Abstract
Current diagnosis of autism spectrum disorder (ASD) is based on assessment of behavioral symptoms, although there is strong evidence that ASD affects multiple organ systems including the gastrointestinal (GI) tract. This study used Fisher discriminant analysis (FDA) to evaluate plasma metabolites from 18 children with ASD and chronic GI problems (ASD + GI cohort) and 20 typically developing (TD) children without GI problems (TD − GI cohort). Using three plasma metabolites that may represent three general groups of metabolic abnormalities, it was possible to distinguish the ASD + GI cohort from the TD − GI cohort with 94% sensitivity and 100% specificity after leave-one-out cross-validation. After the ASD + GI participants underwent Microbiota Transfer Therapy with significant improvement in GI and ASD-related symptoms, their metabolic profiles shifted significantly to become more similar to the TD − GI group, indicating potential utility of this combination of plasma metabolites as a biomarker for treatment efficacy. Two of the metabolites, sarcosine and inosine 5′-monophosphate, improved greatly after treatment. The third metabolite, tyramine O-sulfate, showed no change in median value, suggesting it and correlated metabolites to be a possible target for future therapies. Since it is unclear whether the observed differences are due to metabolic abnormalities associated with ASD or with GI symptoms (or contributions from both), future studies aiming to classify ASD should feature TD participants with GI symptoms and have larger sample sizes to improve confidence in the results.
1. Introduction
It is currently estimated that 1.7% of children in the United States are diagnosed with autism spectrum disorder (ASD) [1]. The diagnosis of ASD is based on assessment of behavioral symptoms, which include major impairments in social communication, stereotyped behaviors, and restricted interests [2]. Although there is strong evidence that ASD often begins prenatally due to a complex interaction of genetic and environmental factors [3,4], diagnosis of ASD postnatally is difficult at early ages since some obvious symptoms are not present in early infancy and other symptoms are difficult to distinguish from normal development. It is important to diagnose children with ASD as young as possible since available interventions are most effective if started early in life [5]. One national prevalence study of eight-year-olds with ASD found that the median age of diagnosis was 46 months for autism and 52 months for ASD [1]; however, this study did not account for children and adults diagnosed at ages above eight years, so the true median age of diagnosis is even higher. Stable diagnoses of ASD have been found in children as young as 18 months [6], representing a significant disconnect between current and ideal outcomes.
Although ASD is currently diagnosed based upon behavior, there are physiological factors affected by or contributing to ASD. Development of a biomarker-based test for ASD, using quantifiable measures rather than qualitative judgment, would assist with screening for and diagnosing ASD earlier in childhood [7]. This, in turn, would indicate if further evaluation is needed and allow for intervention and/or therapy to begin as early as possible. Early intervention may maximize the opportunity for improving neural connectivity while brain plasticity is still high [8], likely helping to reduce the severity of ASD or even prevent it from fully manifesting [9]. A number of intervention models have been demonstrated to be significantly beneficial for many children with ASD [10], such as the Early Start Denver Model that has been found effective when started in early infancy [11]. It is thus important to diagnose children with ASD, or at high risk of ASD, as soon as possible so that intervention can begin.
Besides aiding with diagnosis, ASD-related biomarkers may offer value for evaluating treatment efficacy. These would serve as complements to current behavioral/symptom assessments and help to further elucidate the underlying biological mechanisms contributing to ASD. For example, multivariate statistical analysis of changes in plasma metabolites has been found to offer value for modeling changes in metabolic profiles and adaptive behavior resulting from clinical intervention [12]. Functional neuroimaging biomarkers may also be promising indicators of biological response to treatment [13]. In addition, eye-tracking metrics could represent further avenues for quantifying changes in behavior resulting from intervention [14,15]. As with diagnostic biomarkers, such approaches can help to mitigate subjectivity in treatment assessment arising from the use of behavioral measures.
Beyond its core symptoms, ASD is also associated with a number of co-occurring conditions that contribute to significant heterogeneity in clinical manifestations of the disorder [16]. Gastrointestinal (GI) problems are one such group of conditions that are common in children with ASD [17,18], especially constipation and/or diarrhea, and are strongly correlated with more severe ASD-related symptoms [19]. Chronic GI symptoms may be due to perturbed gut microbiome homeostasis in individuals with ASD [20], with the resulting metabolic abnormalities possibly contributing to altered GI and nervous system function [21]. If the gut microbiome does indeed have roles in ASD pathophysiology, then correcting its abnormalities may offer one therapeutic pathway for alleviating the symptoms of ASD and its co-occurring conditions [22,23].
This work presents the results of a pilot study using multivariate statistical modeling to highlight differences between 18 children with ASD and chronic GI disorders and a group of 20 typically developing peers (TD) without GI symptoms. The classification model was then applied to the same participants with ASD during and after treatment with Microbiota Transfer Therapy (MTT) [24] to validate its use as a marker of metabolic changes due to clinical treatment.
