Abstract
This work presents a molecular level investigation on the nature and mode of binding between imidazolium-based ionic liquids (ILs) ([Cnbim]Br where n = 2, 4, 6) with calf thymus DNA. This investigation offers valuable insight into the mechanisms of interactions that can affect the structural features of DNA and possibly cause the alteration or inhibition of DNA function. To expedite analysis, the study resorted to using molecular docking and COnductor like Screening MOdel for Real Solvents (COSMO-RS) in conjunction with fluorescence spectroscopic data for confirmation and validation of computational results. Both the fluorescence and docking studies consistently revealed a weak interaction between the two molecules, which corresponded to the binding energy of a stable docking conformation in the range of −5.19 to −7.75 kcal mol−1. As predicted, the rod-like structure of imidazolium-based ILs prefers to bind to the double-helix DNA through a minor groove. Interestingly, the occurrence of T-shape π-π stacking was observed between the amine group in adenine that faces the aromatic ring of imidazole. In addition, data of COSMO-RS for the interaction of individual nucleic acid bases to imidazolium-based ILs affirmed that ILs showed a propensity to bind to different bases, the highest being guanine followed by cytosine, thymine, uracil, and adenine.
1. Introduction
In recent years, the study on ionic liquids (ILs) has been a fascinating area of chemical research due to their unique properties such as low toxicity, high thermal and chemical stability, high boiling point, negligible vapor pressure, and excellent conductivity [1]. The applicability and versatility of ILs extend to fields such as organic synthesis [2,3], electrochemistry [4], catalysis and biocatalyst [2,5], separations [6], pharmaceuticals, and medicine [7,8], etc. As a matter of fact, the past decade has seen ILs being utilized in life science applications concerning biomolecules viz. proteins, enzymes, and nucleic acids [9,10].
It is undeniable to say that the utilization of ILs in DNA innovation has been broadly reported in recent years. However, most studies were focused on comprehending the progression and properties of DNA in ILs. Be that as it may, there exist a few conflicting reports with regards to the binding mode between the two molecules. A noteworthy point to highlight here, DNA carries the genome sequence, which plays key physiological roles in the life process of a living being. Interactions between DNA and small molecules, i.e., ILs via covalent or non-covalent interactions can potentially cause the alteration or inhibition of DNA function. Therefore, it is crucial that a molecular level clarification of the binding mode between DNA and ILs is performed to gain insight into the mechanisms of interactions that may affect the structural features of DNA.
Previously, it was reported that the cationic molecule of [Bmim][PF6] formed intercalation binding with DNA, based on resonance light scattering data [11]. Later, Ding et al. [12] proposed an aggregation-induced DNA perturbation by [Bmim][Cl]. Satpathi et al. [13] also described a similar work with the use of guanidinium tris(pentafluoroethyl) trifluorophosphate ionic liquid (Gua-IL). They discovered that the cationic head group of Gua-IL cation interacts with the negatively charged phosphate backbone of DNA, which leads to the compaction of the DNA structure. This work literacy was continued by Liu and co-workers [14] who investigated the impact of different alkyl chain lengths of imidazolium-based ILs towards calf thymus DNA using extensive photophysical methods. Data showed that this class of ILs formed intercalative mode binding with DNA, which corroborated earlier findings by Wang and co-workers [15]. Thus, it is clear that from the aforementioned findings, the mode of binding between ILs and DNA is still ambiguous and can be debatable among researchers.
In this milieu, an expedient way to observe and comprehend mechanisms of interactions between DNA and ILs may be done via a computationally integrated spectroscopic technique. Previously, Sugimoto and co-workers [16,17] used data from melting temperatures in combination with molecular dynamics (MD) simulations to visualize the interaction of cholinium dihydrogenphosphate ([Ch][Dhp]) with oligodeoxynucleotides. Their analytical technique highlighted fascinating information, for instance, the cholinium cation was found to prefer minor-groove instead of the major-groove binding on DNA structure. This led to the consequent stabilization and destabilization of the adenine-thymine (A-T) and guanine-cytosine (G-C) base pairs, respectively. Recently, MD simulation performed along with an experimental study revealed that the duplex DNA was stabilized by electrostatic attractions as the main interaction [18,19].
Due to the fact that DNA is a polyatomic anion with an infinite number of atoms, it is not practical to perform quantum mechanics (QM) calculation. Therefore, in this work, molecular docking and COSMO-RS studies were carried out to assess the binding mode between ILs and calf thymus DNA. The results are then compared with previously reported works on different ILs, thus that a conclusion can be made based on the binding mode of ILs to DNA.
2. Materials and Methods
2.1. Materials
Calf thymus DNA (ctDNA, ~10 kilo base pair (kbp), D1501) and ethidium bromide (EB, > 0.99) were purchased from Sigma-Aldrich (Darmstadt, Germany) and used as received. The purity of DNA was determined by calculating the ratio of absorbance at 260/280 nm. The ratio was estimated as 1.9, which implied the absence of any kind of protein contamination. The concentration of DNA was established using UV-Visible spectroscopy using the Beer-Lambert Law equation (, where represents the absorbance value (no unit), is the extinction coefficient constant (M−1 cm−1), is the concentration of a substance (M) and is the light path length (in cm). An extinction coefficient of 6600 M−1 cm−1 at 260 nm was employed, and the concentration of DNA was expressed in terms of base molarity of the phosphate groups [20]. All other chemicals were of analytical grade and used without further purification unless specified otherwise. All solutions were prepared using deionized water type III (Super Q Millipore system) with a conductivity below 18 μs cm−1. 1-alkyl-3-butylimidazolium bromide ILs ([Cnbim]Br where n = 2, 4, 6) were synthesized and characterized according to a previous method [21]. 1H, 13C NMR, and elemental analyses were used to check the purity of each IL. The ILs assessed here have been successfully used in previous studies [18,21].
