Abstract
The aim of this study was preparation of new derivatives based on 2-((4-chlorophenoxy)methyl)-N-(arylcarbamothioyl)benzamide structure; the new compounds were characterized by IR, NMR (1H, 13C) spectroscopy, and elemental analysis. The obtained compounds were evaluated for their in vitro antimicrobial activity against planktonic and biofilm-embedded microbial cells (Staphylococcus aureus, Enterococcus faecalis, Escherichia coli, Pseudomonas aeruginosa, Candida albicans), by qualitative and quantitative assays. Some of the compounds revealed promising antibacterial and antifungal activities, with low minimum inhibitory concentration values between 0.15 and 2.5 mg/mL and minimal biofilm eradication concentrations of 0.019–2.5 mg/mL. To investigate the potential target of their antibacterial activity, in silico drug-likeness and molecular docking screenings on Staphylococcus aureus DNA gyrase were performed. The compound with the best antibacterial activity (1g) was docked into topoisomerase II DNA gyrase enzymes (PDB ID: 2XCS) and showed valuable interactions with the target protein along with good docking scores, suggesting that it can act by the inhibition of DNA replication. The tested compounds exhibited only a poor antioxidant activity, as revealed by the in vitro assay using 1,1-diphenyl-2-picrylhydrazyl (DPPH) assay.
1. Introduction
The thiourea skeleton is found in the structure of some drugs, such are noxytiolin [1], an anti-infective drug, sulfathiourea [2], a bacteriostatic sulfonamide derivative, loflucarban [3] with a thiocarbanilide structure, having antifungal properties, thiophanate [4] and thiophanate-methyl [5], two drugs with fungicide and nematicidal effect.
Thiocarlide [6] is a thiourea drug known as an effective anti-tuberculosis drug, active against a range of multidrug-resistant strains of Mycobacterium tuberculosis which has been used clinically.
Burimamide [7] and metiamide [8] are histamine antagonists used for the treatment of peptic ulcers, underlying the molecular design of cimetidine.
Representative structures containing a thiourea fragment for non-nucleoside reverse transcriptase inhibitors (NNRTIs) are phenethylthiazolylthiourea [9] and trovirdine [10]. The thiourea derivatives with different pharmacological properties are summarized in Figure 1.
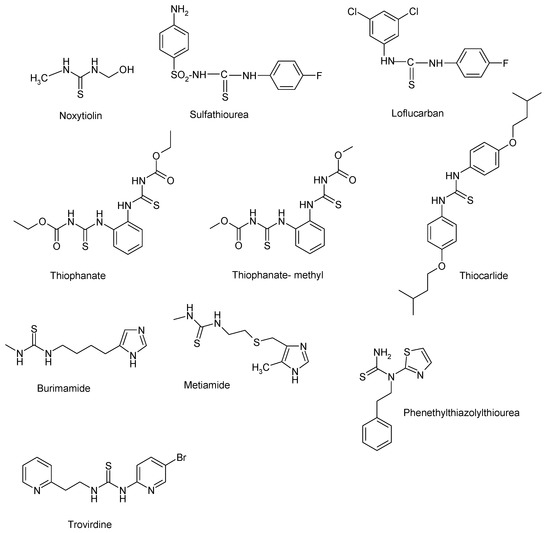
Figure 1.
Thiourea derivatives with pharmacological properties.
Thiourea derivatives can have a wide range of applications in the pharmaceutical field, due to their diverse biological properties, such as antibacterial [11,12,13,14,15], antifungal [16,17,18], antimalarial [19,20], and anthelmintic [21]. These derivatives are also known for their anticancer [22,23,24,25], anticonvulsant [26,27,28], analgesic, anti-inflammatory [29,30], and antioxidant [31,32,33] effects. Their applications are extending beyond the pharmaceutical field, some research paper highlighting their possible use as insecticidal [34], herbicidal [35,36], and plant growth regulatory agents [37].
Also, the thiourea derivatives can be considered as building blocks in the synthesis of heterocyclic compounds [38,39].
In meeting the need to discover new and more effective antioxidant therapies, some of the thiourea derivatives compounds were developed and tested using different in vitro methods: 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenging assay, nitric oxide (NO) radical scavenging assay, 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) radical scavenging assay, superoxide anion scavenging assay in order to evaluate their free-radical scavenging activity.
In a recent study, it was demonstrated that thiourea derivatives of 2,3-dihydro-1H-inden-1-amine with phenyl ring, 4-fluoro-phenyl ring and 4-nitro-phenyl ring exhibit promising antioxidant activity [40] Thiourea derivatives of 6-fluoro-3-(piperidin-4-yl)benzo[d]isoxazole which have the phenyl ring substituted with fluorine atom and trifluoromethyl group (electron withdrawing properties), showed good antibacterial, antifungal and antioxidant activity [41], and some bis(thiourea) compounds have been tested for their antioxidant activity and N,N’-bis(o-fluorobenzamidothiocarbonyl)hydrazine has the high antioxidant activity than ascorbic acid, probably due to the presence of fluorine substituted in ortho position [42].
Antioxidant activities of the Co(II), Ni(II) and Cu(II) complexes of N-((6-methylpyridin-2-yl)carbamothioyl)thiophene-2-carboxamide were determined using DPPH and ABTS assays. Bis(N-((6-methylpyridin-2-yl)carbamothioyl)thiophene-2-carboxamide) cobalt(II) shows good antioxidant properties, better than those of propyl gallate and worse than those of vitamin E [43]. In conclusion, compounds containing the thiourea skeleton in the molecule can be valuable candidates for future chemical modulation in order to achieve different biological actions.
Thiazolidine and pyrrolidine compounds containing a thiourea moiety were tested in antioxidant experiments, by the DPPH and ABTS radical scavenging assays at different concentrations, and were shown to be useful for the prevention of oxidative stress [44].
Based on this research, we consider that thiourea derivatives could prevent or decrease the damage to the human body caused by free radicals, which attack biological molecules by hydrogen donation or by acting as electron donors. We determined the antioxidant potential of the synthesized compounds by the DPPH method, knowing that antioxidants can scavenge DPPH radicals by hydrogen donation, which causes a decrease in DPPH absorbance. The DPPH radicals are stable free radicals commonly used as substrates to evaluate in vitro antioxidant activity and to evaluate the ability of antioxidants to scavenge free radicals, which are known to be a major factor in the biological damage caused by oxidative stress.
As a continuation of our previous research [45,46,47,48], we have synthesized new 2-((4-chlorophenoxy)methyl)-N-(arylcarbamothioyl)benzamides containing fluorine and trifluoromethyl group as substituents on aryl radical. Antimicrobial, antioxidant and in silico drug-likeness and molecular docking studies were performed.
2. Materials and Methods
2.1. Chemistry
The chemicals were purchased from Merck (Darmstadt, Germany) and Sigma Aldrich (St. Louis, MO, USA) and were used as received, excepting acetone and 1,2-dichloroethane which were distilled from the drying agents according to conventional methods and also ammonium thiocyanate was dried by heating at 100 °C.
The melting points were determined on an Electrothermal 9100 capillary melting point apparatus (Bibby Scientific Ltd., Stone, UK) in open capillary tubes and the reported values are uncorrected.
The elemental analyses were performed using a PerkinElmer 2400 Series II CHNS/O Elemental Analyzer (Waltham, MA, USA).
The FT-IR spectra were recorded on Bruker Vertex 70 FT-IR spectrometer (Bruker Corporation, Billerica, MA, USA).
The NMR spectra were recorded on a Varian Gemini 300BB instrument (Varian Medical Systems, Palo Alto, CA, USA), at 300 MHz for 1H NMR and at 75 MHz for 13C NMR, as δ values in parts per million (ppm), using as solvent hexadeuterodimethylsulfoxide and tetramethylsilane as internal standard.
2.1.1. General Synthesis Method of the New 2-((4-Chlorophenoxy)methyl)-N-(Arylcarbamothioyl) Benzamides
The 2-(4-chlorophenoxymethyl)benzoic acid and the 2-(4-chlorophenoxymethyl)benzoyl chloride were synthesized according to the method presented in a previous paper [49]. Only 2-(4-chlorophenoxymethyl)benzoic acid was isolated, the reaction yield being 51% (calculated based on phtalide amount).
In a round- bottom flask, equipped with a heating mantle, water condenser and calcium chloride drying tube on top of the condenser, was placed a solution of 2-(4-chlorophenoxymethyl)benzoyl chloride (0.01 mol) in anhydrous acetone (15 mL). A solution of dried ammonium thiocyanate (0.01 mol) in anhydrous acetone (5 mL) was added to this solution. The reaction mixture was refluxed for 1 h and then cooled at the room temperature. A solution of the aromatic primary amine in anhydrous acetone was added to this mixture. The reaction mixture was heated for one more hour. The resulted benzamide is precipitated by pouring into cold water. The crude product was purified by crystallization from isopropanol, in the presence of charcoal. The reaction yields were calculated based on the used amine amount.