2. Materials and Methods
2.1. Study Population and MTT Treatment
The details of the study population and MTT protocol are outlined in a previous study [24]. Briefly, the study involved 18 children with ASD and chronic GI problems (ASD + GI cohort) and 20 TD children without GI problems (TD − GI cohort), all aged 7–16 years old. ASD + GI participants’ medical records from the previous two years were extensively reviewed by the study physician to determine eligibility for the study. Diagnoses of ASD were then verified using the Autism Diagnostic Interview-Revised through a phone interview with the parents by an evaluator. This was followed by a general physical health examination by the study physician to verify that the children had chronic GI symptoms of moderate to severe severity. Exclusion criteria included antibiotic use in the previous six months or probiotic use in the previous three months, dependence on tube feeding, the presence of life-threatening GI problems, having recent or scheduled surgeries, being severely malnourished or underweight, and being diagnosed with a single-gene disorder, major brain malformation, ulcerative colitis, Crohn’s disease, celiac disease, or eosinophilic esophagitis. TD children were identified as those not having a diagnosed mental disorder including ASD, attention-deficit hyperactivity disorder, depression, or anxiety; in addition, none of the TD children had parents or siblings with ASD.
MTT consisted of two weeks of oral vancomycin (an antibiotic to reduce pathogenic bacteria), one day of fasting and MoviPrep (a bowel cleanse to remove the vancomycin and further reduce levels of intestinal bacteria), one or two days of a high-dose of fecal microbiota (FM), and seven or eight weeks of low-dose FM. The FM consisted of a full spectrum of highly-purified microbiota extracted from stool samples of healthy, carefully-screened donors and prepared as previously described [25]. Prilosec, a stomach acid suppressant, was also administered during eight weeks of treatment to increase the survival of orally administered FM through the stomach.
Improvement of GI symptoms through MTT was primarily assessed by the Gastrointestinal Symptom Rating Scale (GSRS) [26] as completed by parents/guardians. The GSRS contains fifteen questions scored in five domains (abdominal pain, reflux, indigestion, diarrhea, and constipation) for evaluating GI symptoms during the previous week on a seven-point Likert scale. From the beginning to the end of MTT, the average GSRS score decreased 82% compared to baseline; eight weeks after treatment stopped, the average score was still 77% lower than at baseline. Changes in ASD-related symptoms were evaluated by the Childhood Autism Rating Scale (CARS), Social Responsiveness Scale, Aberrant Behavior Checklist, and Parental Global Impressions-III (PGI-III). Compared to baseline, the average CARS score decreased by 22% after MTT and by 24% after the eight weeks of follow-up [24]. A significant negative correlation was also detected between the change in GSRS and PGI-III (Spearman rank correlation coefficient of −0.59) [24].
2.2. Metabolite Measurements
Plasma samples were collected by phlebotomists in the morning from fasting participants. The samples were frozen immediately and stored in a −80 °C freezer. When all samples for the study were collected, they were shipped on dry ice to Metabolon (Durham, NC, USA), where sample preparation and data acquisition were processed to obtain metabolite profiling. Samples were extracted and analyzed by the Metabolon platform with the ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) instruments. The Metabolon platform consists of sample accessioning, sample preparation, quality assurance/quality control, and UPLC-MS/MS measurements, and the detailed information is described by Long et al. [27]. Measurements for a total of 621 plasma metabolites were available for this study.
Plasma samples were collected from all ASD + GI participants at baseline (Week 0), after the administration of oral vancomycin prior to microbiota transfusion (Week 3), and after the end of MTT treatment (Week 10). Plasma samples from TD − GI controls were only collected at Week 0 as these participants did not undergo treatment.
2.3. Statistical Methods
Multivariate analysis was performed with Fisher discriminant analysis (FDA) [28]. The objective of FDA is to determine a linear combination of metabolites that best separates the ASD + GI and TD − GI study cohorts at baseline (pre-treatment). Prior to FDA, each metabolite measurement was rescaled so that the mean value in the combined ASD + GI and TD − GI groups was 0 with a standard deviation of 1. A discriminant score was calculated by FDA for each study participant by multiplying each input metabolite measurement by a calculated parameter value and then summing these products together. The parameters for each metabolite were estimated such that the difference in mean discriminant score between the ASD + GI cohort and TD − GI cohort was maximized, and the variance of scores within each cohort was simultaneously minimized. Further mathematical details of the algorithm are provided in a previous paper by the authors [29]. Although other methods can be used, FDA has been found to be well-suited for this type of research [30].
Performing FDA with all 621 metabolites would lead to model overfitting and minimize generalizability of any findings; thus, it was necessary to identify an optimal subset of measurements for FDA. To facilitate this analysis, the most significant metabolites for classification were determined through a two-step process. First, any metabolites having fewer than fifteen measurements (i.e., 40% of participants) above the detection limit were excluded. The rationale with this step was to focus only on metabolites that had continuous distributions of values across participants, while still allowing for the possibility that a measurement could be almost entirely below the detection limit in one cohort and above the limit in the other cohort. A univariate analysis was then conducted to compute a receiver operating characteristic (ROC) curve for each individual metabolite by plotting the false positive rate against the true positive rate at different ASD + GI/TD − GI classification thresholds. The area under the ROC curve (AUROC) was then calculated to quantify the separation between the ASD + GI and TD − GI cohorts offered by the metabolite. AUROC values typically range between 0.5 and 1.0, with 0.5 reflecting uninformative classification and 1.0 denoting perfect separation. A general understanding is that AUROC values of 0.5–0.6 indicate meaningless classification, 0.6–0.7 is poor, 0.7–0.8 is average, and 0.8–0.9 is good, [31], although interpretations may vary by discipline. AUROC values between 0.9–1.0 reflect excellent classification and are desirable for diagnostic tests. For this study, metabolites yielding an AUROC of at least 0.70 were selected as candidates for multivariate analysis with FDA.
