cherradi, S.-E.; Menegaz, B.A.; Ludwig, J.A.; Mikos, A.G. Flow perfusion effects on three-dimensional culture and drug sensitivity of Ewing sarcoma. Proc. Natl. Acad. Sci. USA 2015, 112, 10304–10309. [Google Scholar] [CrossRef]
- Radisic, M.; Euloth, M.; Yang, L.; Langer, R.; Freed, L.E.; Vunjak-Novakovic, G. High-density seeding of myocyte cells for cardiac tissue engineering. Biotechnol. Bioeng. 2003, 82, 403–414. [Google Scholar] [CrossRef]
- Ramadhan, W.; Ohama, Y.; Minamihata, K.; Moriyama, K.; Wakabayashi, R.; Goto, M.; Kamiya, N. Redox-responsive functionalized hydrogel marble for the generation of cellular spheroids. J. Biosci. Bioeng. 2020, 130, 416–423. [Google Scholar] [CrossRef]
- Tostões, R.M.; Leite, S.B.; Miranda, J.P.; Sousa, M.; Wang, D.I.; Carrondo, M.J.; Alves, P.M. Perfusion of 3D encapsulated hepatocytes-A synergistic effect enhancing long-term functionality in bioreactors. Biotechnol. Bioeng. 2011, 108, 41–49. [Google Scholar] [CrossRef]
- Rebelo, S.P.; Costa, R.; Silva, M.M.; Marcelino, P.; Brito, C.; Alves, P.C.; Brito, C. Three-dimensional co-culture of human hepatocytes and mesenchymal stem cells: Improved functionality in long-term bioreactor cultures. J. Tissue Eng. Regen. Med. 2015, 11, 2034–2045. [Google Scholar] [CrossRef] [PubMed]
- Hwa, A.J.; Fry, R.C.; Sivaraman, A.; So, P.T.; Samson, L.D.; Stolz, D.B.; Griffith, L.G. Rat liver sinusoidal endothelial cells survive without exogenous VEGF in 3D perfused co-cultures with hepatocytes. FASEB J. 2007, 21, 2564–2579. [Google Scholar] [CrossRef] [PubMed]
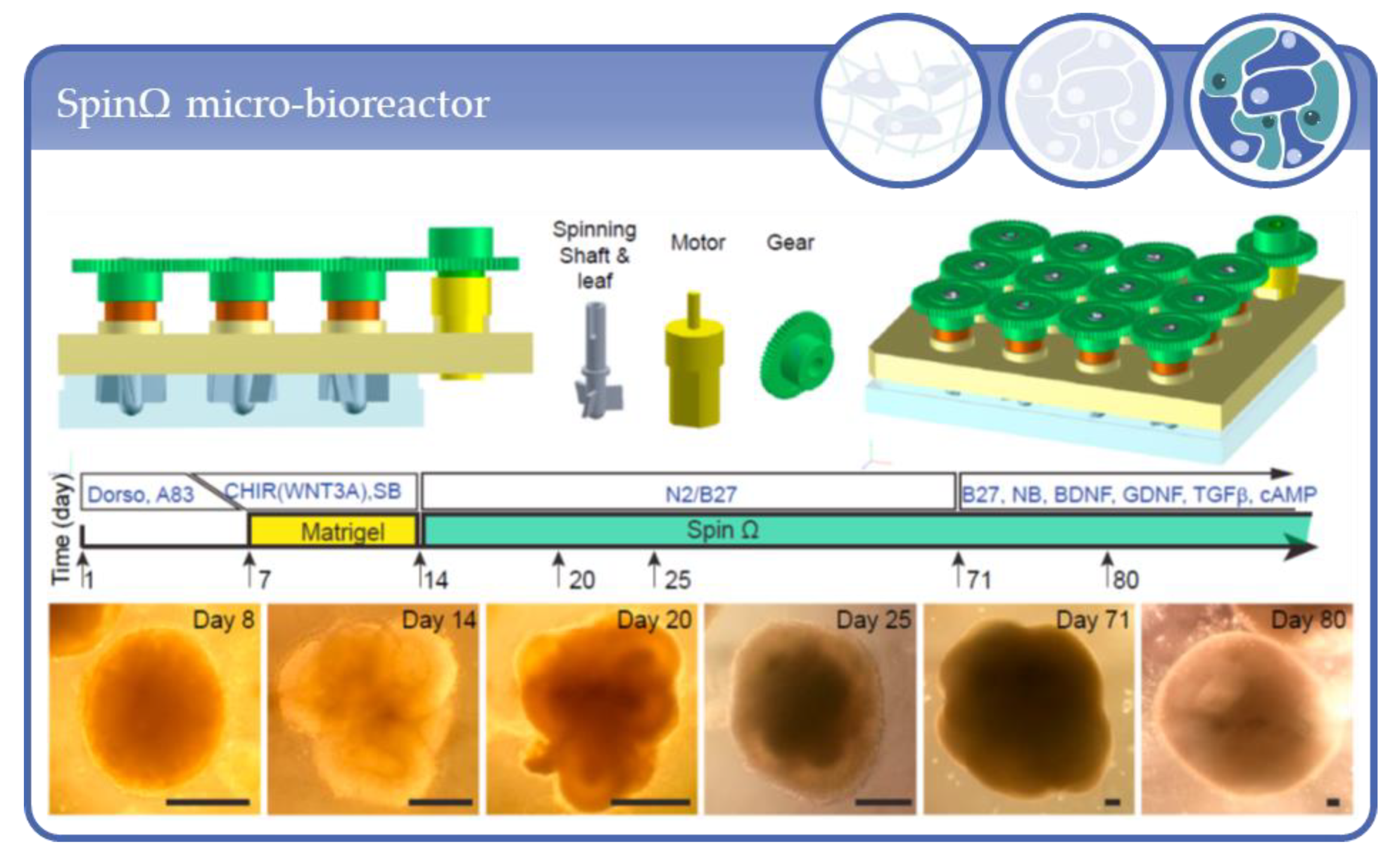
- Qian, X.; Jacob, F.; Song, M.M.; Nguyen, H.N.; Song, H.; Ming, G.-L. Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018, 13, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nat. Cell Biol. 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- DiStefano, T.; Chen, H.Y.; Panebianco, C.; Kaya, K.D.; Brooks, M.J.; Gieser, L.; Morgan, N.Y.; Pohida, T.; Swaroop, A. Accelerated and Improved Differentiation of Retinal Organoids from Pluripotent Stem Cells in Rotating-Wall Vessel Bioreactors. Stem Cell Rep. 2018, 10, 300–313. [Google Scholar] [CrossRef]
- Salerno-Gonçalves, R.; Fasano, A.; Sztein, M.B. Development of a Multicellular Three-dimensional Organotypic Model of the Human Intestinal Mucosa Grown Under Microgravity. J. Vis. Exp. 2016, e54148. [Google Scholar] [CrossRef]
- Devarasetty, M.; Wang, E.; Soker, S.; Skardal, A. Mesenchymal stem cells support growth and organization of host-liver colorectal-tumor organoids and possibly resistance to chemotherapy. Biofabrication 2017, 9, 021002. [Google Scholar] [CrossRef]