2.1.2. Spectral data
2-((4-Chlorophenoxy)methyl)-N-(3-fluorophenylcarbamothioyl)benzamide (1a). Yield 74%; mp 135.1–136.3 °C;
1H-NMR (dmso-d6, δ ppm, J Hz, T = 298K): 12.47 (br s, 1H, NH, deuterable); 11.91 (br s, 1H, NH, deuterable); 7.70 (dt, 3J(F-H18) = 10.7 Hz, 4J(H20-H18) = J(H22-H18) = 1.4 Hz, 1H, H-18); 7.65 (dd, J = 1.3 Hz, J = 7.5 Hz, 1H, H-7); 7.61–7.55 (m, 2H, H-4, H-5); 7.49 (td, J = 1.4 Hz, J = 7.5 Hz, 1H, H-6); 7.43 (dd, 3J(H21-H20) = 6.7 Hz, J(F-H20) = 8.3 Hz, 1H, H-20); 7.31 (d, J = 8.9 Hz, 2H, H-11, H-13); 7.11 (td, J(H20,22-H21) = 7.6 Hz, J(F-H22) = 2.4 Hz, 1H, H-21); 7.01 (d, J = 8.9 Hz, 2H, H-10, H-14); 5.32 (s, 2H, H-8).
13C-NMR (dmso-d6, δ ppm, T = 298 K): 178.97 (C-16); 169.95 (C-1); 161.61 (d, J(F-C19) = 245.3 Hz, C-19); 157.00 (C-9); 139.45 (d, 4J(F-C17) = 11.2 Hz, C-17); 135.30 (Cq); 133.20 (Cq); 124.70 (Cq); 131.13 (CH); 130.23 (d, J(F-C21) = 9.2 Hz, C-21); 129.21 (C-11, C-13); 128.58 (CH); 128.42 (CH); 127.89 (CH); 116.44 (C-10, C-14); 120.15 (C-22); 112.91 (d, J(F-C18) = 21.2 Hz, C-18); 110.06 (d, J(F-C20) = 25.5 Hz, C-20); 67.76 (C-8).
FT-IR (solid in ATR, ν cm−1): 3240 m; 3095 w; 3036 w; 1665 m; 1608 s; 1567 vs; 1527 vs; 1491 vs; 1452 m; 1347 m; 1328 m; 1312 m; 1264 s; 1243 vs; 1155 s; 1031 m; 904 w; 859 w; 814 m; 783 w; 767 w; 730 w; 709 w; 692 m; 672 m.
Anal. calcd. for C21H16ClFN2O2S (414.88): C, 60.80; H, 3.89; N, 6.75; S, 7.73%; Found: C, 60.65; H, 3.97; N, 6.79; S 7.76%.
2-((4-Chlorophenoxy)methyl)-N-(4-fluorophenylcarbamothioyl)benzamide (1b). Yield 56%; mp 137.3–138.6 °C;
1H-NMR (dmso-d6, δ ppm, J Hz, T = 298 K): 12.28 (s, 1H, NH, deuterable); 11.85 (s, 1H, NH, deuterable); 7.62 (dd, J = 1.2 Hz, J = 7.4 Hz, 1H, H-7); 7.60–7.53 (m, 4H, H-4, H-5, H-18, H-22); 7.48 (td, J = 1.4 Hz, J = 7.4 Hz, 1H, H-6); 7.32 (d, J = 9.0 Hz, 2H, H-11, H-13); 7.24 (t, J(F-H19,21) = J(H19,21-H18,22) = 8.5, 2H, H-19, H-21); 7.01 (d, J = 9.0 Hz, 2H, H-10, H-14); 5.31 (s, 2H, H-8).
13C-NMR (dmso-d6, δ ppm, T = 298 K): 179.47 (C-16); 169.94 (C-1); 159.93 (d, J(F-C20) = 243.7 Hz, C-20); 157.00 (C-9); 135.25 (Cq); 133.27 (Cq); 132.17 (d, J(F-C17) = 8.9Hz, C-17); 124.71 (Cq); 131.05 (CH); 129.20 (C-11, C-13); 128.53 (CH); 128.41 (CH); 127.86 (CH); 126.76 (d, 3J(F-C18,22) = 8.3 Hz, C-18, C-22); 116.46 (C-10, C-14); 115.29 (d, J(F-C19,21) = 22.6 Hz, C19, C-21); 67.79 (C-8).
FT-IR (solid in ATR, ν cm−1): 3148 s; 3026 m; 2889 m; 1684 s; 1596 m; 1525 vs; 1508 vs; 1489 vs; 1411 m; 1378 m; 1326 m; 1286 m; 1244 m; 1228 m; 1163 s; 1001 m; 1079 m; 1004 m; 825 m; 794 w; 777 w; 745 m; 718 m; 694 m; 661 m.
Anal. calcd. for C21H16ClFN2O2S (414.88): C, 60.80; H, 3.89; N, 6.75; S, 7.73%; Found: C, 60.97; H, 3.92; N, 6.71; S 7.72%.
2-((4-Chlorophenoxy)methyl)-N-(2,3,4-trifluorophenylcarbamothioyl)benzamide (1c). Yield 79%; mp 134.1–134.9 °C;
1H-NMR (dmso-d6, δ ppm, J Hz, T = 298 K): 12.00 (br s, 2H, NH, deuterable); 7.65 (dd, J = 1.3 Hz, J = 7.5, 1 H, H-7); 7.61–7.55 (m, 2H, H-4, H-5); 7.55 (m, 1H, H-22); 7.49 (td, J = 1.4 Hz, J = 7.5 Hz, 1H, H-6); 7.39 (tdd, J(F20-H21)= J(H22-H21) = 9.2 Hz, J(F18-H21) = 2.6 Hz, J(F19-H21) = 4.8 Hz, 1H, H-21); 7.31 (d, J = 8.9 Hz, 2H, H-11, H-13); 7.01 (d, J = 8.9 Hz, 2H, H-10, H-14); 5.31 (s, 2H, H-8).
13C-NMR (dmso-d6, δ ppm, T = 298K): 181.04 (C-16); 169.98 (C-1); 156.94 (C-9); 148.79 (ddd, 3J(F20-C18) = 2.5 Hz, 2J(F19-C18) = 9.8 Hz, J(F18-C18) = 246.8 Hz, C-18); 145.43 (ddd, J(F18-C20) = 3.7 Hz, J(F19-C20) = 10.8 Hz, J(F20-C20) = 249.9 Hz, C-20); 139.19 (ddd, J(F18(20)-C19) = 14.4 Hz, J(F20(18)-C19) = 16.5 Hz, J(F19-C19) = 248.1 Hz, C-19); 135.26 (Cq); 133.06 (Cq); 124.70 (Cq); 124.22 (dd, J(F19-C17) = 3.8 Hz, J(F18-C17) = 9.2 Hz, C-17); 131.11 (CH); 129.16 (C-11, C-13); 128.61 (CH); 128.49 (CH); 127.88 (CH); 116.44 (C-10, C-14); 122.72 (td, J(F18-C22) = J(F20-C22) = 7.5 Hz, J(F19-C22) = 3.4 Hz, C-22); 111.62 (dd, J(F20-C21) = 18.0 Hz, J(F19-C21) = 3.9 Hz, C-21); 67.81 (C-8).
FT-IR (solid in ATR, ν cm−1): 3136 m; 3067 m; 1681 m; 1531 s; 1514 vs; 1486 vs; 1453 m; 1321 m; 1325 s; 1304 m; 1271 m; 1237 s; 1178 s; 1150 s; 1050 m; 999 m; 903 w; 827 w; 749 s; 731 m; 657 w.
Anal. calcd. for C21H14ClF3N2O2S (450.86): C, 55.94; H, 3.13; N, 6.21; S, 7.11%; Found: C, 55.75; H, 3.11; N, 6.29; S 7.14%.
2-((4-Chlorophenoxy)methyl)-N-(2,4,5-trifluorophenylcarbamothioyl)benzamide (1d). Yield 65%; mp 158.4- 159.2 °C;
1H-NMR (dmso-d6, δ ppm, J Hz, T = 298 K): 12.27 (br s, 1H, NH, deuterable); 12.12 (br s, 1H, NH, deuterable); 8.07 (ddd, 4J(H22-F18) = 6.4 Hz, 4J(H22-F20) = 8.7 Hz, 3J(H22-F21) = 11.7 Hz, 1H, H-22); 7.73 (td, 3J(H19-F18) = 3J(H19-F20) = 10.4 Hz, 4J(H19-F21) = 7.7 Hz, 1H, H-19); 7.63 (br d, J = 7.3 Hz, 1H, H-7); 7.56–7.61 (m, 2H, H-4, H-5); 7.48 (m, 1H, H-6); 7.30 (d, J = 9.0 Hz, 2H, H-11, H-13); 6.99 (d, J = 9.0 Hz, 2H, H-10, H-14); 5.30 (s, 2H, H-8).
13C-NMR (dmso-d6, δ ppm, T = 298 K): 180.40 (C-16); 170.21 (C-1); 156.97 (C-9); 151.27 (ddd, J(F21-C21) = 248.5 Hz, J(F20-C21) = 8.0 Hz, J(F18-C21) = 3.0 Hz, C-21); 147.25 (ddd, J(F20-C20) = 248.3 Hz, J(F21-C20) = 14.2 Hz, J(F18-C20) = 12.4 Hz, C-20); 144.98 (ddd, J(F18-C18) = 241.8 Hz, J(F20-C18) = 13.5 Hz, J(F21-C18) = 3.4 Hz, C-18); 135.31 (Cq); 133.07 (Cq); 131.19 (CH); 129.19 (C-11, C-13); 128.64 (CH); 128.55 (CH); 127.95 (CH); 124.71 (C-12); 122.90 (ddd, J(F20-C17) = 3.9 Hz, J(F21-C17) = 9.2 Hz, J(F18-C17) = 11.3 Hz, C-17); 116.46 (C-10, C-14); 115.45 (ddd, J(F18-C22) = 0.7 Hz, J(F20-C22) = 1.8 Hz, J(F21-C22) = 22.2 Hz, C-22); 106.01 (dd, J(F20-C19) = 22.3 Hz, J(F18-C19) = 26.6 Hz, C-19); 67.83 (C-8).