Classification with FDA first involved using the top candidate metabolites to exhaustively evaluate all combinations of up to five metabolites. For each number of metabolites used (two, three, four, or five), the 1000 combinations producing the highest AUROC from the fitted discriminant scores were retained. The distributions of discriminant scores within each cohort yielded by each top combination in FDA were then estimated with kernel density estimation; this method uses Gaussian kernels to approximate the probability density functions (PDFs) of the discriminant scores. Defining our classification threshold for separating these distributions to be the null hypothesis H0, which states that a given sample belongs to the TD − GI cohort, the Type I (false positive) error is then taken to be the probability of incorrectly diagnosing a TD − GI participant as being ASD + GI. Similarly, the Type II (false negative) error β is defined as the probability of incorrectly diagnosing an ASD + GI participant as being TD − GI. The Type I and Type II errors were calculated based on H0 with respect to the PDFs obtained from model fitting.
The top 1000 combinations for each number of metabolites were evaluated with leave-one-out cross-validation, in which the classification of each participant was predicted using an FDA model fitted to the remaining (n minus 1) participants’ samples. This step is important, as it means that rather than merely fitting to the data, an estimate of the model’s ability to predict new data was obtained. It generally yields lower accuracies than fitting procedures, but the results are more likely to reflect generalizability to larger data sets. To evaluate each candidate model, the cross-validated sensitivity (or true positive rate, TPR, calculated as the number of correctly classified ASD + GI participants divided by the total number of ASD + GI participants) and specificity (or true negative rate, TNR, calculated as the number of correctly classified TD − GI children divided by the total number of TD−GI children) were calculated at values of the classification threshold H0 for which β = 0.01, 0.05, 0.10, and 0.20; modulating H0 in this manner allowed for characterization of the cross-validated performance of each model when placing the threshold at different points along the ASD + GI distribution.
To further evaluate individual FDA models, sample-level classification accuracies (CAs) and misclassification errors (MEs) resulting from leave-one-out cross-validation were also assessed. While holding out each participant in cross-validation, the PDFs of discriminant scores for the remaining n minus 1 participants were estimated. The percent membership of a held-out sample in its own cohort’s PDF (i.e., the probability of being classified in the correct cohort) was taken to be that sample’s CA, while the percent membership in the incorrect cohort’s PDF (i.e., the probability of being classified in the incorrect cohort) was defined to be the sample’s ME. High-confidence models are those having many samples with CA greater than 0.05 and ME less than 0.05.
The FDA model developed at baseline was used to assess changes in the plasma metabolite data for ASD + GI participants at Week 3 and Week 10 of MTT. Data from these time points were rescaled according to the mean and standard deviation parameters used to rescale the Week 0 metabolites. Changes resulting from MTT were quantified by the Type II error, with respect to the threshold H0, associated with the PDF of the ASD + GI cohort’s discriminant scores at each time point. While the goal in hypothesis testing is typically to minimize Type II error, a larger Type II error is desired in this analysis since it is expected that successful treatment will make the ASD + GI cohort’s distribution less distinguishable from that of the TD − GI cohort. An additional metric of the MTT effect was the effect size, calculated at each time point as the median difference in discriminant score from baseline (where each participant’s sample was paired with their baseline sample); the 95% confidence interval (CI) for the effect size was calculated by non-parametric bootstrap resampling with 10,000 resamples [32]. Significance level α = 0.05 was used for all hypothesis testing.
3. Results
3.1. Univariate Analysis
From 621 total metabolites, 45 metabolites were excluded for not having at least fifteen values above the detection limit. Two others (EDTA and HEPES) were excluded for being possible artifacts of sample processing or possibly being misidentified, leaving 574 metabolites for univariate ROC analysis. Of those 574 metabolites, 61 yielded an AUROC of at least 0.70 (Table 1). No single metabolite perfectly separated the ASD + GI and TD − GI cohorts (i.e., had an AUROC of 1.0), but 11 metabolites did have an AUROC greater than 0.80, suggesting modest potential for univariate classification with these measurements.

Table 1.
The 61 plasma metabolites with at least fifteen values above the detection limit and yielding an area under the receiver operating characteristic curve (AUROC) of at least 0.70 on a univariate basis.
3.2. Classification with FDA
3.2.1. Model Development and Selection
The most promising combinations of two, three, four, and five metabolites for FDA are listed in Table 2. These combinations yielded the highest sensitivity and specificity after cross-validation, which also coincided with high AUROC from the fitted models. Perfect separation from fitting (AUROC of 1.0) was observed with as few as three metabolites and perfect prediction from cross-validation (100% sensitivity and specificity) with as few as four metabolites; here we used the term “separation” for indicating the difference between cohorts yielded by model fitting, and “prediction” when presenting cross-validated results. In the interest of balancing high model accuracy and low model complexity, the best three-metabolite model using sarcosine, tyramine O-sulfate, and inosine 5′-monophosphate (IMP) as inputs (hereafter referred to as the PM3, or plasma model with three metabolites) was considered for further analysis rather than the best four- and five-metabolite models. Assessing model accuracy at different classification thresholds of β provided an indication of the optimal cut-off between the ASD + GI and TD − GI distributions, which was determined to be at β = 0.05 for the PM3.

Table 2.
Fitting and cross-validation results for the combinations of two, three, four, and five metabolites yielding the highest fitted area under the receiver operating characteristic curve (AUROC) when used in the Fisher discriminant analysis. The cross-validated sensitivity (or true positive rate, TPR) and specificity (or true negative rate, TNR) are shown for classification thresholds associated with different values of the Type II error (β) calculated from the fitted probability density functions.