- Skardal, A.; Devarasetty, M.; Rodman, C.; Atala, A.; Soker, S. Liver-Tumor Hybrid Organoids for Modeling Tumor Growth and Drug Response In Vitro. Ann. Biomed. Eng. 2015, 43, 2361–2373. [Google Scholar] [CrossRef]
- Barrila, J.; Yang, J.; Crabbé, A.; Sarker, S.F.; Liu, Y.; Ott, C.M.; Nelman-Gonzalez, M.A.; Clemett, S.J.; Nydam, S.D.; Forsyth, R.J.; et al. Three-dimensional organotypic co-culture model of intestinal epithelial cells and macrophages to study Salmonella enterica colonization patterns. NPJ Microgravity 2017, 3, 1–12. [Google Scholar] [CrossRef]
- Crabbé, A.; Sarker, S.F.; Van Houdt, R.; Ott, C.M.; Leys, N.; Cornelis, P.; Nickerson, C.A. Alveolar epithelium protects macrophages from quorum sensing-induced cytotoxicity in a three-dimensional co-culture model. Cell. Microbiol. 2010, 13, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, T.J.; McCarthy, M.; Cohrs, R.J.; Kaufer, B.B. 3D tissue-like assemblies: A novel approach to investigate virus–cell interactions. Methods 2015, 90, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.C.; Mellody, M.; Meneses, L.K.; Hope, A.C.; Dunn, B.S.; Gomperts, B.N. Development of a Three-Dimensional Bioengineering Technology to Generate Lung Tissue for Personalized Disease Modeling. Curr. Protoc. Stem Cell Biol. 2018, 46, e56. [Google Scholar] [CrossRef] [PubMed]
- Schepers, A.; Li, C.; Chhabra, A.; Seney, B.T.; Bhatia, S.N. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip 2016, 16, 2644–2653. [Google Scholar] [CrossRef]
- Ghiaseddin, A.; Pouri, H.; Soleimani, M.; Vasheghani-Farahani, E.; Tafti, H.A.; Hashemi-Najafabadi, S. Cell laden hydrogel construct on-a-chip for mimicry of cardiac tissue in-vitro study. Biochem. Biophys. Res. Commun. 2017, 484, 225–230. [Google Scholar] [CrossRef]
- Goldman, S.M.; Barabino, G. Spatial Engineering of Osteochondral Tissue Constructs Through Microfluidically Directed Differentiation of Mesenchymal Stem Cells. BioResearch Open Access 2016, 5, 109–117. [Google Scholar] [CrossRef]
- Mauleon, G.; Fall, C.P.; Eddington, D.T. Precise Spatial and Temporal Control of Oxygen within In Vitro Brain Slices via Microfluidic Gas Channels. PLoS ONE 2012, 7, e43309. [Google Scholar] [CrossRef]
- Chung, S.; Sudo, R.; Mack, P.J.; Wan, C.-R.; Vickerman, V.; Kamm, R.D. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip 2009, 9, 269–275. [Google Scholar] [CrossRef]
- Van Midwoud, P.M.; Groothuis, G.M.; Merema, M.T.; Verpoorte, E. Microfluidic biochip for the perifusion of precision-cut rat liver slices for metabolism and toxicology studies. Biotechnol. Bioeng. 2010, 105, 184–194. [Google Scholar] [CrossRef]