FT-IR (solid in ATR, ν cm−1): 3243 w; 3071 w; 2962 w; 1676 m; 1568 vs; 1526 s; 1490 vs; 1439 m; 1322 s; 1238s; 1217 s; 1158 s; 1150 s; 1033 m; 870 m; 707 m; 731 m.
Anal. calcd. for C21H14ClF3N2O2S (450.86): C, 55.94; H, 3.13; N, 6.21; S, 7.11%; Found: C, 56.11; H, 3.13; N, 6.19; S 7.13%.
2-((4-Chlorophenoxy)methyl)-N-(2-trifluoromethylphenylcarbamothioyl)benzamide (1e). Yield 59%; mp 125–126.2 °C;
1H-NMR (dmso-d6, δ ppm, J Hz, T = 298 K): 12.41 (br s, 1H, NH, deuterable); 12.14 (br s, 1H, NH, deuterable); 7.79 (dq, J = 7.9, 1H, H-19); 7.76–7.66 (m, 2H, H-21, H-22); 7.64 (br d, J = 7.4 Hz, 1H, H-7); 7.62–7.45 (m, 4H, H-4, H-5, H-19, H-6); 7.33 (d, J = 9.0 Hz, 2H, H-11, H-13); 7.00 (d, J = 9.0 Hz, 2H, H-10, H-14); 5.30 (s, 2H, H-8).
13C-NMR (dmso-d6, δ ppm, T = 298 K): 181.41 (C-16); 170.60 (C-1); 156.99 (C-19); 135.71 (q, C-17, J(3F-C17) = 2.8 Hz); 135.23 (Cq); 133.17 (Cq); 132.72 (CH); 131.22 (CH); 130.63 (CH); 129.23 (C-11, C-13); 128.80 (CH); 128.67 (CH); 128.05 (CH); 127.72 (CH); 126.14 (q, J(3F-C19) = 3.6 Hz, C-19); 124.45 (q, J(3F-C18) = 30.7 Hz, C-18); 123.89 (q, J(3F-C) = 270.3 Hz, CF3); 116.42 (C-10, C-14); 67.81 (C-8).
FT-IR (solid in ATR, ν cm−1): 3173 m; 3028 m; 1662 m; 1525 s; 1490 vs; 1453 m; 1315 s; 1246 s; 1155 s; 1117 vs; 1035 m; 954 w; 817 m; 758 m; 736 m; 683 m.
Anal. calcd. for C22H16ClF3N2O2S (464.89): C, 56.84; H, 3.47; N, 6.03; S, 6.90%; Found: C, 56.71; H, 3.43; N, 6.09; S 6.87%.
2-((4-Chlorophenoxy)methyl)-N-(3-trifluoromethylphenylcarbamothioyl)benzamide (1f). Yield 61%; mp 114.4–115.9 °C;
1H-NMR (dmso-d6, δ ppm, J Hz, T = 298 K): 12.46 (br s, 1H, NH, deuterable); 11.96 (br s, 1H, NH, deuterable); 8.06 (br s, 1H, H-18); 7.80 (dq, J(H20-H21) = 6.8 Hz, J(3H-H20) = 2.4 Hz, 1H, H-20); 7.67–7.54 (m, 4H, H-22, H-21, H-4, H-5, H-7); 7.49 (m, 1H, H-6); 7.30 (d, J = 9.0 Hz, 2H, H-11, H-13); 7.02 (d, J = 9.0 Hz, 2H, H-10, H-14); 5.32 (s, 2H, H-8).
13C-NMR (dmso-d6, δ ppm, T = 298K): 179.56 (C-16); 169.92 (C-1); 157.03 (C-19); 138.69 (C-17); 135.30 (Cq); 133.26 (Cq); 129.17 (q, J(3F-C19) = 32.1 Hz, C-19); 124.73 (C-12); 123.86 (q, J(3F-C) = 272.7 Hz, CF3); 131.11 (CH); 129.79 (CH); 129.19 (C-11, C-13); 128.65 (CH); 128.54 (CH); 128.44 (CH); 127.92 (CH); 116.43 (C-10, C-14); 122.77 (q, J(3F-C18) = 3.8 Hz, C-18); 121.06 (q, J(3F-C20) = 4.0 Hz, C-20); 67.82 (C-8).
FT-IR (solid in ATR, ν cm−1): 3174 m; 3019 m; 2882 w; 1679 m; 1523 s; 1490 s; 1450 m; 1381 m; 1331 s; 1236 m; 1156 vs; 1116 vs; 1068 m; 1003 m; 893 w; 825 m; 746 s; 693 m; 644 m
Anal. calcd. for C22H16ClF3N2O2S (464.89): C, 56.84; H, 3.47; N, 6.03; S, 6.90%; Found: C, 56.98; H, 3.48; N, 5.99; S 6.90%.
2-((4-Chlorophenoxy)methyl)-N-(4-trifluoromethylphenylcarbamothioyl)benzamide (1g). Yield 67%; mp 160.3–161.9 °C;
1H-NMR (dmso-d6, δ ppm, J Hz, T = 298 K): 12.57 (br s, 1H, NH, deuterable); 11.97 (br s, 1H, NH, deuterable); 7.88 (d, J = 8.5 Hz, 2H, H-19, H-21); 7.76 (d, J = 8.5 Hz, 2H, H-18, H-22); 7.65 (br d, J = 7.2 Hz, 1H, H-7); 7.61–7.54 (m, 2H, H-4, H-5); 7.48 (m, 1H, H-6); 7.30 (d, J = 9.0 Hz, 2H, H-11, H-13); 7.01 (d, J = 9.0 Hz, 2H, H-10, H-14); 5.32 (s, 2H, H-8).
13C-NMR (dmso-d6, δ ppm, T=298K): 179.26 (C-16); 169.99 (C-1); 157.03 (C-19); 141.51 (C-17); 135.37 (Cq); 133.18 (Cq); 131.18 (CH); 129.24 (C-11, C-13); 128.63 (CH); 128.44 (CH); 127.90 (CH); 126.19 (q, J(3F-C20) = 31.8 Hz, C-20); 125.73 (q, J(3F-C19, 21) = 3.8 Hz, 2C, C-19, C-21); 124.75 (C-12); 124.51 (C-18, C-22); 124.00 (q, J(3F-C) = 271.2 Hz, CF3); 116.46 (C-10, C-14); 67.79 (C-8).
FT-IR (ATR in solid, ν cm−1): 3342 m; 3005 m; 2937 w; 1679 m; 1599 s; 1517 s; 1488 vs; 1414 m; 1318 m; 1228 s; 1145 s; 1109 vs; 1064 m; 1028 m; 823 m; 735 m; 669 m; 512 w.
Anal. calcd. for C22H16ClF3N2O2S (464.89): C, 56.84; H, 3.47; N, 6.03; S, 6.90%; Found: C, 56.67; H, 3.50; N, 5.97; S 6.92%.
2.1.3. In Silico Drug-Likeness Analysis and Molecular Docking Screening
Properties Computations
Molecular, topological, conformational characteristics and quantitative structure-activity/property relationships (QSAR/QSPR) analysis on 3D benzamides optimized structures were performed using DFT (density functional theory) method, B3LYP algorithm with 6-31+G (d, p), basis set, for equilibrium geometry, at ground state [50]. The computations were performed using Spartan’18 Wavefunction, Inc. Irvine, CA, U.S.A [51].
In Spartan software, molecular mechanics force field (MMFF, developed at Merck Pharmaceuticals) [52] was used to account for conformer energetics in a multi-step procedure.
Molecular Docking Simulations
Molecular docking approach, using CLC Drug Discovery Workbench Software, was conducted on seven ligands in order to achieve accurate predictions about structure and interactions of the studied benzamide derivatives in complex with DNA gyrase subunit from Staphylococcus aureus, PDB ID 2XCS [53] in order to assess their potential as antimicrobial agents.
In the docking simulation, the ligands (compounds 1a–1g) are fitted into an expected binding site on the surface of a protein target. CLC Drug Discovery Workbench employed also MMFF94 (MMFF) force field when generate 3D structure on import. Different conformations are generated by rotation about rotatable bonds and conformation changes. Thus, the ligand optimizer was achieved by geometry minimization using MMFF94 force field. Also, the molecule is minimized with regard to the binding pocket geometry.
The protein–ligand interaction is scored, and the best scoring binding mode is returned for each ligand, together with the score. The ligand binding mode search is realized inside the binding site (green sphere with a radius large enough to comprise all ligands docked to the receptor protein). After the import of the protein receptor from PDB bank, the next step is the preparation of the protein receptor: Selected protein chains B and D, selected nucleic acid chain F, excluded water molecules, excluded the cofactors. The next step is the setup binding pockets; binding pockets are necessary to guide the docking simulation. After the setup the binding site and the binding pocket, the co-crystallized was extracted and was redocking in the active binding site of the protein receptor, for the validation of the method and of the docking parameters obtained from the molecular docking studies.