Univariate separation between the ASD + GI and TD − GI cohorts provided by each PM3 metabolite is visualized in Figure 1a, which shows that the cohorts separate well when accounting for the three metabolites together. The ROC curves for each PM3 metabolite (Figure 1b) also indicate an ability to classify the two cohorts with average to good accuracy individually (the AUROC values for sarcosine, tyramine O-sulfate, and IMP were 0.83, 0.71, and 0.87, respectively), albeit not as accurately as with the multivariate model where the fitted AUROC value was 1.00 (Table 2).
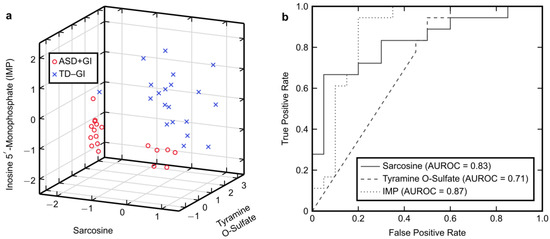
Figure 1.
Univariate characteristics of the plasma model using three metabolites (PM3). (a) Three-dimensional plot for visualizing separation between the autism spectrum disorder with gastrointestinal symptoms (ASD + GI) cohort and the typically developing without gastrointestinal symptoms (TD − GI) cohort at baseline provided by the (rescaled) PM3 metabolites. (b) Receiver operating characteristic (ROC) curves and the area under these curves (AUROC) for each PM3 metabolite.
Many other combinations of metabolites also had results comparable to those presented, with the lowest fitted AUROC being 0.97 among the top 1000 combinations of three metabolites. All top 61 metabolites appeared in at least five top combinations (Figure 2), with cysteinylglycine used the least frequently and appearing in only five combinations (0.5%) total. However, 19 metabolites appeared in at least 5% of combinations. The most frequently used metabolites were IMP (52% of combinations) and 3-phosphoglycerate (32%). Besides IMP, the remaining metabolites comprising the PM3, sarcosine and tyramine O-sulfate, appeared in only 9.3% and 7.2% of combinations, respectively.
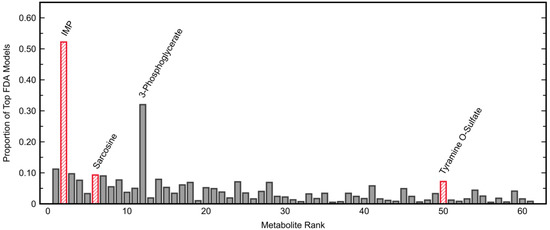
Figure 2.
Frequency of appearance of each top plasma metabolite in the top 1000 three-metabolite Fisher discriminant analysis models. Metabolites are ranked from highest to lowest area under the receiver operating characteristic curve (AUROC) as indicated in Table 1, with the metabolites of the plasma model using three metabolites (PM3) highlighted separately. The two metabolites most commonly appearing in the top combinations were inosine 5′-monophosphate (IMP; 52%) and 3-phosphoglycerate (32%).
The large number of combinations providing good separation between ASD + GI and TD − GI cohorts was likely due to many of the metabolites within the top 61 being highly correlated with each other (Table 3). Sarcosine had the most significant correlations with other metabolites, with the magnitude of the correlation coefficient being as high as 0.96. Tyramine O-sulfate and IMP (maximum correlation coefficients of 0.54 and 0.81, respectively) also had significant correlations with other metabolites. As expected, the PM3 metabolites were not significantly correlated with each other (i.e., each one likely represents a different set of metabolic differences in the ASD + GI cohort). It is worth noting that IMP and 3-phospholgycerate, the metabolites used most frequently in the top combinations (Figure 2), were highly correlated.

Table 3.
The ten metabolites most correlated with each of the metabolites used in the plasma model with three metabolites (PM3), and the correlations between the PM3 metabolites.
3.2.2. Model Fitting and Cross-Validation
Model fitting with the PM3 metabolites provided good separation between the discriminant scores of the ASD + GI and TD − GI cohorts (Figure 3a). Setting the classification threshold at β = 0.05 (i.e., Type II error = 5%) based on the estimated PDFs yielded a Type I error of 2.0% (Figure 3b). After leave-one-out cross-validation, there was still good separation in the predicted discriminant scores (Figure 3c). By classifying samples according to the shown threshold H0, the PM3 achieved 94% sensitivity and 100% specificity after cross-validation (Figure 3d). Positive and negative predictive values were also high for this classification task. In total, the PM3 incorrectly predicted only one ASD + GI participant while correctly predicting all TD − GI participants. Most samples also had high sample-level CA and low sample-level ME after cross-validation (Figure 4), indicating overall high confidence for model predictions.
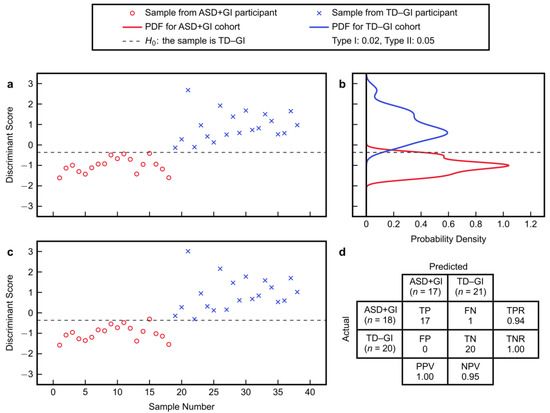
Figure 3.