- Van Midwoud, P.M.; Merema, M.T.; Verpoorte, E.; Groothuis, G.M.M. Microfluidics Enables Small-Scale Tissue-Based Drug Metabolism Studies With Scarce Human Tissue. J. Lab. Autom. 2011, 16, 468–476. [Google Scholar] [CrossRef]
- Visone, R.; Talò, G.; Occhetta, P.; Cruz-Moreira, D.; Lopa, S.; Pappalardo, O.A.; Redaelli, A.; Moretti, M.; Rasponi, M. A microscale biomimetic platform for generation and electro-mechanical stimulation of 3D cardiac microtissues. APL Bioeng. 2018, 2, 046102. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sun, H.; Zhang, J.; Zhang, H.; Meng, F.; Cui, Z. Development of In Vitro 3D TissueFlex® Islet Model for Diabetic Drug Efficacy Testing. PLoS ONE 2013, 8, e72612. [Google Scholar] [CrossRef] [PubMed]
- Trietsch, S.J.; Israëls, G.D.; Joore, J.; Hankemeier, T.; Vulto, P. Microfluidic titer plate for stratified 3D cell culture. Lab Chip 2013, 13, 3548–3554. [Google Scholar] [CrossRef] [PubMed]
- Rieke, M.; Gottwald, E.; Weibezahn, K.-F.; Layer, P.G. Tissue reconstruction in 3D-spheroids from rodent retina in a motion-free, bioreactor-based microstructure. Lab Chip 2008, 8, 2206. [Google Scholar] [CrossRef]
- Gottwald, E.; Nies, C.; Wuchter, P.; Saffrich, R.; Truckenmüller, R.; Giselbrecht, S. A Microcavity Array-Based 3D Model System of the Hematopoietic Stem Cell Niche. Bioinform. MicroRNA Res. 2019, 2017, 85–95. [Google Scholar] [CrossRef]
- Radisic, M.; Marsano, A.; Maidhof, R.; Wang, Y.; Vunjak-Novakovic, G. Cardiac tissue engineering using perfusion bioreactor systems. Nat. Protoc. 2008, 3, 719–738. [Google Scholar] [CrossRef]
- Cheng, M.; Moretti, M.; Engelmayr, G.C.; Freed, L.E. Insulin-like Growth Factor-I and Slow, Bi-directional Perfusion Enhance the Formation of Tissue-Engineered Cardiac Grafts. Tissue Eng. Part A 2009, 15, 645–653. [Google Scholar] [CrossRef]
- Kenar, H.; Kose, G.T.; Toner, M.; Kaplan, D.L.; Hasirci, V. A 3D aligned microfibrous myocardial tissue construct cultured under transient perfusion. Biomaterials 2011, 32, 5320–5329. [Google Scholar] [CrossRef]
- Zhou, W.; Chen, Y.; Roh, T.; Lin, Y.; Ling, S.; Zhao, S.; Lin, J.D.; Khalil, N.; Cairns, D.M.; Manousiouthakis, E.; et al. Multifunctional Bioreactor System for Human Intestine Tissues. ACS Biomater. Sci. Eng. 2018, 4, 231–239. [Google Scholar] [CrossRef]
- Hoerstrup, S.P.; Zund, G.; Sodian, R.; Schnell, A.M.; Grünenfelder, J.; Turina, M.I. Tissue engineering of small caliber vascular grafts. Eur. J. Cardio-Thoracic Surg. 2001, 20, 164–169. [Google Scholar] [CrossRef]
- Pekor, C.; Gerlach, J.C.; Nettleship, I.; Schmelzer, E. Induction of Hepatic and Endothelial Differentiation by Perfusion in a Three-Dimensional Cell Culture Model of Human Fetal Liver. Tissue Eng. Part C Methods 2015, 21, 705–715. [Google Scholar] [CrossRef] [PubMed]