2.2. Steady-State Fluorescence
Steady state fluorescence was carried out using a Cary Eclipse Fluorescence Spectrophotometer (Agilent, Santa Clara, CA, USA). The measurements were carried out using a 5.0 nm fixed band slits through a 1.0 cm path length quartz cuvette. All spectra were corrected for background intensities using a solution without DNA. The experiments were performed by continuous addition of 1 µL of 0.3 mM calf thymus DNA solution into a fixed 0.5 M [Cnbim]Br (where n = 2, 4, 6) ILs solution for each titration, and the emission intensity was observed. All triplicate titrations were recorded at 25 °C. The excitation wavelength and maximum emission for each [Cnbim]Br ILs were detected at 320 and 358 nm, respectively.
2.3. Molecular Docking Studies
The crystal structure of calf thymus DNA (PDB ID: 425D) was downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (https://www.rcsb.org/pdb) [22]. The TmoleX package (Version 4.1.1) (COSMOlogic, Leverkusen, Germany) was employed for geometry optimization of 3 imidazolium-based ILs’ cations using the Hartree-Fock theory with 6-31G* basic set [23]. A geometry optimization calculation using the Hartree-Fock level provided more meaningful, accurate values. The * accounts for the polarization effect of the species and was further optimized using the Density Functional Theory (DFT) level with the BP function that consisted of resolution of identity approximation and a triple-ξ valence polarized basis set. A similar optimization method has been employed previously [24,25]. The required pdbqt file for the DNA and cations were prepared using Auto Dock Tools (ADT) 1.5.4 [26]. Partial atomic charges for each atom in DNA were adopted from the Optimized Potential for Liquid Simulations (OPLS) force field [27], while the atomic charges for ILs were obtained from a previous study [18]. Parameters of the optimized ILs structures were successfully used in previous MD simulations and provided reliable data against the experimental evidence. The grid size for the x-, y- and z-direction was set to 100, 56, and 52, respectively, while the center of the search space was set to 29.58, 29.82, and 29.71 for the x-, y- and z-direction. AutoDock4 using Lamarckian Genetic Algorithm (LGA) was used for the docking studies between the DNA and the imidazolium-cation [28], and default values were used for other setting parameters. In this study, a total of 10 different conformations with different binding energies were generated. The conformation that exhibited the lowest energy value and most stable conformation (obtained from the clustering analysis) was identified as the best binding mode in each docking study. The 3D model of the docking results was visualized using PyMOL [29].
2.4. COSMO-RS Modelling
COnductor like Screening MOdel for Real Solvents (COSMO-RS) is the integration of the COSMO model used to predict thermos-physico-chemical properties of fluids in its pure and mixture state. Details on the calculation and procedure to estimate the excess enthalpy were obtained from the literature [30]. Initially, the nucleic acid bases (A, C, G, T, and U) were geometry optimized using the TmoleX package (Version 4.1.1). Then, COSMO-RS calculated the thermodynamic properties of fluid mixtures based on the 3D polarized charge distribution (σ, sigma) on the molecular surface of the individual bases, obtained from the quantum chemical calculations [31]. A 3D screening of the charge distribution on the molecular surface can be easily visualized on the histogram σ-profile. The information was useful for a qualitative description of the molecule and anticipated possible interactions of the compounds in a fluid mixture. Conversely, ILs were modeled as an equimolar mixture of cation and anion, considering that they were composed of 2 distinct ions, rather than of a single ionic molecule that conceptually resembled the actual situation. The thermodynamic calculation of the molecular interaction in this work was conducted using COSMOtherm software using the BP_TZVP_C30_1301 parameter file [24,25,31].
3. Results and Discussion
3.1. Steady State Fluorescence
fSteady state fluorescence spectroscopy is an effective technique to study the interaction of macromolecules and small compounds. In this work, the fluorescence spectroscopic method was applied to the imidazolium-based ILs as the compounds are known for non-negligible absorption, as described by Paul et al. [32] who observed the emission intensities of imidazolium-based ILs. They discovered fascinating fluorescence behaviors by the ILs at wavelengths beyond 300 nm, in which the [Cnbim]Br was excitation wavelength-dependent, exhibiting the highest peak at an excitation wavelength of 320 nm.
The type of binding can be identified by observing changes in the spectrum. According to Nakamoto et al. [33], the formation of a covalent bond tended to cause a red shift and hyperchromism, whereas intercalative binding contributed to bathochromism and hypochromism [34]. Moreover, electrostatic interaction was characterized by the lower hypochromicity without a bathochromic shift. Figure 1 illustrates the emission intensity of fixed concentration of 0.5 M [C2bim]Br during continuous addition of ctDNA. Most importantly, elevation in fluorescence intensity suggested that the ILs were capable of binding with the DNA.
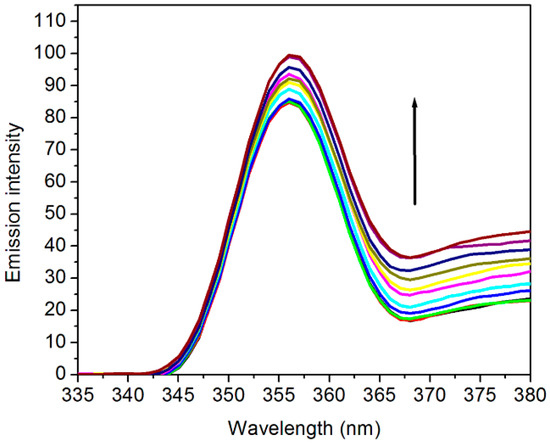
Figure 1.