Fisher discriminant analysis using the best combination of three metabolites to separate the autism spectrum disorder with gastrointestinal symptoms (ASD + GI) cohort and the typically developing with no gastrointestinal symptoms (TD − GI) cohort: (a) fitted discriminant scores, (b) probability density functions (PDFs) of the fitted scores, (c) cross-validated scores, and (d) confusion matrix from cross-validation based on classification with the shown threshold. The confusion matrix summarizes the number of true positive (TP), false negative (FN), false positive (FN), and true negative (TN) predictions. The true positive rate is TPR = TP/(TP + FN), the true negative rate is TNR = TN/(TN + FP), the positive predictive value is PPV = TP/(TP + FP), and the negative predictive value is NPV = TN/(TN + FN).
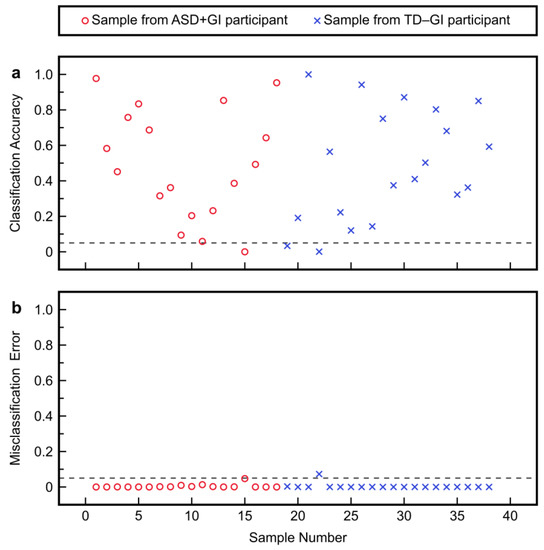
Figure 4.
Cross-validation results with the plasma model with three metabolites (PM3) to separate the autism spectrum disorder with gastrointestinal symptoms (ASD + GI) cohort from the typically developing with no gastrointestinal symptoms (TD − GI) cohort: (a) sample-level classification accuracies (CAs) and (b) sample-level misclassification errors (MEs).
3.2.3. Model Application to MTT Time Points
Application of the PM3 to the plasma metabolite data at Week 3 and Week 10 of MTT revealed an overall shift of the treated ASD + GI participants towards the TD − GI distribution (Figure 5). After three weeks of treatment, median concentrations of two PM3 metabolites (sarcosine and IMP) changed from 15% and 41%, respectively, of the median TD − GI values to 100% and 89% of the median TD − GI values; after ten weeks of treatment, the medians were at 97% and 102% of the median TD − GI values. In other words, MTT therapy rapidly changed the abundance of these metabolites to be more similar to samples from the TD − GI cohort, and they remained similar to the TD − GI values after ten weeks of therapy (Table 4, Figure 6). In contrast, the third PM3 metabolite (tyramine O-sulfate) started at 34% of the median TD − GI value and overall remained unchanged at Week 3 and Week 10 (with the majority of values being below the detection limit), although a small number of samples shifted further into the TD − GI range at later time points. From a multivariate classification standpoint, the Type II error at Week 3 increased to 80% after being just 5% at baseline, but the effect size for this shift was not significant (Table 4). At Week 10, the Type II error increased further to 94% and the effect size became statistically significant (indicated by the 95% CI not containing zero). These increases in Type II error can be interpreted as the ASD + GI cohort after treatment becoming more metabolically similar to the TD − GI cohort. It should be noted that one participant from the ASD + GI cohort at Week 10 had an exceptionally large discriminant score (17.9) and is not shown in the plot (since it is off the scale) but was still factored into the numerical calculations.
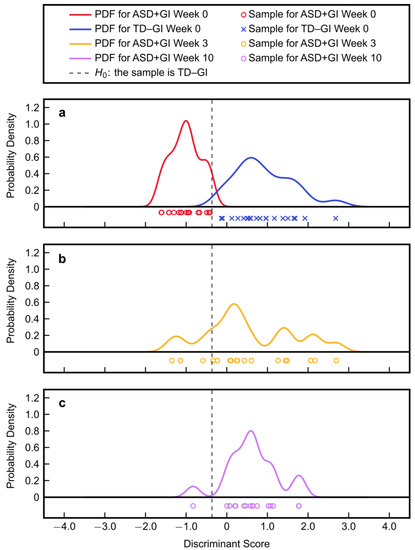
Figure 5.
Discriminant scores and their probability density functions (PDFs) for (a) the baseline Week 0 autism spectrum disorder with gastrointestinal symptoms (ASD + GI) cohort and typically developing with no gastrointestinal symptoms (TD − GI) cohort, (b) the Week 3 ASD + GI cohort, and (c) the Week 10 ASD + GI cohort. It should be noted that one ASD + GI sample at Week 10 had a score of 17.9 and is not plotted since it is off the scale (strongly in the TD − GI range).

Table 4.
Changes in key metabolite concentrations, discriminant scores, Type II errors, and effect sizes at different time points of Microbiota Transfer Therapy (MTT) for individuals with autism spectrum disorder and gastrointestinal symptoms (ASD + GI cohort). Metabolite concentrations presented here are normalized such that the median value is 1 in the typically developing with no gastrointestinal symptom (TD − GI) group at Week 0. Discriminant scores are from the plasma model with three metabolites (PM3) fitted to Week 0 data and then applied to Week 3 or Week 10 data. Type II error was calculated based on the determined threshold for H0 (the null hypothesis that an individual is in the TD − GI group). The effect size was the median change in discriminant score at each MTT time point with respect to baseline, where each individual’s score after treatment was paired with their baseline score.