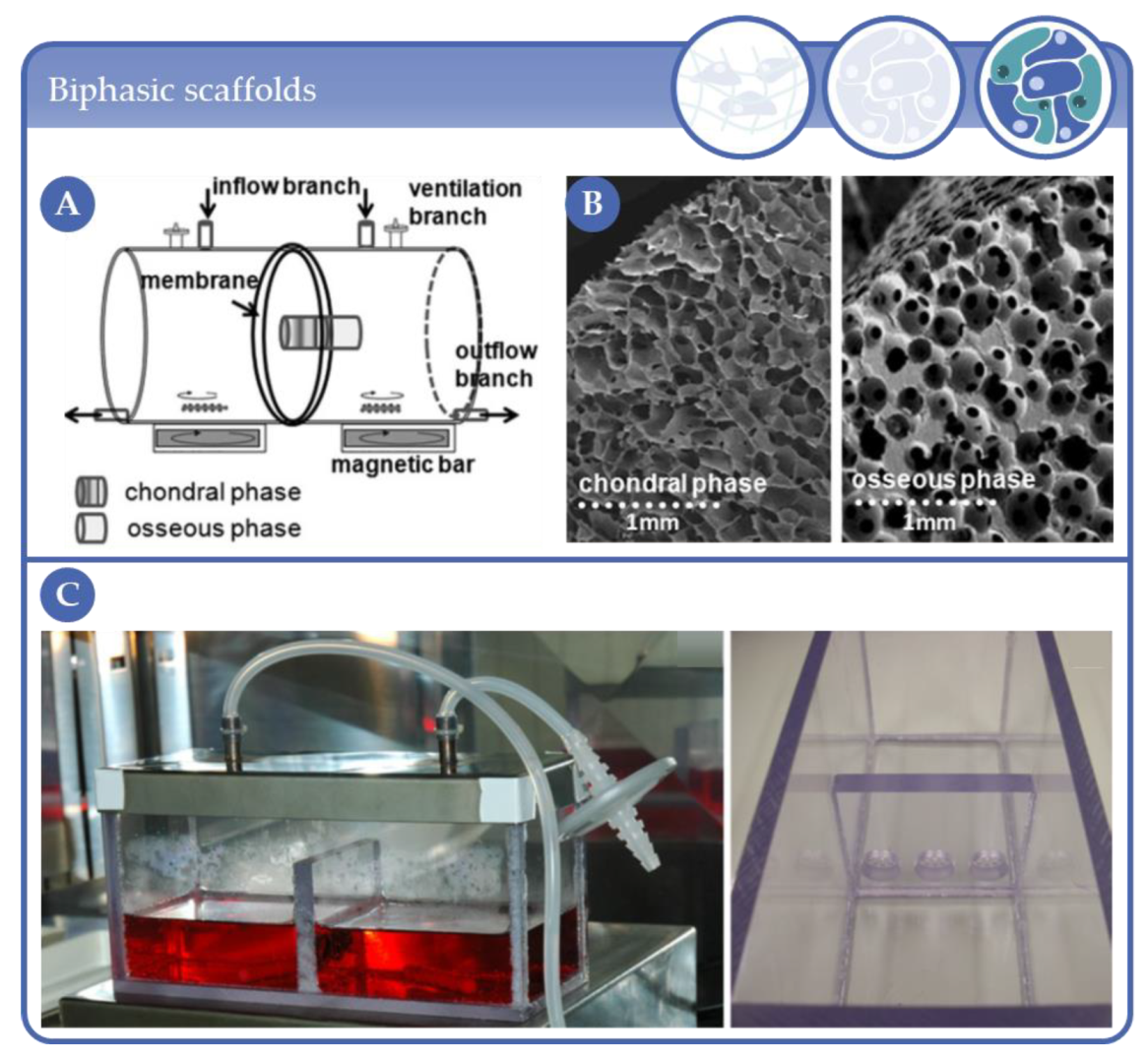
- Liu, X.-G.; Jiang, H.-K. Preparation of an osteochondral composite with mesenchymal stem cells as the single-cell source in a double-chamber bioreactor. Biotechnol. Lett. 2013, 35, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudifar, N.; Doran, P.M. Osteogenic differentiation and osteochondral tissue engineering using human adipose-derived stem cells. Biotechnol. Prog. 2012, 29, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, N.J.; Wang, Q.G.; Cartmell, S.H. A Perfusion Co-Culture Bioreactor for Osteochondral Tissue Engineered Plugs. J. Biomater. Tissue Eng. 2014, 4, 162–171. [Google Scholar] [CrossRef]
- Daley, E.L.; Kuttig, J.; Stegemann, J.P. Development of Modular, Dual-Perfused Osteochondral Constructs for Cartilage Repair. Tissue Eng. Part C Methods 2019, 25, 127–136. [Google Scholar] [CrossRef]
- Daneshgar, A.; Tang, P.; Remde, C.; Lommel, M.; Feufel, M.A.; Kertzscher, U.; Klein, O.; Weinhart, M.; Pratschke, J.; Sauer, I.M.; et al. Teburu—Open source 3D printable bioreactor for tissue slices as dynamic three-dimensional cell culture models. Artif. Organs 2019, 43, 1035–1041. [Google Scholar] [CrossRef]
- Song, J.J.; Guyette, J.P.; E Gilpin, S.; Gonzalez, G.A.; Vacanti, J.P.; Ott, H.C. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat. Med. 2013, 19, 646–651. [Google Scholar] [CrossRef]
- Song, L.; Zhou, Q.; Duan, P.; Guo, P.; Li, D.; Xu, Y.; Li, S.; Luo, F.; Zhang, Z. Successful Development of Small Diameter Tissue-Engineering Vascular Vessels by Our Novel Integrally Designed Pulsatile Perfusion-Based Bioreactor. PLoS ONE 2012, 7, e42569. [Google Scholar] [CrossRef]
- Aizawa, M.; Matsuura, T.; Zhuang, Z. Syntheses of Single-Crystal Apatite Particles with Preferred Orientation to the a- and c-Axes as Models of Hard Tissue and Their Applications. Biol. Pharm. Bull. 2013, 36, 1654–1661. [Google Scholar] [CrossRef]
- Gerlach, J.; Mutig, K.; Sauer, I.M.; Schrade, P.; Efimova, E.; Mieder, T.; Naumann, G.; Grunwald, A.; Pless, G.; Mas, A.; et al. Use of primary human liver cells originating from discarded grafts in a bioreactor for liver support therapy and the prospects of culturing adult liver stem cells in bioreactors: A morphologic study. Transplantation 2003, 76, 781–786. [Google Scholar] [CrossRef]
- Esch, E.W.; Bahinski, A.; Huh, D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015, 14, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, Q.; Zourob, M. Organ-on-a-chip engineering: Toward bridging the gap between lab and industry. Biomicrofluidics 2020, 14, 041501. [Google Scholar] [CrossRef]