Molecular docking study was started for the prediction of the antibacterial activity of the designed compounds 1a–g against Staphylococcus aureus, who is the most dangerous of all the many common staphylococcal bacteria.
2.2. Biological Assay
2.2.1. Qualitative Assessment of the Antimicrobial Activity
The antimicrobial assays were performed on reference (bearing the ATCC code) microbial strains, i.e., Gram-positive (Staphylococcus aureus ATCC 25923, Enterococcus feacalis ATCC29212) and Gram- negative (Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853) bacteria, as well as the fungal strain Candida albicans ATCC 10231.
The qualitative evaluation of the antimicrobial activity was performed by the adapted disk diffusion, as previously reported [54], using Mueller–Hinton Agar (MHA) medium for bacteria and Yeast Peptone Glucose (YPG) in case of yeast. The compounds were solubilized in dimethylsulfoxide (DMSO) and the starting stock solution concentration was of 10 mg/mL.
2.2.2. Quantitative Assessment of the Antimicrobial Activity
The quantitative assay of the antimicrobial activity was performed by the liquid medium microdilution method, in 96 multi-well plates, in order to establish the MIC and MBEC values.
Binary microdilution assays in 96-well plates were carried out for quantitative assessment of the MIC of the tested compounds against the same reference microbial strains such as those used in the qualitative assay. The microbial cultures were maintained in nutrient agar slant at 4 °C. The working cultures were obtained by subculturing onto nutrient agar.
Briefly, serial binary dilutions of the compound stock solution in DMSO (10 mg/mL) were performed in a volume of 100 μL broth. In the first well, a volume of 100 μL broth and 100 μL of chemical compound solution were added. Thereafter, 100 μL from the first well were transferred in the second 100 μL from the second well were transferred in the third and so on until the tenth well, from which 100 μL were discarded. Afterwards, each well was inoculated with 20 μL microbial suspension with a density of MacFarland 0.5, equivalent to 1.5 × 108 microbial cells/mL. Microbial suspension was realized in sterile physiological serum from 24 h solid cultures obtained on nutrient agar. For each testing a growth control (a well containing broth inoculated with the corresponding microbial strain), a standard drug (tobramycin for bacterial strains and fluconazole for the yeast strain) and a negative broth control (a well containing only broth) were used. The inoculated plates were incubated for 24 h at 37 °C and the absorbance at 620 nm measured spectrophotometrically. The lowest concentration of the chemical agent that inhibited the growth of a microorganism was regarded/interpreted as the MIC (mg/mL). The experiments were performed in duplicate.
2.2.3. The Influence of the Tested Compounds on the Biofilm Development
The microtiter method was used for investigation of the influence of the compounds on biofilm formation. After MICs determination, the plates were emptied and each well was washed three times with sterile phosphate buffered saline (PBS) to remove the microbial planktonic cells. The biofilms formed on the bottom and wall of the wells were fixed with cold methanol for 5 min and stained with 0.1% aqueous solution of crystal violet, for 20 min, at room temperature. Thereafter, acetic acid 33% was added for solubilization of the biofilms and the absorbance at 490 nm of the resulting colored solution was measured using a spectrophotometer. The lowest concentration of the chemical agent that inhibited the adherence of a microorganism was regarded/interpreted as the MBEC (mg/mL).
2.3. Antioxidant Activity of the New Compounds
For TAC measurements, a 2 × 10−4 M solution of DPPH in methanol was prepared. The benzamides derivatives were dissolved also in methanol solution at a 0.1 mg/mL concentration. 1.8 mL DPPH solution was mixed with 0.2 mL solution of each compound and the resulting mixture was kept in dark for 30 min, then the absorbance of the mixture was measured at 517 nm. The blank sample was obtained from 1.8 mL DPPH solution and 0.2 mL methanol. TAC values were calculated according to Equation (1).
3. Results
3.1. Chemistry
The new compounds (1a–g) with the thiourea skeleton were prepared according to the Figure 2, from 2-(4-chlorophenoxymethyl)benzoyl isothiocyanate (2) and primary amines. The isothiocyanate was obtained by reaction of 2-(4-chlorophenoxymethyl)benzoyl chloride (3) with ammonium thiocyanate. Also, the acid chloride was obtained from 2-(4-chlorophenoxymethyl)benzoic acid (4) by refluxing with thionyl chloride in anhydrous 1,2-dichloroethane. The acid (4) resulted by treating with hydrochloric acid the potassium salt of 2-(4-chlorophenoxymethyl)benzoic acid (5) obtained from reaction of phthalide (6) with potassium p-chlorophenoxide in xylene.
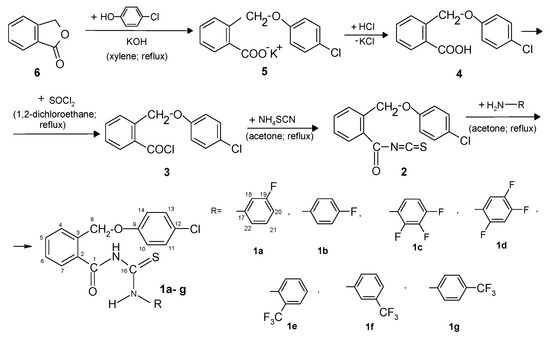
Figure 2.
The synthetic route of the new benzamides 1a–g.
The chemical structures of the new compounds were established on the basis of their spectroscopic data and elemental analysis.
The IR bands are given as w—weak, m—medium, s—strong, vs—very strong. The νN-H of the amide group can be observed in the infrared spectrum between 3342 and 3136 cm−1. The IR spectra show a strong absorption band in the 1684–1662 cm−1 region due to νC=O. The frequencies observed at 1527 and 1514 cm−1 are described to the δN–H amide. The antisymmetric vibration of the alkyl-aryl ether group is assigned at 1246–1228 cm−1 and the symmetric vibration of the same group is found at 1050–1001 cm−1. Absorption bands due to νC=S are observed in the 1163–1145 cm−1 region.
Significant peaks in the 1H-NMR are shown in the order: chemical shifts, multiplicity (s-singlet, d-doublet, t-triplet, q-quartet, m-multiplet, dd-double doublet, dt-double triplet, dq-double quartet, td-triple doublet, ddd-double double doublet, tdd-triple double doublet, and br-broad signal), coupling constant(s) in Hertz (Hz), number of protons, and signal/atom attribution.
The 1H-NMR spectra of 1a–g show signals in the range 12.57–12.00 ppm and 12.14–11.85 ppm assigned to the thiourea NH protons. The singlet observed between 5.32–5.30 ppm is assigned to the methylene group attached to the oxygen atom. The 13C-NMR data are reported in the following order: chemical shifts, coupling constant (where there) and signal/atom attribution (Cq-quaternary carbon).
The 13C-NMR chemical shifts were observed in the regions, δ 181.41–178.97 ppm for C=S and δ 170.60–169.92 ppm for C=O. The signal of carbon from -CH2O-group can be observed in the range of δ 67.83–67.76 ppm.
3.2. In Silico Drug-Like and Molecular Docking Screening
3.2.1. Ligand Preparation
A computational study to compare molecular properties, QSAR properties and mechanics calculations for the new compound 1g has been conducted using density functional theory (DFT). The 3D structure used for calculations was generated and its geometry has been optimized by energy minimization, in order to obtain the most stable conformer (Figure 3).
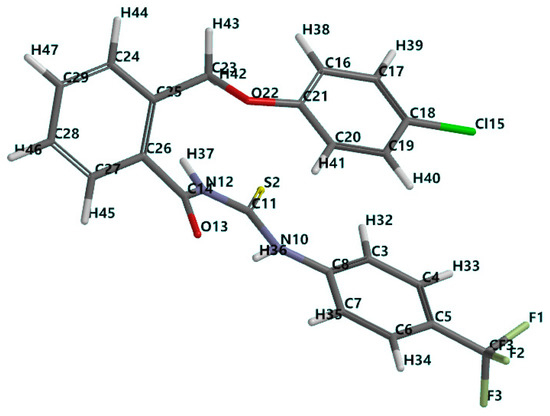
Figure 3.
Tube representation of the optimized molecular structure of 2-((4-chlorophenoxy)methyl)-N-(4-trifluoromethylphenylcarbamothioyl)benzamide (1g). (the numbering of the atoms was done according to the software).