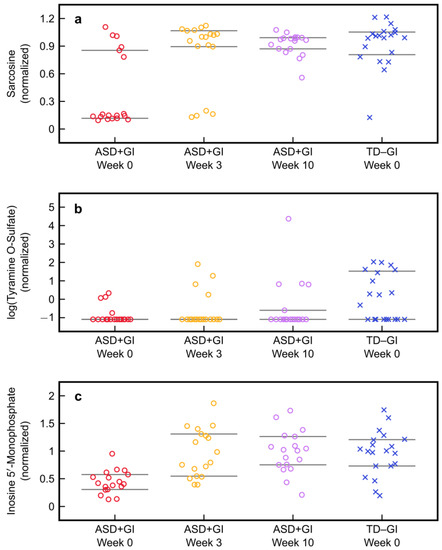
Figure 6.
Normalized values of the metabolites used in the plasma model with three metabolites (PM3) in the typically developing with no gastrointestinal symptoms (TD − GI) cohort at baseline and the autism spectrum disorder with gastrointestinal symptoms (ASD + GI) cohort before and after Microbiota Transfer Therapy: (a) sarcosine, (b) tyramine O-sulfate, and (c) inosine 5′-monophosphate. Each metabolite was normalized such that the median value was 1.0 in the TD − GI cohort. The 25th and 75th percentiles for each group’s metabolite values are indicated by the grey bars. Note that values for tyramine O-sulfate are plotted on a logarithmic scale.
4. Discussion
Univariate analysis of plasma metabolites revealed that many individual metabolites could modestly differentiate between the ASD + GI and TD − GI cohorts, with 61 metabolites yielding an AUROC of at least 0.7 and the highest AUROC of 0.89 associated with nicotinamide riboside. No individual metabolite could classify ASD + GI with an AUROC value greater than 0.9, which is in contrast to multivariate modeling with FDA that was able to identify at least 1000 combinations of metabolites that could classify with AUROC of 0.97 or greater through model fitting. One of the most promising combinations of metabolites, the PM3, was able to classify ASD + GI with 94% sensitivity and 100% specificity after cross-validation. This multivariate approach achieved a level of separation between the ASD + GI and TD − GI cohorts that could not be attained from the metabolites individually.
Many top metabolites were found to be significantly correlated with each other, possibly due to these metabolites coming from the same or closely connected metabolic pathways [33]. Multivariate approaches such as FDA are appropriate for addressing correlations in biological networks [34] and do not require that the relationships between measurements be specified or well-defined. By identifying metabolites for the PM3 that were largely uncorrelated, it is possible to maximize the amount of discriminating information (i.e., metabolic patterns separating the ASD + GI and TD − GI cohorts) with a minimal number of metabolites. Further investigation of the biological significance of these metabolites and the metabolites they are correlated with is warranted.
Classification performance of the PM3 was evaluated with leave-one-out cross-validation, which supported the classifier’s ability to generalize to independent data sets. Although implementing other methods of cross-validation such as k-fold cross-validation may help to further support these conclusions (especially given the large panel of metabolites involved), the small sample size introduces limitations with respect to how much the data set can be partitioned without approaching very small sample sizes in those partitions. A true validation set containing new ASD + GI and TD − GI participants (without treatment) would help to further evaluate the PM3 and alleviate potential concerns of overfitting, which is still not completely ruled out here given the small sample size and large initial number of available metabolites. Applying the model to the MTT Week 3 and Week 10 data for ASD + GI participants suggested that it may be a useful biomarker of treatment efficacy, and that major changes in metabolites associated with ASD and/or GI symptoms did occur; this is consistent with reported improvements in GI and ASD-related symptoms after treatment [24] and with a recent study that found mice colonized with the gut microbiota of children with ASD to show significantly different metabolic and behavioral profiles from mice colonized with the gut microbiota of TD children [35]. It is also worth highlighting that the large initial metabolic shift observed at Week 3 reflects the effect of vancomycin by itself, while the later shift observed at Week 10 reflects the effect of vancomycin in addition to MTT. Future studies may aim to include additional sample collection time points to evaluate the contributions of the individual treatment steps and better characterize the metabolic changes brought about by MTT.
For the PM3 metabolites, it is interesting to note that sarcosine at baseline had a bimodal distribution, with most ASD + GI participants having very low levels (15% of the TD − GI median) but with several in the normal range. After MTT they were all in the normal range. Conversely, the distribution of IMP was unimodal and broadly low in the ASD + GI group but clearly improved after treatment. The majority of values for tyramine O-sulfate (78%) were below the detection limit in the ASD + GI cohort at baseline, and at Week 3 and Week 10 there were still 78% and 72% of samples below the detection limit, respectively. In contrast, less than half of the values for tyramine O-sulfate (45%) were below the detection limit at baseline in the TD − GI cohort. The lack of improvement in this metabolite after MTT may indicate it (and/or correlated metabolites) as a target for future interventions to further improve the metabolic profiles of children with ASD and GI symptoms; however, it is also possible that its production/consumption are host-driven and not microbial, and thus is less likely to be responsive to MTT. Additionally, since so many values were below the detection limit in this study, it would be beneficial to measure this metabolite with greater accuracy in future studies to increase the reliability of the measurements for classification and evaluation of treatment efficacy.