Fluorescence emission spectra of [C2bim]Br in the absence (bottom curve) and presence of ctDNA in deionized water. The arrow indicates that the emission intensity increases with the increasing addition of DNA. The excitation wavelength was set at 320 nm.
The study predicted that the smaller sized cation molecules with shorter hydrocarbon chain lengths could enter the DNA grooves to create a more hydrophobic microenvironment between the base pairs. Figure 2 represents the DNA bases showing numerous recognition sites of C-G and A-T pairs. Adenine of the C-6 amine acts as a hydrogen bond donor, while Adenine N-7 and thymine C-4 carbonyl in the A-T base pair act as hydrogen bond acceptors. In contrast, the thymine methyl group acts as a stabilizer for van der Waals interaction. In the C-G base pair, cytosine C-4 amine can function as a hydrogen bond donor, whereas both guanine C-6 carbonyl and guanine N-7 can act as hydrogen bond acceptors. Non-covalent interactions are usually formed between a small molecule, for instance, the rod-shaped imidazolium cation, and DNA molecules at the DNA bases boundary and the grooves. Since the fluorescence emission is very sensitive, any changes in the local environment of ILs can invoke changes in the intensity of the signal. In this perspective, the increasing intensity of the fluorescence signifies changes in the local environment of the imidazolium cation from being well-solvated in aqueous solution to a hydrophobic surrounding.
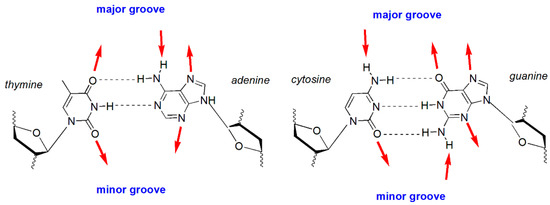
Figure 2.
Hydrogen bond recognition sites accessible from the DNA major and minor grooves. The outward facing arrows on the major and minor grooves represent the hydrogen bond receptors, while the inward facing arrows represent hydrogen bond donors.
3.2. Docking Studies
Docking is one of the well-founded methods to observe molecular level interactions between biomolecules and ligands. In this work, the binding energy was predicted to result from the interaction between the different alkyl chain length of imidazolium cation ([Cnbim]+ where n = 2, 4, and 6) and ctDNA. The searching grid space was set to cover the whole duplex DNA in order to get reliable data. By referring to Figure 3, the docking study analysis revealed that the cation of ILs preferably binds to the minor groove of DNA. Pertinently, the results of the docking study and fluorescence spectra agreed well with the observed increase in emission intensity of ILs.
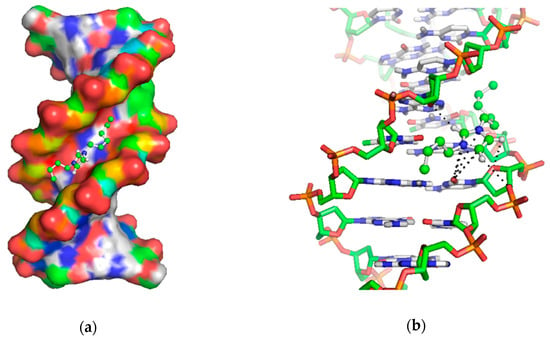
Figure 3.
(a) Docking pose of imidazolium-cation with calf thymus DNA; (b) close up of the predicted inter-molecular interaction between cation and DNA-bases.
Table 1 shows the tabulated data for the binding energy of docked structures between cations and DNA. Based on the predicted results, the change in alkyl chain lengths clearly influenced the binding energy () between the ILs and ctDNA. The value of binding energy () decreased as the length of the alkyl chain was increased. [C6bim]+ had the lowest energy (7.75 kcal mol−1), which corresponded to a binding constant of 3.44 × 103 L mol−1, with van der Waals and desolvation energies being the main sources of energy. In this scenario, the electrostatic energy was insignificant for the interaction. It is important to note that the desolvation energy in this study referred to the hydrophobic energy that resulted from the hydrophobic interaction between the hydrocarbon length of the cation and non-polar sites of the bases inside the groove. An earlier work that assessed the binding of the morpholinium bromide ionic liquid ([Mor1,2][Br]) to DNA also described a similar observation [35]. They reported that total energies of both van der Waals and desolvation were more negative than the electrostatic energy.

Table 1.
The binding energy of imidazolium cation to calf thymus DNA obtained from the docking simulation study.
In terms of binding energy, the value obtained in this study (5.19 to 7.75 kcal mol−1) was marginally higher compared to [Mor1,2][Br] (where = 4.57 kcal mol−1) [33]. By comparing to Haque et al. [36], their estimated binding energy of the [Ch][Cl]-DNA complex was 3.86 kcal mol−1, which was two times lower as compared to ours. Possible reasoning for this discrepancy was the existence of a planar geometry in the imidazolium ring that offered multiple sites of interaction in the minor grooves. In addition, the non-bonded cation-π interaction can be established from the charge delocalization among polar atoms in the planar group [37]. In our case, such an interaction occurred between the oxygen atom in cytosine that faced the aromatic imidazole cation (Figure 3b). Conversely, the binding between the cholinium-based ILs and DNA was found to be weak—largely due to the steric hindrance of the bulky choline cation. Similarly, morpholinium-based ILs exhibited slightly lower binding energy than imidazolium-based ILs as the ring of morpholinium cation was non-planar and large.