In Table 1, there are given the calculated molecular properties and common descriptors used in quantitative structure and activity relationship (QSAR) analysis, based in the electron density and the electrostatic potential: weight, energy, area, volume, ovality, polarizability, water-octanol partition coefficient (logP), polar surface area (PSA), dipole moment, energy of the frontier molecular orbitals (FMOs)-the highest occupied molecular orbital (HOMO), and lowest unoccupied molecular orbital (LUMO), along with their energy gap (ΔE). These characteristics were discussed in order to assess the drug-likeness in terms of oral bioavailability according Lipinski’s rule of five (RO5) [55] and are useful for further molecular docking studies, to assess the flexibility and the binding ability of studied conformers to bind to the receptor protein. Calculation were performed on the isolated molecules, in gas, at equilibrium geometry in ground state, for the lowest energy conformer, that present an energy minimum in all dimensions. Area, volume, polar surface area (PSA), and ovality are resulted from a space-filling model. Area and volume increase as expected, with the molecular weight of the compounds. Ovality represents a measure of deviation from a spherical shape, where 1.0 is associated with the sphere model and values >1.0 indicate deviation. The magnitude of the dipole moment is also reported. Greatest dipole moments suggest larger separation of charges. Polarizability and logP are descriptors based on the electron density surface. The smaller value of dipole moment exhibits the compound 1e (4.15) and the greater, 1g (7.92). Differences in structures are done by the number of fluorine atoms and their positions. This fact is reflected also by the obtained values of logP for compounds 1a-b, 1c-d, and 1f-g, respectively. Same similarity in values are obtained for polarizability, also. The difference in energies between the HOMO and LUMO (ΔE) is of great interest when evaluate the reactivity of molecules and their kinetic stability. Lower energy gap conducts to enhanced stability of the molecule. The smallest values for frontier molecular orbitals present the compound 1e, followed by 1d, suggesting their greatest kinetic stability among the others. Also, these compounds, have shown no interactions with amino acids residues of the active site where docked.

Table 1.
Molecular properties and descriptors for 1a–g compounds, calculated with Spartan’18 software.
Lipinski’s rule of five is used to assess oral bioavailability of drug candidates, when designing new therapeutically potential agents. This rule imposes several molecular restrictions: Maximum 5 hydrogen bond donors (HBD), maximum 10 hydrogen bond acceptors (HBA), a molecular mass that does not exceed the value of 500 Da and the octanol-water partition coefficient (log P) values lower than 5. To be effective, a therapeutic agent must be able of transportation to its target. Orally administrated drugs must cross cell membranes in the intestine or blood-brain barrier in case of neural drugs. To evaluate these abilities, some molecular features such as polar surface area (PSA) (resulted from the nitrogen and oxygen atoms together with its attached hydrogens), electrostatic potential map and hydrophobicity/hydrophilicity measure given by logP, are considered.
In Figure 4 there are illustrated three of the most commonly used graphical models, exemplified for the compound 1g: The electrostatic potential map for elucidating molecular charge distributions, and the local ionization potential and LUMO maps for anticipating electrophilic and nucleophilic reactivity, respectively.
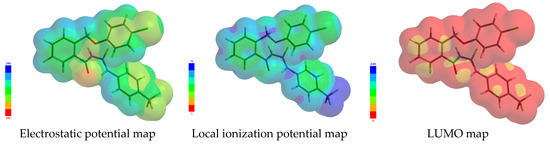
Figure 4.
The most commonly-used graphical models, for 1g compound.
Also, the theoretical calculation showed that the antibacterial activity seems to depend on the LUMO energy value, a lower LUMO value determining a higher antibacterial activity [56].
3.2.2. Docking Studies
The docking studies have been also carried out. The score and hydrogen bonds formed with the amino acids from group interaction atoms are used to predict the binding modes, the binding affinities and the orientation of the docked compound in the active site of the protein-receptor. The protein–ligand complex has been realized based on the X-ray structure of Staphylococcus aureus DNA gyrase, downloaded from the Protein Data Bank (PDB ID: 2XCS). RXV(6-methoxy-4-(2-{4-[([1,3]oxathiolo[5,4-c]pyridin-6-ylmethyl)amino]piperidin-1-yl}ethyl) quinoline-3-carbonitrile interacting with amino acid residues of the active site, are shown in Figure 5a. Co-crystallized reveals docking score −61.50 (RMSD 2.49 Å) and shows the occurrence of one hydrogen bond with ASP 1083(B) (2.902 Å). The docking pose of the co-crystallized interacting with the amino acids residues is presented in Figure 5b.The docking studies revealed that 1g, which has a good in vitro activity against Staphylococcus aureus ATCC 25923 (MIC = 0.62 mg/mL) reveals docking score −45.93 (RMSD 0.75 Å) and shows the occurrence of one hydrogen bond with ASP 1083(D) (2.917 Å) (Table 2, Figure 6a). Also, the 1f compound, reveals a good docking score −45.02 (RMSD 0.40 Å) and shows the occurrence of two hydrogen bonds with ASP 1083(D) (3.115 Å) and with ARG 1122(B) (3.191 Å) (Figure 7a). The compound with the best activity against Staphylococcus aureus ATCC 25923 (MIC = 0.31 mg/mL), 1a, reveals docking score −44.96 (RMSD 0.15 Å) and shows the occurrence of two hydrogen bonds with ASP 1083(D) (3.077 and 3.154 Å) (Figure 8a). The docking pose of the ligands 1g, 1f and 1a interacting with the amino acids residues is presented in Figure 6b, Figure 7b and Figure 8b and in Table 2. The better score docking has been obtained from compound 1e (docking score: −47.64; RMSD 0.05 Å). But in the case of this compound, it was observed that it does not show the occurrence of some hydrogen bonds with some amino acid residues from the binding site of the 2XCS. Same situation has been observed for 1c compound (docking score: −42.89; RMSD 0.12) and for 1d compound (docking score: −44.28; RMSD 0.06). These three compounds show weak affinity for the active site of the protein receptor. This could be explained by the nature of the substituent R (compound 1a–g, Figure 2). For the ligands who realized hydrogen bonds with the amino acid residues, the docking score decreases in order: 1g (R is 4-trifluoromethylphenyl group) > 1f (R is 3-trifluoromethylphenyl group) > 1a (R is 3-fluorophenyl group) > 1b (R is 4-fluorophenyl group). In the case of the compounds who do not realized hydrogen bonds with amino acid residues from the binding site of the protein receptor, 1e (R is 3-trifluoromethylphenyl group) > 1d (R is 3,4,6-trifluorophenyl group) > 1c (R is 2,3,4-trifluorophenyl group). It was observed that the bulk, the position and the number of the substituent groups on the benzene ring from R substituent has an influence on the binding modes, the binding affinities and the orientation of the docked ligands in the active site of the protein-receptor.
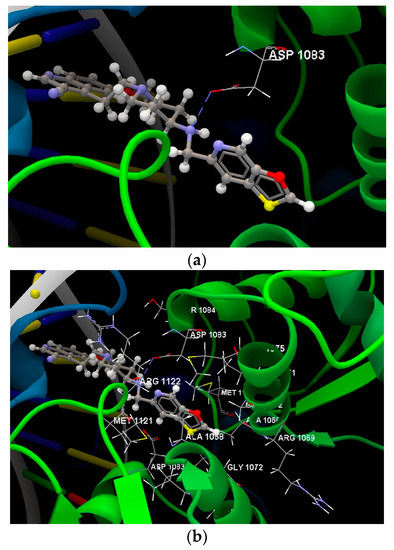
Figure 5.
(a) The hydrogen bond between the amino acid residue of the ASP 1083(B) and the co-crystallized was displayed in dashed lines. (b) Docking pose of the co-crystallized ligand interacting with amino acids residues.

Table 2.
The list of intermolecular interactions between the ligand molecules docked with 2XCS using CLC Drug Discovery Workbench Software.
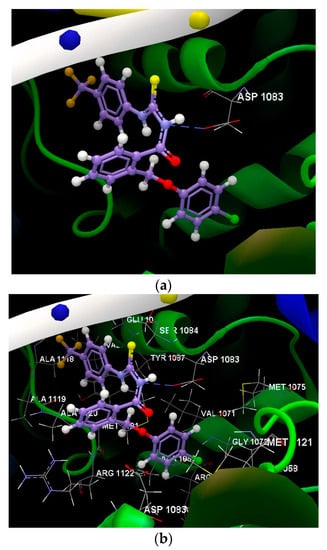
Figure 6.
(a) The hydrogen bond between the amino acid residue of the ASP 1083(D) and the compound 1g was displayed in dashed lines. (b) Docking pose of the 1g ligand interacting with amino acids residues.
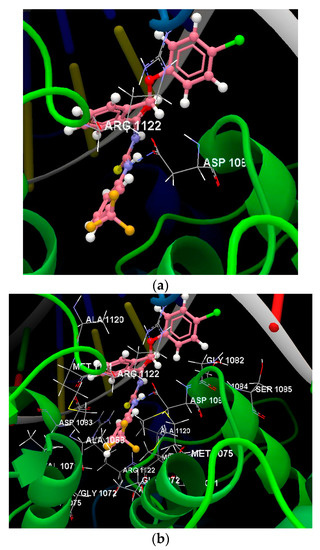
Figure 7.
(a) The hydrogen bonds between the amino acid residues of the ASP 1083(D), ARG 1122(B) and the compound 1f was displayed in dashed blue lines. (b) Docking pose of the 1f ligand interacting with amino acids residues.
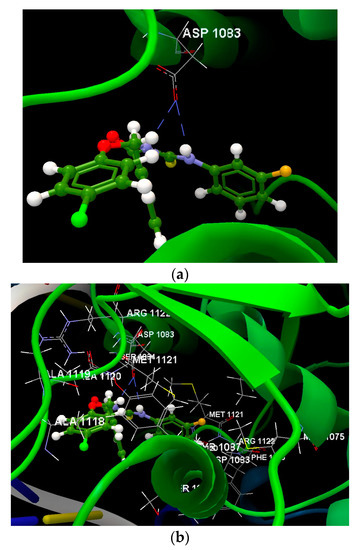
Figure 8.