Previous work by the authors revealed that blood markers of DNA methylation and oxidative stress from the folate-dependent one-carbon metabolism (FOCM) and transsulfuration (TS) pathways could be used to predict ASD status with 98% sensitivity and 96% specificity [36] with subsequent validation in a follow-up study [30]. Multivariate analysis of FOCM/TS markers has also been found to provide an indication of metabolic and behavioral improvement resulting from clinical interventions [12]. The current panel of metabolites is not directed specifically at these pathways but was used to achieve (preliminary) results comparable to those obtained using targeted FOCM/TS measurements, albeit in individuals with known gastrointestinal symptoms. That being said, several metabolites appearing in the top 61 metabolites, such as sarcosine, cysteinylglycine, and glutamate, do have roles in FOCM and/or TS, and sarcosine was further included in the PM3. Future plasma metabolomics studies may wish to target additional FOCM/TS markers to further explore their validity for accurately classifying ASD.
There have been many other studies of metabolites in plasma or serum of children with ASD [37,38,39,40,41,42,43,44,45,46]. Several of these studies attempted to classify ASD versus TD children, with some of the most successful studies including Anwar et al. [37] (four-metabolite model with sensitivity/specificity of 92%/84%) and Momeni et al. [40] (three-peptide model with sensitivity/specificity of 95%/85%). Among these studies, none of them included cross-validation or validation in another study, unlike the aforementioned studies analyzing FOCM/TS metabolites [30,36]. Similarly, in this paper we demonstrated that a three-metabolite model was able to distinguish ASD + GI from TD − GI with 94% sensitivity and 100% specificity after leave-one-out cross-validation. Only one metabolomics paper [47] specifically investigated the subset of children with ASD who had GI problems (using urinary metabolites) and it was found that the ASD group with GI problems had four gut bacterial metabolites that were significantly different. Additionally, none of the previous metabolite studies assessed changes after treatment to determine if the model could be used as a biomarker of treatment efficacy.
In studies such as this one where the prevalence of ASD in the study sample did not match the prevalence of ASD in the overall population, the classifier’s positive and negative predictive values may be incorrectly represented with respect to its true clinical values [48]. To gain an indication of the classifier’s true clinical utility, the Bayes’ adjusted positive and negative predictive values should be calculated by incorporating the true population prevalence. Without adjusting for prevalence, the positive and negative predictive values of the PM3 were 100% and 95%, respectively. After adjusting for ASD population prevalence, assuming the current U.S. prevalence estimate of 1.7% [1], the positive predictive value remains 100% and the negative predictive value increases to 99%. There is thus minimal mismatch between the predictive values of the PM3 in the study population and the adjusted estimates for the general population.
A major limitation of our study was its small sample size. Although the PM3 was able to accurately classify a small number of participants, it remains to be seen whether the metabolic patterns used for classification would hold up for a larger study population. The results thus require validation on larger cohorts beyond what our cross-validation procedure was able to accomplish. Moreover, potential subgroups in the ASD + GI cohort should be considered, since previous studies have reported the presence of subgroups in ASD [49,50]. The small sample size might also influence the presence of outliers in the post-treatment ASD + GI participants (as seen at Week 10). It would have been valuable to validate the PM3 on later time points for TD − GI participants, but plasma samples were only collected from the TD − GI cohort at baseline. Additionally, this study compared individuals with ASD and GI symptoms to TD individuals without GI issues, meaning the classification of ASD versus TD and subsequent assessment of ASD at later time points was confounded by the presence of GI issues in the ASD + GI cohort. The observed metabolic shifts at Week 3 and Week 10 might not be due solely to improvement in ASD-related symptoms and might be influenced by improvements in GI-related symptoms. The PM3 should be further developed for a more general population of individuals, regardless of GI (or other co-occurring condition) status and would also ideally be generalizable to individuals with or without single-gene disorders.
5. Conclusions
This small pilot study resulted in the development of many promising metabolic markers for distinguishing children with ASD and GI symptoms from TD children without GI symptoms. In particular, the combination of the three metabolites sarcosine, tyramine O-sulfate, and IMP was one of the most promising. The model developed from these three metabolites was applied to the ASD + GI group after three and ten weeks of Microbiota Transfer Therapy, and it was found that during and after treatment there was much less difference between the PM3 discriminant scores of the two groups, consistent with significant improvements in GI and ASD-related symptoms in the ASD + GI treatment group. Two of the PM3 metabolites, sarcosine and IMP, improved substantially during and after MTT, but one of them (tyramine O-sulfate) did not change notably, and hence it (and the metabolites it is correlated with) may be a target for future therapies. A larger study is needed to validate the PM3 and should include TD children with GI symptoms.
Author Contributions
All authors conceived of the current study and contributed to its design and coordination. J.B.A., D.-W.K., and R.K.-B. collected and interpreted the clinical data. T.V. analyzed the data and generated the tables and figures. J.B.A., T.V., and J.H. interpreted the statistical results. J.B.A and T.V. drafted the manuscript and revised it with contributions from J.H., D.-W.K., and R.K.-B. All authors read and approved of the submitted manuscript.
Funding
The authors gratefully acknowledge partial financial support from the National Institutes of Health (grant 1R01AI110642).