Based on the stable docked complex in Figure 3, the cation had a higher tendency to bind to A-T rich regions of residues DA19-DT6 and DT18-DA7. The binding occurred between the polar hydrogen atoms (C-H) in the imidazolium ring and oxygen atom (O4) at the backbone of DNA. Based on previous work [37], such an interaction was the consequence of the electrostatic attraction between the ring of imidazole and the backbone of DNA. Data of the docking study demonstrated that ILs with similar structures to the [Cnbim]+ cations tended to have a similar binding pattern. Notably, the addition of a hydrocarbon chain in the imidazolium cation could favorably improve the binding of ILs to DNA. On the contrary, the employment of increasingly longer alky chains reduced the binding energy, thereby inferring the formation of stronger hydrophobic interactions. The findings in this study are consistent with an earlier MD simulation that described a duplex DNA in hydrated imidazolium-based ILs that was stabilized by an electrostatic attraction and groove binding mechanism [21].
3.3. COSMO-RS Studies
3.3.1. Sigma Profiles of the Individual Compounds (Nucleic Acid Bases and ILs Molecules)
COSMO-RS was a reliable tool due to its ability to produces results with good agreement to the experimental data. COSMO-RS was utilized in this study to estimate the interaction energy in the ternary blend of water + IL with nucleic acid bases in a mixture state. The COSMO-RS theory prescribed that the σ-profile of molecules ranged from −0.03 to 0.03 e/Å2, which was the probability of the relative amount of surface segments with polarity, σ, on the surface of the molecule. The COSMO-RS histogram of σ-profile can be qualitatively divided into three main regions according to the cut-off values: The hydrogen bond donor (HBD) (σ < 0.008 e/Å2), the hydrogen bond acceptor (HBA) (σ > +0.008 e/Å2), and the nonpolar regions (0.008 < σ < +0.008 e/Å2). Figure 4 and Figure 5 depicts the σ-profile of the nucleic acid bases (A, C, G, T, and U) and the ions of ILs (i.e., [C2bim]+, [C4bim]+, and [C6bim]+ cations, and [Br]− anion).
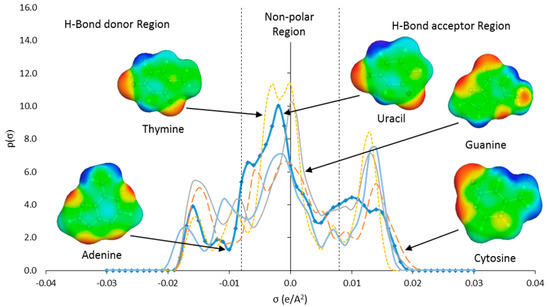
Figure 4.
Sigma profile of nucleic acid bases (adenine, cytosine, guanine, thymine, and uracil) predicted by COSMO-RS.
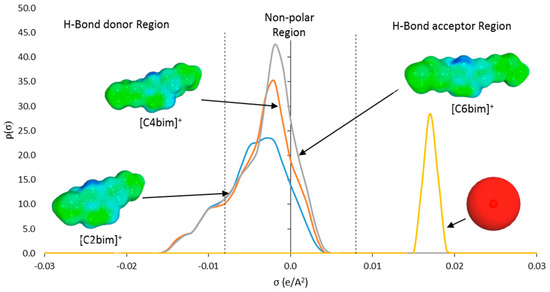
Figure 5.
Sigma profile of ILs ions ([C2bim]+, [C4bim]+, and [C6bim]+ cations, and [Br]− anion) predicted by COSMO-RS.
Based on the σ-profiles (Figure 4), all nucleic acid bases exhibited peaks in both HBD and HBA regions. These peaks can be interpreted as the locations where H-bonds may be formed between the base pairs, which resulted in the double helix structure. The peaks in the HBD region originated from polar H atoms attached to the N or O atom in the DNA structure (light blue colored sigma surface). Whereas, peaks in the HBA region identify polar N or O atoms in the structure of the bases (red colored sigma surface). The aromatic structure of the bases produced broad peaks in the nonpolar region (green colored sigma surface).
The main peak in the nonpolar region corresponding to charge distribution of 0.003 e/Å2 represented the non-polar attribute that originated from the aliphatic alkyl chain and the aromatic head group (green colored sigma surface) (Figure 5). Peak with charge distribution of 0.013 e/Å2 in the HBD region represented the acidic hydrogen atoms in the aromatic ring (light blue colored sigma surface). The increasing peak intensity in the nonpolar region denoted the increasing lengths of the aliphatic alkyl chain. In contrast, the single dominant peak in the HBA region signified the [Br]− anion at +0.018 e/Å2 (red shaded sigma surface) (Figure 5).
3.3.2. Prediction of Excess Enthalpies and Evaluation of Molecular Interactions in Ternary Mixture of Water + ILs with DNA Bases Systems Using COSMO-RS
Excess enthalpy, refers to the change of enthalpy following the introduction of DNA into the mixture of water and ILs. This value is useful to predict the strength of the interaction between nucleic acid bases and ILs in the mixture. The predicted whole composition for the water and IL mixtures is illustrated in Figure 6 and Figure A1 (Appendix A), respectively. Results showed that ILs bind significantly to various bases, the negative value indicated a favorable interaction between aqueous IL with free nucleic acid bases. A similar trend was observed in the aqueous mixture of [Ch]Cl + DNA bases [36]. The order of interaction for all types of ILs and the bases were ranked as follows: Guanine > cytosine > thymine > uracil > adenine, thus consistent with findings of another similar study [37]. A similar order of interaction was also obtained based on the excess Gibbs energy as shown in Figure 7 and Figure A2 in Appendix A.