(a) The hydrogen bonds between the amino acid residue of the ASP 1083(D) and the compound 1a was displayed in dashed blue lines. (b) Docking pose of the 1a ligand interacting with amino acids residues.
The calculated properties with CLC Drug Discovery Workbench Software, are presented in Table 2 (the number of flexible bonds, the number of hydrogen bond donors, the number of hydrogen bond acceptors and log P). These parameters can predict if a molecule possesses properties that might turn it into an active drug, according to the Lipinski’s rule of five. The number of violations of the Lipinski rules allows to evaluate drug-likeness for a molecule.
Regarding properties calculated with Spartan’18 (Table 1) and screening of ligands with CLC Drug Discovery Workbench (Table 3) we constant that all studied structures show one deviation from this rule. The violation of the Lipinski’s statements refers to water-octanol partition coefficient values, calculated with the second software (grater then 5, results in one Lipinski’s violation). The difference between the obtained values for log P arises from the different methods of calculation of this coefficient used by the two software. Spartan Software uses for LogP estimation, the method of Ghose, Pritchett and Crippen [58] while in CLC Software this parameter is calculated according XLOGP3-AA method [59].

Table 3.
Calculated properties of ligands properties with CLC Drug Discovery Workbench Software.
3.3. Antimicrobial Activity Against Planktonic Cells
3.3.1. Qualitative Assay
The qualitative evaluation of the antimicrobial activity performed by the adapted disk diffusion has shown that the tested compounds inhibited the bacterial and fungal growth. Pseudomonas aeruginosa ATCC 27853 was susceptible to inhibitory activity of most of the tested compounds, exception being 1a and 1d. The other Gram- negative bacillus tested (Escherichia coli ATCC 25922) was more resistant to the benzamides activity, except 1g. The Gram-positive bacteria were inhibited only by one compound (Staphylococcus aureus ATCC 25923-1g) and two (Enterococcus faecalis ATCC 29212-1f, 1g) respectively. The fungal strain was sensitive to the action of 1b and 1g.
2-((4-Chlorophenoxy)methyl)-N-(4-trifluoromethylphenylcarbamothioyl)benzamide (1g) had the highest inhibitory activity, influencing the growth of all the tested strains. This activity might be due to presence of the -CF3 in para position. Moreover, the compounds with this group were the most active compounds against the tested microbes. Compounds 1a and 1d showed no inhibitory activity on all the tested strains (Table 4).

Table 4.
The qualitative antimicrobial activity evaluation of the new compounds.
3.3.2. Quantitative Assay
The tested compounds exhibited a good inhibitory growth effect against the reference microbial strains, being more active against Gram-positive bacteria (Staphylococcus aureus ATCC 25923 and Enterococcus faecalis ATCC 27853). The most susceptible microorganism demonstrated to be Enterococcus faecalis, with MIC values ranging from 0.15 mg/mL (compounds 1a, 1f) to 0.62 mg/mL (compounds 1c, 1e). Compound 1a was the most active against all the tested microbial strains, with MIC values ranging from 0.15 mg/mL (Enterococcus faecalis ATCC 29212) to 0.62 mg/mL (Pseudomonas aeruginosa ATCC 27853). From the tested compounds, only 1b, 1e, and 1g showed an antimicrobial effect against the yeast reference strain. The MIC values are presented in Table 5.

Table 5.
Results of the quantitative assay of MIC for the compounds 1a–g (MIC values expressed in mg/mL).
3.4. Anti-Biofilm Activity
The investigated compounds demonstrated an inhibitory effect against the adherence of microbial cells to the inert substratum.
The compounds showed a good anti-biofilm activity against Candida albicans ATCC 10231, with MBEC values of 0.019 mg/mL (compound 1b, 1g), 0.15 mg/mL (compound 1d) and 0.31 (compounds 1a, 1c, 1f). Regarding the bacterial biofilms, those formed by Gram-positive bacteria (Staphylococcus aureus ATCC 25923 and Enterococcus faecalis ATCC 27853) were the most susceptible to the tested compounds, with MBEC values ranging from 0.15 mg/mL (compound 1g) to 1.25 mg/mL. Compound 1g exhibited the most evident inhibitory activity on Staphylococcus aureus ATCC 25923 biofilm (0.15 mg/mL). The most active compound on Enterococcus faecalis ATCC 27853 biofilm was 1d (0.31 mg/mL). Among the Gram-negative biofilms, the Escherichia coli ATCC 25922 biofilm was susceptible to the action of the new compounds, the most active being 1c and 1f (0.62 mg/mL). The biofilms formed by the Gram-negative strain Pseudomonas aeruginosa ATCC 27853 were not affected by the tested chemical agents (Table 6).

Table 6.
Results of the anti-biofilm assay of MBEC for the compounds 1a–g. (MBEC values expressed in mg/mL).
3.5. Antioxidant Activity
DPPH stable free radical is used as a substrate to evaluate the antioxidant in vitro activity and also to evaluate the ability of antioxidant agents to eliminate free radicals, which have a major contributes to the biological damage caused by oxidative stress [60].
The total antioxidant activity (TAC) reflects the property of tested compounds to protect the organism from the damage caused by oxidative stress and free radicals, being thus an important property of the biological active substances. Several methods to measure the TAC are reported in literature [60]. In this study we used the DPPH method, which is based on the employing of the stable free radical, 2,2-diphenyl-1-picrylhydrazyl (DPPH). The TAC values (%) of the compounds 1a–g were calculated using the Equation (1):
where Absini is the initial absorption of DPPH radical (measured at 517 nm) and Abs30min is the absorbance recorded at the same wavelength, after 30 min.
% Inhibition = Absini−Abs30min/Absini × 100
All tested new compounds exhibited only a poor antioxidant activity (Table 7). However, the antioxidant activity of the new 1a and 1c proved to be superior to that of the 1b, 1d–f. The best antioxidant activity was obtained for the compound 1a (12.89%), followed by compound 1c (12.53%).

Table 7.
Antioxidant activity of 1a–g.
4. Discussion
Fluoro/trifluoromethyl-substituted acylthiourea derivatives have been successfully synthesized and characterized.
Many research studies point out the advantages of introduction of a fluorine moiety in a drug that the judicious incorporation of the fluorine atom(s) or the trifluoromethyl group(s) into a drug molecule represents a good strategy to improve its pharmacokinetics and pharmacodynamics properties, and therefore, it biological activity (absorption, drug transport, affinity to the target protein and metabolic stability).
The considerable stability of the carbon-fluorine bond contributes to the delay or inhibition of metabolism or the prolongation of the drug half-life.
The fact that the carbon–fluorine bond is even more hydrophobic than the carbon-hydrogen bond causes the lipophilicity of the molecule to increase most of the times and, consequently, its bioavailability [61].
Therefore, fluoro/trifluoromethyl-anilines were subjected to the in situ reaction with 2-(4-chlorophenoxymethyl) benzoyl isothiocyanate to obtain the new compounds.
In the first stage of the in silico drug-likeness and molecular docking screening, the 3D structure has been generated. The most stable conformer was obtained by geometry optimization and energy minimization. A series of topological, conformational characteristics and QSAR properties, important to assess the flexibility and the ability of studied conformer to bind to the protein receptor, was determined and analyzed. The studied properties were the basis for assessing the conformer’s ability to bind to the receiver. The docking studies have been carried out to predict the binding modes, the binding affinities and the orientation of the docked compounds at the active site of the protein.
The molecular docking study, used to understand ligand-protein interactions, suggests a moderate activity as inhibitors of Staphylococcus aureus DNA gyrase, which makes possible the use of these compounds as scaffolds for new potent drugs, after further research.
The results of the antimicrobial activity assay by the qualitative method, showed that the most active compound was 1g, which inhibited the growth of all the tested strains. The results of the quantitative assay highlighted the best activity of compound 1a. The good anti-biofilm activity of the tested compounds could be probably explained by the the inhibition of cell-to-cell communication through the mechanism of quorum-sensing or of cell-to-surface attachment, thus preventing the initial phases of biofilm development or the disruption of the protective biofilm matrix. The anti-quorum sensing agents, despite their low or absent microbicidal activity, could thus exert a good anti-biofilm activity [62].
The anti-biofilm properties of these compounds may allow their use to cover the surface of biomaterials or medical devices. The best anti-biofilm activity was recorded for the compound 1g. The results showed that the presence of a fluorine atom or a trifluoromethyl group in the structure is sufficient to obtain a good antimicrobial activity. The results also concluded that the position of fluorine atom(s) or trifluoromethyl group as substituent(s) on the phenyl radical attached to thiourea nitrogen influenced significantly the biological activity.
The new compounds exhibited a poor DPPH-radical scavenging ability.
5. Conclusions
New 2-((4-chlorophenoxy)methyl)-N-(arylcarbamothioyl)benzamides have been designed, synthesized by the nucleophilic addition reaction of various primary amines with fluorine atom(s) or trifluoromethyl group to 2-(4-chlorophenoxymethyl)benzoyl isothiocyanate and their potential as antimicrobial and antioxidant agents was investigated. The structures of the synthesized compounds were elucidated on the basis of their elemental analysis and spectroscopic data. Results of a predictive computational study were reported to achieve a drug-likeness assessment of seven benzamide derivatives. Physico-chemical properties and common molecular descriptors important for the QSAR analysis, were obtained and discussed. As a result of docking simulations, the score and hydrogen bonds formed with the amino acids residues were used to predict the binding modes, the binding affinities and the orientation of the docked 2-((4-chlorophenoxy)methyl)-N-(arylcarbamothioyl)benzamides in the active site of the protein-receptor 2XCS. A correlation of the predicted data obtained by molecular docking study with the experimental data obtained from the evaluation of the antimicrobial activity against Staphylococcus aureus ATCC 25923 of compounds was observed.