- Baert, Y.; Ruetschle, I.; Cools, W.; Oehme, A.; Lorenz, A.; Marx, U.; Goossens, E.; Maschmeyer, I. A multi-organ-chip co-culture of liver and testis equivalents: A first step toward a systemic male reprotoxicity model. Hum. Reprod. 2020, 35, 1029–1044. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.-R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59. [Google Scholar] [CrossRef]
- Ahadian, S.; Civitarese, R.; Bannerman, D.; Mohammadi, M.H.; Lu, R.; Wang, E.; Davenport-Huyer, L.; Lai, B.; Zhang, B.; Zhao, Y.; et al. Organ-On-A-Chip Platforms: A Convergence of Advanced Materials, Cells, and Microscale Technologies. Adv. Heal. Mater. 2018, 7, 7. [Google Scholar] [CrossRef]
- Park, D.; Lee, J.; Chung, J.J.; Jung, Y.; Kim, S.H. Integrating Organs-on-Chips: Multiplexing, Scaling, Vascularization, and Innervation. Trends Biotechnol. 2020, 38, 99–112. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 2018, 22, 310–324. [Google Scholar] [CrossRef]
- Sosa-Hernández, J.E.; Villalba-Rodríguez, A.M.; Romero-Castillo, K.D.; Aguilar-Aguila-Isaías, M.A.; García-Reyes, I.E.; Hernández-Antonio, A.; Ahmed, I.; Sharma, A.; Parra-Saldívar, R.; Iqbal, H.M. Organs-on-a-Chip Module: A Review from the Development and Applications Perspective. Micromachines 2018, 9, 536. [Google Scholar] [CrossRef]
- Terrell, J.A.; Jones, C.G.; Kabandana, G.K.M.; Chen, C. From cells-on-a-chip to organs-on-a-chip: Scaffolding materials for 3D cell culture in microfluidics. J. Mater. Chem. B 2020, 8, 6667–6685. [Google Scholar] [CrossRef] [PubMed]
- Azizipour, N.; Avazpour, R.; Rosenzweig, D.H.; Sawan, M.; Ajji, A. Evolution of Biochip Technology: A Review from Lab-on-a-Chip to Organ-on-a-Chip. Micromachines 2020, 11, 599. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Abouleila, Y.; Si, L.; Ortega-Prieto, A.M.; Mummery, C.L.; Ingber, D.E.; Mashaghi, A. Human Organs-on-Chips for Virology. Trends Microbiol. 2020, 28, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Klak, M.; Bryniarski, T.; Kowalska, P.; Gomolka, M.; Tymicki, G.; Kosowska, K.; Cywoniuk, P.; Dobrzanski, T.; Turowski, P.; Wszoła, M. Novel Strategies in Artificial Organ Development: What is the Future of Medicine? Micromachines 2020, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Allwardt, V.; Ainscough, A.J.; Viswanathan, P.; Sherrod, S.D.; McLean, J.A.; Haddrick, M.; Pensabene, V. Translational Roadmap for the Organs-on-a-Chip Industry toward Broad Adoption. Bioengineering 2020, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Materne, E.-M.; Ramme, A.P.; Terrasso, A.P.; Serra, M.; Alves, P.M.; Brito, C.; Sakharov, D.A.; Tonevitsky, A.; Lauster, R.; Marx, U. A multi-organ chip co-culture of neurospheres and liver equivalents for long-term substance testing. J. Biotechnol. 2015, 205, 36–46. [Google Scholar] [CrossRef]