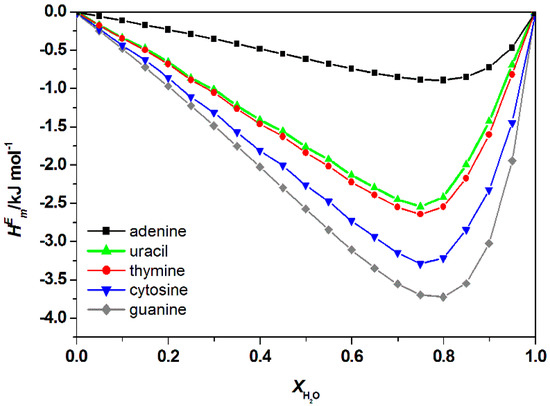
Figure 6.
Calculated total excess enthalpies, , of water + [C2bim]Br + nucleic acid bases at 298.15 K by COSMO-RS.
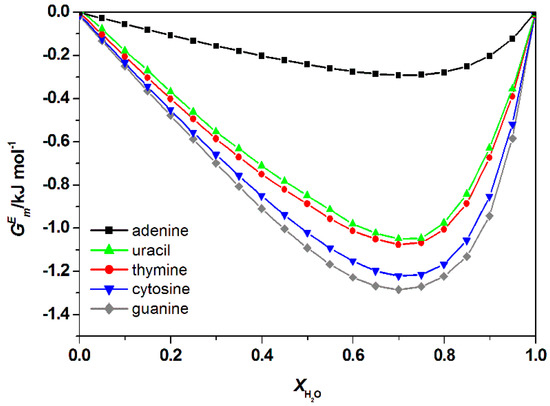
Figure 7.
Calculated total excess Gibb’s Free energy, , of water + [C2bim]Br + nucleic acid bases at 298.15 K by COSMO-RS.
Figure 8 shows the special contribution for aqueous [C2bim]Br with the guanine at 298.15 K at = 0.8 (for other studied ILs see Appendix A Figure A3, Figure A4 and Figure A5). Using COSMO-RS model, specific contribution to the can be estimated. is the sum of specific molecular interaction, which represents the electrostatic-misfit (), hydrogen bonds () and van der Waals forces (). The total contribution of each specific interaction can be summarized by the following equation:
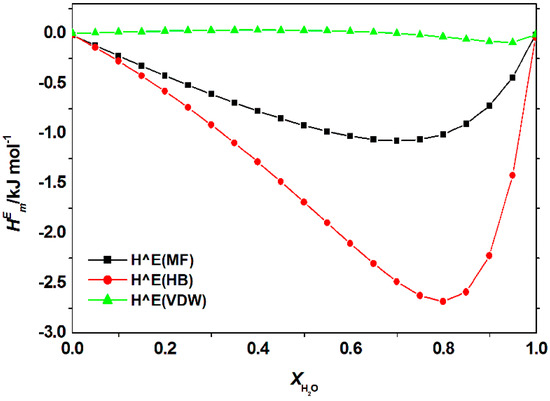
Figure 8.
Contribution of electrostatic-misfit interaction (MF); hydrogen bonds interaction (HB) and van der Waals forces (VDW) to the excess enthalpy, of the mixture of [C2bim]Br + water + nucleic acid base (guanine) at 298.15 K predicted by COSMO-RS.
Figure 9 illustrates the calculated for aqueous [C2bim]Br, [C4bim]Br and [C6bim]Br with guanine at 298.15 K at = 0.8 (for other studied nucleic acid bases see Appendix A Figure A6. The data revealed the negative value decreased with a further increase in the alkyl chain length of the cation for all studied systems. This trend explains the formation of a higher number of van der Waals interactions between the aqueous IL and guanine. For all aqueous solutions of ILs, the dominant interaction was the hydrogen bonding, which contributed to the exothermicity of the mixtures. The order of interaction based on for all ILs with nucleic acid bases were as follow: [C2bim]Br > [C4bim]Br > [C6bim]Br. A similar interaction order was also obtained based on excess Gibbs energy, as shown in Figure 10. Based on the COSMO-RS model, the most favorable interaction between aqueous ILs + nucleic acid bases mixtures is aqueous [C2bim]Br + guanine. The more negative the excess Gibbs energy, the more attractive the interaction between the ILs and the nucleic acid bases.
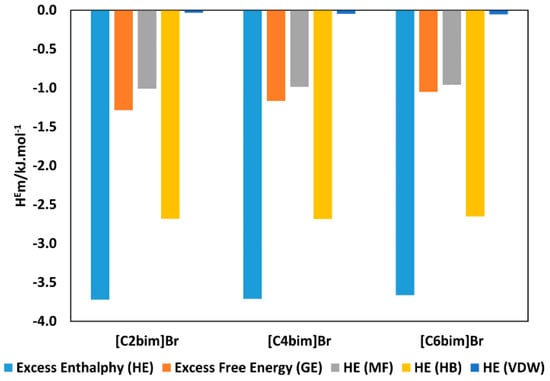
Figure 9.
Comparison of excess enthalpy, (blue bars) and excess free Gibbs energy, (orange bars), of aqueous [C2bim]Br with guanine mixture at 298.15 K predicted by COSMO-RS at = 0.8 and contribution of (gray bars), (yellow bars), and (purple bars) to the total excess enthalpy.
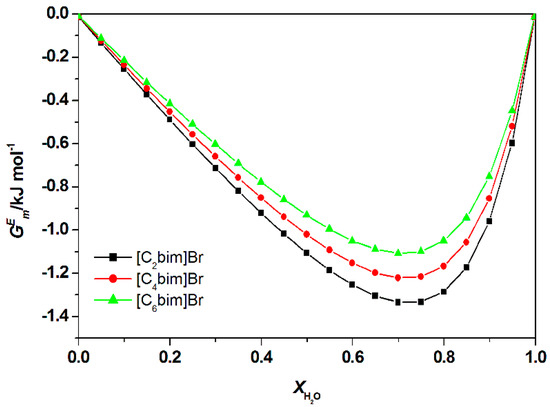
Figure 10.