Taken together, the bioevaluation and in silico analyses recommend the compound 1g as a promising antimicrobial agent, active both against planktonic and biofilm embedded bacterial cells and presumably acting by the inhibition of DNA replication.
Author Contributions
Conceptualization, C.L., D.C.N., A.V.M., L.P., I.Z., I.R.P. and M.C.C.; methodology, C.L., R.R., F.D., I.Z., M.P., A.L.A., D.I.U. software, R.R., L.P., A.S., validation, C.L., M.T.C., F.D., writing—original draft preparation, C.L.; writing—review and editing, I.R.P., C.L., visualization, M.C.C., supervision, C.L., C.E.D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This paper was financially supported by “Carol Davila” University of Medicine and Pharmacy through Contract no. 23PFE/17.10.2018 funded by the Ministry of Research and Innovation within PNCDI III, Program 1–Development of the National RD system, Subprogram 1.2–Institutional Performance–RDI excellence funding projects.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- DrugBank. Available online: https://www.drugbank.ca/drugs/DB13838 (accessed on 18 February 2020).
- DrugBank. Available online: https://www.drugbank.ca/drugs/DB13699 (accessed on 18 February 2020).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Loflucarban (accessed on 18 February 2020).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Thiophanate (accessed on 18 February 2020).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Thiophanate-methyl (accessed on 18 February 2020).
- DrugBank. Available online: https://www.drugbank.ca/drugs/DB13608 (accessed on 18 February 2020).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Burimamide (accessed on 18 February 2020).
- DrugBank. Available online: https://www.drugbank.ca/drugs/DB08805 (accessed on 18 February 2020).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Phenethylthiazolylthiourea (accessed on 18 February 2020).
- Pedersen, O.S.; Pedersen, E.B. Non-nucleoside reverse transcriptase inhibitors: The NNRTI boom. Antivir. Chem. Chemother. 1999, 10, 285–314. [Google Scholar] [CrossRef]
- Maalik, A.; Rahim, H.; Saleem, M.; Fatima, N.; Rauf, A.; Wadood, A.; Malik, M.I.; Ahmed, A.; Rafique, H.; Zafar, M.N.; et al. Synthesis, antimicrobial, antioxidant, cytotoxic, antiurease and molecular docking studies of N-(3-trifluoromethyl)benzoyl-N′-aryl thiourea derivatives. Bioorg. Chem. 2019, 88. [Google Scholar] [CrossRef] [PubMed]
- Alimohammadi, A.; Mostafavi, H.; Mahdavi, M. Thiourea derivatives based on the dapsone-naphthoquinone hybrid as anticancer and antimicrobial agents: In vitro screening and molecular docking studies. Chem. Select. 2020, 5, 847–852. [Google Scholar] [CrossRef]
- Bielenica, A.; Kedzierska, E.; Fidecka, S.; Maluszynska, H.; Miroslaw, B.; Koziol, A.E.; Stefanska, J.; Madeddu, S.; Giliberti, G.; Sanna, G.; et al. Synthesis, antimicrobial and pharmacological evaluation of thiourea derivatives of 4H-1,2,4-triazole. Lett. Drug. Des. Discov. 2015, 12, 251–252. [Google Scholar] [CrossRef]
- Naz, S.; Zahoor, M.; Umar, M.N.; Ali, B.; Ullah, R.; Shahat, A.A.; Mahmood, H.M.; Sahibzada, M.U.K. Enzyme inhibitory, antioxidant and antibacterial potentials of synthetic symmetrical and unsymmetrical thioureas. Drug Des. Dev. Ther. 2019, 13, 3485–3495. [Google Scholar] [CrossRef]
- Tatar, E.; Karakuş, S.; Küçükgüzel, Ș.G.; Okullu, S.O.; Ünübol, N.; Kocagöz, T.; De Clercq, E.; Andrei, G.; Snoeck, R.; Pannecouque, C.; et al. Design, synthesis, and molecular docking studies of a conjugated thiadiazole–thiourea scaffold as antituberculosis agents. Biol. Pharm. Bull. 2016, 39, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhai, Z.-W.; Shi, Y.-X.; Tan, C.-X.; Weng, J.-Q.; Han, L.; Li, B.-J.; Liu, X.-H. Novel trifluoromethylpyrazole acyl thiourea derivatives: Synthesis, antifungal activity and docking study. Lett. Drug. Des. Discov. 2019, 16. [Google Scholar] [CrossRef]
- Larik, A.F.; Saeed, A.; Faisal, M.; Channar, P.A.; Azam, S.S.; Ismail, H.; Dilshad, E.; Mirza, B. Synthesis, molecular docking and comparative efficacy of various alkyl/aryl thioureas as antibacterial, antifungal and α-amylase inhibitors. Comput. Biol. Chem. 2018, 77, 193–198. [Google Scholar] [CrossRef]
- Larik, F.A.; Saeed, A.; Channar, P.A.; Ismail, H.; Dilshad, E.; Mirza, B. New 1-octanoyl-3-aryl thiourea derivatives: Solvent-free synthesis, characterization and multi-target biological activities. Bangladesh J. Pharmacol. 2016, 11, 894–902. [Google Scholar] [CrossRef]
- Pingaew, R.; Sinthupoom, N.; Mandi, P.; Prachayasittikul, V.; Cherdtrakulkiat, R.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Synthesis, biological evaluation and in silico study of bis-thiourea derivatives as anticancer, antimalarial and antimicrobial agents. Med. Chem. Res. 2017, 26, 3136–3148. [Google Scholar] [CrossRef]
- El Bissati, K.; Redel, H.; Ting, L.-M.; Lykins, J.D.; McPhillie, M.J.; Upadhya, R.; Woster, P.M.; Yarlett, N.; Kim, K.; Weiss, L.M. Novel synthetic polyamines have potent antimalarial activities in vitro and in vivo by decreasing intracellular spermidine and spermine concentrations. Front. Cell. Infect. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Vajragupta, O.; Pananookooln, S.; Tuntiwachwuttikul, P.; Foye, W. Ovicidal activity of 6-substituted-2- and -4-aminoquinolines and their mono and bis(thiourea)derivatives against human hookworm. J. Sci. Soc. Thailand 1998, 14, 51–59. [Google Scholar] [CrossRef]
- Ansari, M.M.; Deshmukh, S.P.; Khan, R.; Musaddiq, M. Synthesis antimicrobial and anticancer evaluation of 1-aryl-5-(o-methoxyphenyl)-2-S-benzyl Isothiobiurets. Int. J. Med. Chem. 2014. [Google Scholar] [CrossRef]
- Viswas, R.S.; Pundir, S.; Lee, H. Design and synthesis of 4-piperazinyl quinoline derived urea/thioureas for anti-breast cancer activity by a hybrid pharmacophore approach. J. Enzyme Inhib. Med. Chem. 2019, 34, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Al-Harbi, R.A.K.; El-Sharief, M.A.M.S.; Abbas, Y. Synthesis and anticancer activity of bis-benzo[d][1,3]dioxol-5-yl thiourea derivatives with molecular docking study. Bioorg. Chem. 2019, 90, 103088. [Google Scholar] [CrossRef]
- Zhang, R.-Z.H.B.; Huang, X.-C.; Liang, G.-B.; Qin, J.-M.; Pan, Y.-M.; Liao, Z.-X.; Wang, H.-S. Synthesis and biological evaluation of terminal functionalized thiourea-containing dipeptides as antitumor agents. RSC Adv. 2017, 7, 8866. [Google Scholar]
- Çelen, A.O.; Kaymakçioùlu, B.; Gümrü, S.; Toklu, H.Z.; Aricioùlu, F. Synthesis and anticonvulsant activity of substituted thiourea derivatives. Marmara Pharm. J. 2011, 15, 43–47. [Google Scholar] [CrossRef]
- Kumar, P.; Sharma, A.; Gupta, S.K. Synthesis, characterization and anticonvulsant activity on novel thiourea derivatives. Int. J. Pharm. Med. Res. 2014, 2, 116–122. [Google Scholar]
- Thakur, A.S.; Deshmukh, R.; Jha, A.K.; Kumar, P.S. Molecular docking study and anticonvulsantactivity of synthesized 4-((4,6-dimethyl-6H-1,3-thiazin-2-yl)phenylsulfonyl)urea/ thiourea derivatives. J. King. Saud. Univ. Sci. 2018, 30, 330–336. [Google Scholar] [CrossRef]
- Alagarsamy, V.; Meena, S.; Ramseshu, K.V.; Solomon, V.R.; Thirumurugan, K.; Dhanabal, K.; Murugan, M. Synthesis, analgesic, anti-inflammatory, ulcerogenic index and antibacterial activities of novel 2-methylthio-3-substituted-5,6,7,8-tetrahydrobenzo(b)thieno[2,3-d]pyrimidin-4(3H)-ones. Eur. J. Med. Chem. 2006, 41, 1293–1300. [Google Scholar] [CrossRef]