- Schimek, K.; Frentzel, S.; Luettich, K.; Bovard, D.; Rütschle, I.; Boden, L.; Rambo, F.; Erfurth, H.; Dehne, E.-M.; Winter, A.; et al. Human multi-organ chip co-culture of bronchial lung culture and liver spheroids for substance exposure studies. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- van den Berg, A.; Mummery, C.L.; Passier, R.; Van Der Meer, A.D. Personalised organs-on-chips: Functional testing for precision medicine. Lab Chip 2018, 19, 198–205. [Google Scholar] [CrossRef]
- Geraili, A.; Jafari, P.; Hassani, M.S.; Araghi, B.H.; Mohammadi, M.H.; Ghafari, A.M.; Tamrin, S.H.; Modarres, H.P.; Kolahchi, A.R.; Ahadian, S.; et al. Controlling Differentiation of Stem Cells for Developing Personalized Organ-on-Chip Platforms. Adv. Heal. Mater. 2017, 7, 1700426. [Google Scholar] [CrossRef]
- Smirnova, L.; Kleinstreuer, N.; Corvi, R.; Levchenko, A.; Fitzpatrick, S.C.; Hartung, T. 3S—Systematic, systemic, and systems biology and toxicology. ALTEX 2018, 35, 139–162. [Google Scholar] [CrossRef]
- Ferrari, E.; Palma, C.; Vesentini, S.; Occhetta, P.; Rasponi, M. Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems. Biosensors 2020, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2020, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fetah, K.L.; DiPardo, B.J.; Kongadzem, E.; Tomlinson, J.S.; Elzagheid, A.; Elmusrati, M.; Khademhosseini, A.; Ashammakhi, N. Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration. Small 2019, 15, e1901985. [Google Scholar] [CrossRef] [PubMed]
- Swayden, M.; Soubeyran, P.; Iovanna, J. Upcoming Revolutionary Paths in Preclinical Modeling of Pancreatic Adenocarcinoma. Front. Oncol. 2020, 9, 1443. [Google Scholar] [CrossRef]
- Williams, K.A.; Saini, S.; Wick, T.M. Computational Fluid Dynamics Modeling of Steady-State Momentum and Mass Transport in a Bioreactor for Cartilage Tissue Engineering. Biotechnol. Prog. 2002, 18, 951–963. [Google Scholar] [CrossRef]
- Sucosky, P.; Osorio, D.F.; Brown, J.B.; Neitzel, G.P. Fluid mechanics of a spinner-flask bioreactor. Biotechnol. Bioeng. 2004, 85, 34–46. [Google Scholar] [CrossRef]
- Yu, P.; Lee, T.; Zeng, Y.; Low, H. A 3D analysis of oxygen transfer in a low-cost micro-bioreactor for animal cell suspension culture. Comput. Methods Programs Biomed. 2007, 85, 59–68. [Google Scholar] [CrossRef]