Calculated total excess Gibb’s Free energy, , of water + ILs + guanine at 298.15 K by COSMO-RS.
4. Conclusions
The study performed a systematic assessment of the mode and strength of binding between calf thymus DNA and imidazolium-based ILs. Data on molecular docking revealed that ILs and calf thymus DNA formed a weak interaction with G in the range of 5.19 to 7.75 kcal mol−1. Aside from electrostatic attraction, the occurrence of T-shape π-π stacking between the bases and the aromatic ring of imidazole was also observed in the stable docking conformation. Despite that ILs interacted weakly with calf thymus DNA, the preparations exhibited potential affinity towards individual nuclei acid bases, as predicted by COSMO-RS. Most importantly, data of the molecular docking study corroborated the results of the fluorescence experiments. The study effectively demonstrated the mode of binding of imidazolium-based ILs to bases in which the binding preference increases from adenine, uracil, thymine, cytosine, and guanine. The findings of this study also emphasize the role of [Cnbim]Br ILs cation absorption for long term/high-temperature stabilization of DNA material. The present work demonstrated the role of the developed [Cnbim]Br ILs as solvents to stabilize DNA stored at room temperature. It was made apparent by the preferable binding of smaller cations of [Cnbim]Br ILs to the minor groove of ct-DNA, causing a formidable change in the microenvironment that to be more hydrophobic to expel water from immediate contact with DNA. This dehydration effect can extend the lifetime of ct-DNA by circumventing a hydrolytic attack that instigates rapid deamination and depurination while increasing the thermal stability of the base pairs. Notwithstanding safe storage and extending the lifetime of genetic blueprints, it is envisioned that the applicabilities of [Cnbim]Br ILs can be extended to the manipulation of nucleic acids and the fine-tuning of DNA microenvironment for compaction and possible applications in nanotechnology.
Author Contributions
Conceptualization, K.J., M.A.K., and M.B.A.R.; methodology, K.J. and M.A.K.; software, K.J.; validation, N.M.Y., H.A., and R.A.W.; formal analysis, K.J. and M.A.K.; investigation, N.M.Y., H.A., and R.A.W.; resources, K.J.; writing—original draft preparation, K.J. and M.A.K.; writing—review and editing, H.A., M.B.A.R., and R.A.W.; visualization, K.J., M.A.K., and N.M.Y.; supervision, M.B.A.R., H.A., and R.A.W.; funding acquisition, K.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Short Term Internal Research Funding, Universiti Teknologi PETRONAS, grant number: STIRF 0153AA-F21.
Acknowledgments
The authors would like to acknowledge AP Cecilia Devi Wilfred from Centre of Research in Ionic Liquids (CORIL), Universiti Teknologi PETRONAS for providing the TmoleX and COSMO-RS softwares.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
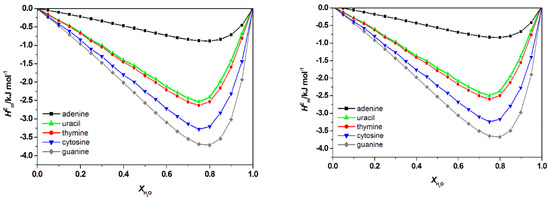
Figure A1.
Calculated total excess enthalpies , of water + [C4bim][Br] + nucleic acid bases (left) and , of water + [C6bim][Br] + nucleic acid bases (right) at 298.15 K by COSMO-RS. There is no significant differences on the total excess enthalpies , of nucleic acid bases in [Cnbim][Br ILs (where n = 2, 4, and 6).
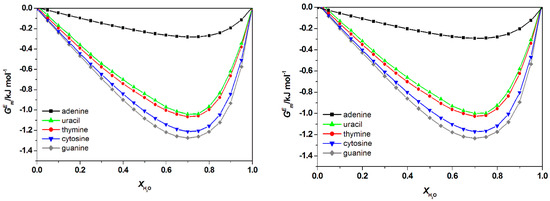
Figure A2.
Calculated total excess Gibb’s Free energy, , water + [C4bim][Br] + nucleic acid bases (left) and water + [C6bim][Br] + nucleic acid bases (right) at 298.15 K by COSMO-RS. There is no significant differences on total Gibb’s Free energy , of nucleic acid bases in [Cnbim][Br ILs (where n = 2, 4, and 6).
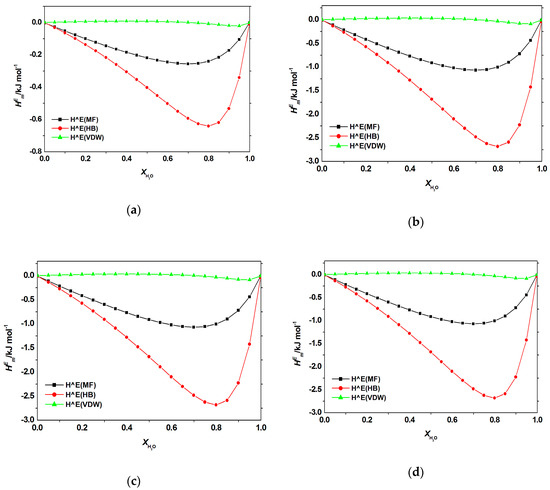
Figure A3.
Contribution of electrostatic-misfit interaction (MF); hydrogen bonds interaction (HB); van der Waals forces (VDW) to the excess enthalpy of aqueous mixtures of [C2bim]Br with (a) adenine; (b) cytosine; (c) thymine, and (d) uracil at 298.2 K predicted by COSMO-RS.