- Sheorey, R.V.; Thangathiruppathy, A.; Alagarsamy, V. Synthesis, analgesic and anti-inflammatory activities of 3-ethyl-2-substituted amino-3H-quinazolin-4-ones. Trop. J. Pharm. Res. 2013, 12, 583–589. [Google Scholar] [CrossRef]
- Sudzhaev, A.R.; Rzaeva, I.A.; Nadzhafova, R.A.; Safarov, Y.A.; Allakhverdiev, M.A. Antioxidant properties of some thiourea derivatives. Russ. J. Appl. Chem. 2011, 84, 1394. [Google Scholar]
- Hameed, A.; Jafri, L.; Sheikh, M.A. Effect of thiourea on proteins, catalase, guaiacol-peroxidase and protease activities in wheat leaves under H2O2 induced oxidative stress. Plant Physiol. 2013, 4, 857–864. [Google Scholar]
- Kalaiyarasi, A.; Haribabu, J.; Gayathri, D.; Gomathi, K.; Bhuvanesh, N.S.P.; Karvembu, R.; Biju, V.M. Chemosensing, molecular docking and antioxidant studies of 8-aminoquinoline appended acylthiourea derivatives. J. Mol. Struct. 2019, 1185, 450–460. [Google Scholar] [CrossRef]
- Wang, B.; Ma, Y.; Xiong, L.; Li, Z. Synthesis and insecticidal activity of novel N-pyridylpyrazole carbonyl thioureas. Chin. J. Chem. 2012, 30, 815–821. [Google Scholar] [CrossRef]
- Yunyun, G.; Wei, H.; Xinghai, L.; Jianquan, V.; Chengxia, T. Synthesis and herbicidal activities of 1-methylcyclohexyl-formayl thioureas. Chin. J. Org. Chem. 2013, 33, 2396–2401. [Google Scholar]
- Wua, A.-B.; Duana, L.-P. Transition metal complexes of N-(4,6-dimethoxypyrimidin-2-ylcarbamothioyl)benzamide: Design, synthesis and herbicidal activity. J. Chin. Chem. Soc. 2009, 56, 539–542. [Google Scholar] [CrossRef]
- Lin, Q.; Yao, H.; Wei, T.-B.; Zhang, Y.-M. Synthesis and plant growth regulatory activity of o-nitrobenzoylthiourea derivatives. Indian J. Chem. 2009, 48, 124–127. [Google Scholar]
- Kachhadia, V.V.; Patel, M.R.; Joshi, H.S. Heterocyclic systems containing S/N regioselective nucleophilic competition: Facile synthesis, antitubercular and antimicrobial activity of thiohydantoins and iminothiazolidinones containing the benzo[b]thiophene moiety. J. Serb. Chem. Soc. 2005, 70, 153–161. [Google Scholar] [CrossRef]
- Sun, C.; Huang, H.; Feng, M.; Shi, X.; Zhang, X.; Zhou, P. A novel class of potent influenza virus inhibitors: Polysubstituted acylthiourea and its fused heterocycle derivatives. Bioorg. Med. Chem. Lett. 2005, 16, 162–166. [Google Scholar] [CrossRef]
- Chandrasekhar, M.; Syam Prasad, G.; Venkataramaiah, C.; Umapriya, K.; Raju, C.N.; Seshaiah, K.; Rajendra, W. In silico and in vitro antioxidant and anticancer activity profiles of urea and thiourea derivatives of 2,3-dihydro-1H-inden-1-amine. J. Recept. Signal. Transduct. Res. 2020, 40, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Sudhamani, H.; Basha, S.T.; Venkateswarlu, N.; Vijaya, T.; Raju, C.N. Synthesis and characterization of new thiourea and urea derivatives of 6-fluoro-3-(piperidin-4-yl)benzo[d]isoxazole: In vitro antimicrobial and antioxidant activity. J. Chem. Sci. 2015, 127, 1739–1746. [Google Scholar] [CrossRef]
- Firdausiah, S.; Hasbullah, S.A.; Yamin, B.M. Synthesis, structurale elucidation and antioxidant study of ortho-substituted N,N’-bis(benzamidothiocarbonyl)hydrazine derivatives. J. Phys. Conf. Ser. 2018, 979, 012010. [Google Scholar] [CrossRef]
- Yeşilkaynak, T. Synthesis and characterization of N-((6-methylpyridin-2-yl)carbamothioyl)thiophene-2-carboxamide and its Co(II), Ni(II) and Cu(II) complexes: Calculation of the molecular orbitals and antioxidant and antitumor activities. J. Turkish Chem. Soc. Sect. Chem. 2016, 3, 1–14. [Google Scholar] [CrossRef]
- Da Silva, T.L.; Forain Miolo, L.M.; Sousa, F.S.S.; Brod, L.M.P.; Savegnago, L.; Schneider, P.H. New thioureas based on thiazolidines with antioxidant potential. Tetrahedron Lett. 2015, 56, 6674–6680. [Google Scholar] [CrossRef]
- Nacea, V.; Boscenco, R.; Missir, A.V.; Limban, C.; Bărbuceanu, S. Sinteza şi caracterizarea unor noi combinaţii complexe ale Cu (I) şi Cu (II) cu noi tioureide N,N’-disubstituite. Rev. Chim. Buchar. 2005, 56, 68–71. [Google Scholar]
- Limban, C.; Missir, A.V.; Chiriţă, I.C.; Bădiceanu, C.D.; Drăghici, C.; Balotescu, M.C.; Stamatoiu, O. New thioureides of 2-(4-methyl-phenoxymethyl)-benzoic and 2-(4-methoxy-phenoxymethyl)-benzoic acids with biological activity. Rev. Roum. Chim. 2008, 53, 595–602. [Google Scholar]
- Limban, C.; Missir, A.V.; Grumezescu, A.M.; Oprea, A.E.; Grumezescu, V.; Vasile, B.S.; Socol, G.; Trușcă, R.; Caproiu, M.T.; Chifiriuc, M.C.; et al. Bioevaluation of novel anti-biofilm coatings based on PVP/Fe3O4 nanostructures and 2-((4-ethylphenoxy)methyl)-N-(arylcarbamothioyl)benzamides. Molecules 2014, 19, 12011–12030. [Google Scholar] [CrossRef]
- Limban, C.; Missir, A.V.; Nuţă, D.C.; Căproiu, M.T.; Papacocea, M.T.; Chiriţă, C. Synthesis of some new 2-((4-chlorophenoxy)methyl)-N-(arylcarbamothioyl)benzamides as potential antifungal agents. Farmacia 2016, 64, 775–779. [Google Scholar]
- Limban, C.; Balotescu Chifiriuc, M.C.; Missir, A.V.; Chiriţă, I.C.; Bleotu, C. Antimicrobial activity of some new thioureides derived from 2-(4-chlorophenoxymethyl)benzoic acid. Molecules 2008, 13, 567–580. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle- Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Hehre, W.J. A Guide to Molecular Mechanics and Quantum Chemical Calculations; Wavefunction, Inc.: Irvine, CA, USA, 2003. [Google Scholar]
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Bax, D.B.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.; Hennessy, A.; Hibbs, M.; et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 2010, 466, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Calu, L.; Badea, M.; Chifiriuc, M.C.; Bleotu, C.; David, G.-I.; Ioniţă, C.; Măruţescu, L.; Lazăr, V.; Stanică, N.; Soponaru, I.; et al. Synthesis, spectral, thermal, magnetic and biological characterization of Co(II), Ni(II), Cu(II) and Zn(II) complexes with a Schiff base bearing a 1,2,4-triazole pharmacophore. J. Therm. Anal. Calorim. 2015, 120, 375–386. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Misral, H.; Sapari, S.; Rahman, T.; Ibrahim, N.; Yamin, B.M.; Hasbulla, S.A. Evaluation of novel N-(dibenzylcarbamothioyl)benzamide derivatives as antibacterial agents by using DFT and drug-likeness assessment. J. Chem. 2018. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical scoring functions for advanced protein-ligand docking with plants. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef]
- Ghose, A.K.; Pritchett, A.; Crippenic, G.M. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships III: Modeling hydrophobic interactions. J. Comput. Chem. 1988, 9, 80–90. [Google Scholar] [CrossRef]
- Cheng, T.; Zhao, Y.; Li, X.; Lin, F.; Xu, Y.; Zhang, X.; Li, Y.; Wang, R. Computation of octanol-water partition coefficients by guiding an additive model with knowledge. J. Chem. Inf. Model. 2007, 47, 2140–2148. [Google Scholar] [CrossRef]
- Prior, R.L.; Wu, X.; Schaich, K. Standardized methods for the determination of antioxidant capacity and phenolics in food and dietary supplements. J. Agric. Food Chem. 2005, 53, 4290–4302. [Google Scholar] [CrossRef]
- Shah, P.; Westwell, A.D. The role of fluorine in medicinal chemistry. J. Enzyme Inhib. Med. Chem. 2007, 22, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Algburi, A.; Comito, N.; Kashtanov, D.; Dicks, L.M.T.; Chikindas, M.L. Control of biofilm formation: Antibiotics and beyond. Appl. Environ. Microbiol. 2017, 83, e02508–e02516. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).