- Tajsoleiman, T.; Abdekhodaie, M.J.; Gernaey, K.V.; Krühne, U. Efficient Computational Design of a Scaffold for Cartilage Cell Regeneration. Bioengineering 2018, 5, 33. [Google Scholar] [CrossRef]
- Magrofuoco, E.; Elvassore, N.; Doyle, F.J. Theoretical analysis of insulin-dependent glucose uptake heterogeneity in 3D bioreactor cell culture. Biotechnol. Prog. 2012, 28, 833–845. [Google Scholar] [CrossRef]
- Porter, B.; Zauel, R.; Stockman, H.; Guldberg, R.; Fyhrie, D. 3-D computational modeling of media flow through scaffolds in a perfusion bioreactor. J. Biomech. 2005, 38, 543–549. [Google Scholar] [CrossRef]
- Raimondi, M.T.; Moretti, M.; Cioffi, M.; Giordano, C.; Boschetti, F.; Laganà, K.; Pietrabissa, R. The effect of hydrodynamic shear on 3D engineered chondrocyte systems subject to direct perfusion. Biorheology 2006, 43, 215–222. [Google Scholar] [PubMed]
- Hyndman, L.; McKee, S.; Mottram, N.J.; Singh, B.; Webb, S.D.; McGinty, S. Mathematical modelling of fluid flow and solute transport to define operating parameters for in vitro perfusion cell culture systems. Interface Focus 2020, 10, 20190045. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Eckert, H.; Hess, R.; Gelinsky, M.; Rancourt, D.; Krawetz, R.; Cuniberti, G.; Scharnweber, D. Developing a Customized Perfusion Bioreactor Prototype with Controlled Positional Variability in Oxygen Partial Pressure for Bone and Cartilage Tissue Engineering. Tissue Eng. Part C Methods 2017, 23, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Nam, J.; Sun, W. Direct Cell Writing of 3D Microorgan for In Vitro Pharmacokinetic Model. Tissue Eng. Part C Methods 2008, 14, 157–166. [Google Scholar] [CrossRef]
- Zhao, F.; Chella, R.; Ma, T. Effects of shear stress on 3-D human mesenchymal stem cell construct development in a perfusion bioreactor system: Experiments and hydrodynamic modeling. Biotechnol. Bioeng. 2006, 96, 584–595. [Google Scholar] [CrossRef]
- Hassan, C.R.; Qin, Y.; Komatsu, D.E.; Uddin, S.M.Z. Utilization of Finite Element Analysis for Articular Cartilage Tissue Engineering. Materials 2019, 12, 3331. [Google Scholar] [CrossRef]
- Raies, B.A.; Bajic, V.B. In silicotoxicology: Computational methods for the prediction of chemical toxicity. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2016, 6, 147–172. [Google Scholar] [CrossRef]
- Jean-Quartier, C.; Jeanquartier, F.; Jurisica, I.; Holzinger, A. In silico cancer research towards 3R. BMC Cancer 2018, 18, 1–12. [Google Scholar] [CrossRef]








| µCT | Micro Computed Tomography |
|---|---|
| 5-FU | 5-fluoruracil |
| ALP | alkaline phosphatase |
| BBB | blood-brain-barrier |
| BCT | bioartificial cardiac tissue |