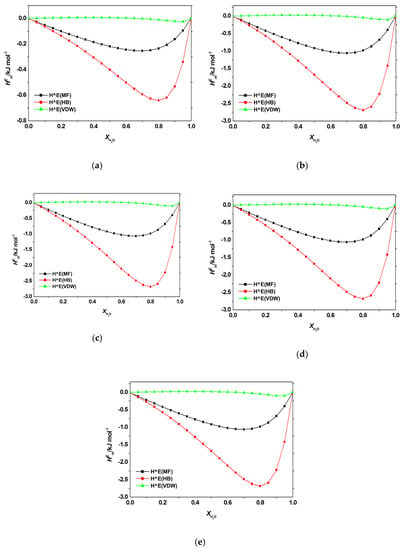
Figure A4.
Contribution of electrostatic-misfit interaction (MF); hydrogen bonds interaction (HB); van der Waals forces (VDW) to the excess enthalpy of aqueous mixtures of [C4bim]Br with (a) adenine; (b) cytosine; (c) guanine, (d) thymine, (e) uracil at 298.2 K predicted by COSMO-RS.
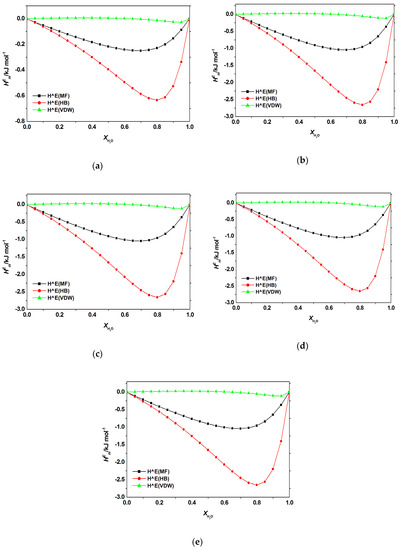
Figure A5.
Contribution of electrostatic-misfit interaction (MF); hydrogen bonds interaction (HB); van der Waals forces (VDW) to the excess enthalpy of aqueous mixtures of [C6bim]Br with (a) adenine; (b) cytosine; (c) guanine, (d) thymine, (e) uracil at 298.2 K predicted by COSMO-RS.
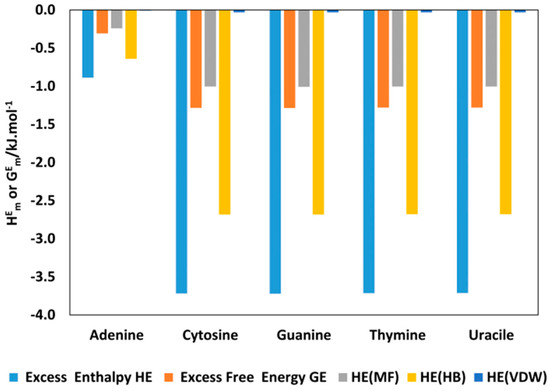
Figure A6.
Comparison of excess enthalpy, (blue bars) and excess free Gibbs energy, (orange bars), of aqueous [C2bim]Br with nucleic acid bases mixture at 298.15 K predicted by COSMO-RS at = 0.8 and contribution of (gray bars), (yellow bars), and (purple bars) to the total excess enthalpy.
References
- Hallett, J.P.; Welton, T. Room-temperature ionic liquids: Solvents for synthesis and catalysis. Chem. Rev. 2011, 111, 3508–3576. [Google Scholar] [CrossRef] [PubMed]
- van Rantwijk, F.; Sheldon, R.A. Biocatalysis in ionic liquids. Chem. Rev. 2007, 107, 2757–2785. [Google Scholar] [CrossRef] [PubMed]
- Greaves, T.L.; Drummond, C.J. Protic ionic liquids: Properties and applications. Chem. Rev. 2008, 108, 20–237. [Google Scholar] [CrossRef]
- Macfarlane, D.R.; Forsyth, M.; Howlett, P.C.; Pringle, J.M.; Sun, J.; Annat, G.; Neil, W.; Izgorodina, E.I. Ionic liquids in electrochemical devices and processes: Managing interfacial electrochemistry. Acc. Chem. Res. 2007, 40, 1165–1173. [Google Scholar] [CrossRef]
- Barthel, J.; Gores, H.; Mamantov, G.; Papov, A. Chemistry of Nonaqueous Solutions: Current Progress, 1st ed.; Wiley: New York, NY, USA, 1994. [Google Scholar]
- Plechkova, N.V.; Seddon, K.R. Applications of ionic liquids in the chemical industry. Chem. Soc. Rev. 2008, 37, 123–150. [Google Scholar] [CrossRef]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological activity of ionic liquids and their application in pharmaceutics and medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef]
- Giernoth, R. Task-specific ionic liquids. Angew. Chem. Int. Ed. 2010, 49, 2834–2839. [Google Scholar] [CrossRef]
- Vijayaraghavan, R.; Izgorodin, A.; Ganesh, V.; Surianarayanan, M.; MacFarlane, D.R. Long-term structural and chemical stability of DNA in hydrated ionic liquids. Angew. Chem. Int. Ed. 2010, 49, 1631–1633. [Google Scholar] [CrossRef]
- Figueiredo, A.M.; Sardinha, J.; Moore, G.R.; Cabrita, E.J. Protein destabilisation in ionic liquids: The role of preferential interactions in denaturation. Phys. Chem. Chem. Phys. 2013, 15, 19632–19643. [Google Scholar] [CrossRef]
- Cheng, D.H.; Chen, X.W.; Wang, J.H.; Fang, Z.L. An abnormal resonance light scattering arising from ionic-liquid/DNA/ethidium interactions. Chem. Eur. J. 2007, 13, 4833–4839. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, L.; Xie, J.; Guo, R. Binding characteristics and molecular mechanism of interaction between ionic liquid and DNA. J. Phys. Chem. B 2010, 114, 2033–2043. [Google Scholar] [CrossRef] [PubMed]
- Satpathi, S.; Sengupta, A.; Hridya, V.M.; Gavvala, K.; Koninti, R.K.; Roy, B.; Hazra, P. A green solvent induced DNA package. Sci. Rep. 2015, 5, 9137. [Google Scholar] [CrossRef]
- Liu, H.; Dong, Y.; Wu, J.; Chen, C.; Liu, D.; Zhang, Q.; Du, S. Evaluation of interaction between imidazolium-based chloride ionic liquids and calf thymus DNA. Sci. Total Environ. 2016, 566, 1–7. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Zhang, S. Binding Gibbs energy of ionic liquids to calf thymus DNA: A fluorescence spectroscopy study. Phys. Chem. Chem. Phys. 2011, 13, 3906–3910. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Sugimoto, N. Structure, stability and behaviour of nucleic acids in ionic liquids. Nucleic Acids Res. 2014, 42, 8831–8844. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Sugimoto, N. A-T base pairs are more stable than G-C base pairs in a hydrated ionic liquid. Angew. Chem. Int. Ed. 2012, 51, 1416–1419. [Google Scholar] [CrossRef]