| CFD | computational fluidic dynamics |
| cfDNA | cell free DNA |
| CVB | coxsackievirus B |
| ECM | extracellular matrix |
| FEA | finite element analysis |
| HARV | high aspect ratio vessel |
| hASCs | human adipose stem cells |
| hBMCs | human bone marrow cells |
| HCV | hepatitis C virus |
| hEB | human embryoid bodies |
| hMSCs | human mesenchymal stem cells |
| HNCs | human nasal chondrocytes |
| HSPCs | hematopoietic stem/progenitor cells |
| HTS | high-throughput-screening |
| IDCCM | integrated dynamic cell culture microchip |
| iPSC | induced pluripotent stem cells |
| LB | Lattice-Boltzmann |
| LSEC | liver sinusoidal endothelial cells |
| MAP | microfluidic analysis platform |
| MBR | micro-bioreactor |
| MOAB | magnetic optically assessible bioreactor |
| MPPS | micro-pathophysiological systems |
| MRI | magnetic resonance imaging |
| NASA | national aeronautics and space administration |
| OoC | organ-on-a-chip |
| PBM | pipe based microbioreactor |
| PDMS | polydimethylsiloxan |
| PEG | polyethylen glycol |
| PEGDA | poly(ethylene glycol) diacrylate |
| PET | polyethylene terephtalate |
| PGA | polyglycolic acid |
| PGS | poly(glycerol sebacetate) |
| PIC | perfusion incubator liver chip |
| PLA | poly(lactic acid) |
| PLGA | poly(lactic-co-glycolic acid) |
| PLLA | poly(L-lactic acid) |
| PTFE | polytetrafluorethene |
| PU | polyurethane |
| RCCS | rotary cell culture system |
| ROS | reactive oxygen species |
| RWV | rotating wall vessel |
| SEM | scanning electron microscope |
| SEVA-C | blend of starch with ethylene vinyl alcohol |
| SPCL | blend of starch with poly(ε-caprolactone) |
| STLV | slow turning lateral vessel |
| TE | tissue engineering |
| TLA | tissue-like assemblies |
| ZIKV | Zika virus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altmann, B.; Grün, C.; Nies, C.; Gottwald, E. Advanced 3D Cell Culture Techniques in Micro-Bioreactors, Part II: Systems and Applications. Processes 2021, 9, 21. https://doi.org/10.3390/pr9010021
Altmann B, Grün C, Nies C, Gottwald E. Advanced 3D Cell Culture Techniques in Micro-Bioreactors, Part II: Systems and Applications. Processes. 2021; 9(1):21. https://doi.org/10.3390/pr9010021
Chicago/Turabian StyleAltmann, Brigitte, Christoph Grün, Cordula Nies, and Eric Gottwald. 2021. "Advanced 3D Cell Culture Techniques in Micro-Bioreactors, Part II: Systems and Applications" Processes 9, no. 1: 21. https://doi.org/10.3390/pr9010021
APA StyleAltmann, B., Grün, C., Nies, C., & Gottwald, E. (2021). Advanced 3D Cell Culture Techniques in Micro-Bioreactors, Part II: Systems and Applications. Processes, 9(1), 21. https://doi.org/10.3390/pr9010021