- Jumbri, K.; Abdul Rahman, M.B.; Abdulmalek, E.; Ahmad, H.; Micaelo, N.M. An insight into structure and stability of DNA in ionic liquids from molecular dynamics simulation and experimental studies. Phys. Chem. Chem. Phys. 2014, 16, 14036–14046. [Google Scholar] [CrossRef]
- Chandran, A.; Ghoshdastidar, D.; Senapati, S. Groove binding mechanism of ionic liquids: A key factor in long-term stability of DNA in hydrated ionic liquids? J. Am. Chem. Soc. 2012, 134, 20330–20339. [Google Scholar] [CrossRef]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P. A further examination of the molecular weight and size of desoxypentose nucleic acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Jumbri, K.; Ahmad, H.; Abdulmalek, E.; Abdul Rahman, M.B. Binding energy and biophysical properties of ionic liquid-DNA complex: Understanding the role of hydrophobic interactions. J. Mol. Liq. 2016, 223, 1197–1203. [Google Scholar] [CrossRef]
- Rozenberg, H.; Rabinovich, D.; Frolow, F.; Hegde, R.S.; Shakked, Z. Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets. Proc. Natl. Acad. Sci. USA 1998, 95, 15194–15199. [Google Scholar] [CrossRef] [PubMed]
- Micaelo, N.M.; Baptista, A.M.; Soares, C.M. Parametrization of 1-butyl-3-methylimidazolium hexafluorophosphate/nitrate ionic liquid for the GROMOS force field. J. Phys. Chem. B 2006, 110, 14444–14451. [Google Scholar] [CrossRef] [PubMed]
- Gonfa, G.; Muhammad, N.; Bustam, M.A. Probing the interactions between DNA nucleotides and biocompatible liquids: COSMO-RS and molecular simulation study. Sep. Purif. Technol. 2018, 196, 237–243. [Google Scholar] [CrossRef]
- Rashid, Z.; Wilfred, C.D.; Iyyaswami, R.; Arunagiri, A.; Thanabalan, M. Investigating the solubility of Petroleum Asphaltene in ionic liquids and their interaction using COSMO-RS. J. Ind. Eng. Chem. 2019, 79, 194–203. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Schrodinger, L.L.C. The PyMOL Molecular Graphics System, Version 1.8; Schrödinger: New York, NY, USA, 2015. [Google Scholar]
- Kurnia, K.A.; Coutinho, J.A.P. Overview of the excess enthalpies of the binary mixtures composed of molecular solvents and ionic liquids and their modelling using COSMO-RS. Ind. Eng. Chem. Res. 2013, 52, 13862–13874. [Google Scholar] [CrossRef]
- Eckert, F.; Klamt, A. COSMOtherm Version C2. 1 Release 01.08; COSMOlogic GmbH & Co. KG: Leverkusen, Germany, 2006. [Google Scholar]
- Paul, A.; Mandal, P.K.; Samanta, A. On the optical properties of the imidazolium ionic liquids. J. Phys. Chem. B 2005, 109, 9148–9153. [Google Scholar] [CrossRef]
- Nakamoto, K.; Tsuboi, M.; Strahan, G.D. Drug-DNA Interactions: Structures and Spectra; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Indumathy, R.; Kanthimathi, M.; Weyhermuller, T.; Nair, B.U. Cobalt complexes of terpyridine ligands: Crystal structure and nuclease activity. Polyhedron 2008, 27, 3443–3450. [Google Scholar] [CrossRef]
- Pabbathi, A.; Samanta, A. Spectroscopic and molecular docking study of the interaction of DNA with a morpholinium ionic liquid. J. Phys. Chem. B 2015, 119, 11099–11105. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Khan, I.; Hassan, S.I.; Khan, M.S. Interaction studies of cholinium-based ionic liquids with calf thymus DNA: Spectrophotometric and computational methods. J. Mol. Liq. 2017, 237, 201–207. [Google Scholar] [CrossRef]
- Jumbri, K.; Micaelo, N.M.; Abdul Rahman, M.B. Solvation free energies of nucleic acid bases in ionic liquids. Mol. Sim. 2017, 43, 19–27. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).