Acknowledgments
We thank Sharon McDonough-Means, Thomas Borody, Alex Khoruts, Michael Sadowsky, Alessio Fasano, Devon Coleman, and Elena Pollard for their help with the treatment portion of the Microbiota Transfer Therapy study. We also thank Crestovo (now part of Finch Therapeutics) for their financial support of the metabolomics measurements. The authors would also like to acknowledge Kathryn Hollowood Jones and Uwe Kruger for their initial contributions to the analyses.
Conflicts of Interest
The authors have filed an invention disclosure on the results presented here.
References
- Baio, J.; Wiggins, L.; Christensen, D.L.; Maenner, M.J.; Daniels, J.; Warren, Z.; Kurzius-Spencer, M.; Zahorodny, W.; Robinson Rosenberg, C.; White, T.; et al. Prevalence of autism spectrum disorder among children aged 8 years-Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2014. MMWR Surveill. Summ. 2018, 67, 1–23. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-V, 5th ed.; American Psychiatric Publishing: Washington, DC, USA, 2013; ISBN 978-0-89042-555-8. [Google Scholar]
- Gaugler, T.; Klei, L.; Sanders, S.J.; Bodea, C.A.; Goldberg, A.P.; Lee, A.B.; Mahajan, M.; Manaa, D.; Pawitan, Y.; Reichert, J.; et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014, 46, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Mandy, W.; Lai, M.-C. Annual Research Review: The role of the environment in the developmental psychopathology of autism spectrum condition. J. Child Psychol. Psychiatry 2016, 57, 271–292. [Google Scholar] [CrossRef] [PubMed]
- Fein, D.; Barton, M.; Dumont-Mathieu, T. Optimizing outcome in autism spectrum disorders. Policy Insights Behav. Brain Sci. 2017, 4, 71–78. [Google Scholar] [CrossRef]
- Zwaigenbaum, L.; Bryson, S.E.; Brian, J.; Smith, I.M.; Roberts, W.; Szatmari, P.; Roncadin, C.; Garon, N.; Vaillancourt, T. Stability of diagnostic assessment for autism spectrum disorder between 18 and 36 months in a high-risk cohort. Autism Res. 2016, 9, 790–800. [Google Scholar] [CrossRef] [PubMed]
- McPartland, J.C. Considerations in biomarker development for neurodevelopmental disorders. Curr. Opin. Neurol. 2016, 29, 118–122. [Google Scholar] [CrossRef]
- Pierce, K.; Courchesne, E.; Bacon, E. To screen or not to screen universally for autism is not the question: Why the task force got it wrong. J. Pediatr. 2016, 176, 182–194. [Google Scholar] [CrossRef]
- Orinstein, A.J.; Helt, M.; Troyb, E.; Tyson, K.E.; Barton, M.L.; Eigsti, I.-M.; Naigles, L.; Fein, D.A. Intervention history of children and adolescents with high-functioning autism and optimal outcomes. J. Dev. Behav. Pediatr. 2014, 35, 247–256. [Google Scholar] [CrossRef]
- French, L.; Kennedy, E.M.M. Annual Research Review: Early intervention for infants and young children with, or at-risk of, autism spectrum disorder: A systematic review. J. Child Psychol. Psychiatry 2018, 59, 444–456. [Google Scholar] [CrossRef]
- Ryberg, K.H. Evidence for the implementation of the Early Start Denver Model for young children with autism spectrum disorder. J. Am. Psychiatr. Nurses Assoc. 2015, 21, 327–337. [Google Scholar] [CrossRef]
- Vargason, T.; Kruger, U.; Roth, E.; Delhey, L.M.; Tippett, M.; Rose, S.; Bennuri, S.C.; Slattery, J.C.; Melnyk, S.; James, S.J.; et al. Comparison of three clinical trial treatments for autism spectrum disorder through multivariate analysis of changes in metabolic profiles and adaptive behavior. Front. Cell. Neurosci. 2018, 12, 503. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Pelphrey, K.A.; Sukhodolsky, D.G.; Crowley, M.J.; Dayan, E.; Dvornek, N.C.; Venkataraman, A.; Duncan, J.; Staib, L.; Ventola, P. Brain responses to biological motion predict treatment outcome in young children with autism. Transl. Psychiatry 2016, 6, e948. [Google Scholar] [CrossRef] [PubMed]
- Fletcher-Watson, S.; Hampton, S. The potential of eye-tracking as a sensitive measure of behavioural change in response to intervention. Sci. Rep. 2018, 8, 14715. [Google Scholar] [CrossRef] [PubMed]
- Murias, M.; Major, S.; Davlantis, K.; Franz, L.; Harris, A.; Rardin, B.; Sabatos-DeVito, M.; Dawson, G. Validation of eye-tracking measures of social attention as a potential biomarker for autism clinical trials. Autism Res. 2018, 11, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Soke, G.N.; Maenner, M.J.; Christensen, D.; Kurzius-Spencer, M.; Schieve, L.A. Prevalence of co-occurring medical and behavioral conditions/symptoms among 4- and 8-year-old children with autism spectrum disorder in selected areas of the United States in 2010. J. Autism Dev. Disord. 2018, 48, 2663–2676. [Google Scholar] [CrossRef]
- Holingue, C.; Newill, C.; Lee, L.-C.; Pasricha, P.J.; Daniele Fallin, M. Gastrointestinal symptoms in autism spectrum disorder: A review of the literature on ascertainment and prevalence. Autism Res. 2018, 11, 24–36. [Google Scholar] [CrossRef]
- Vargason, T.; McGuinness, D.L.; Hahn, J. Gastrointestinal symptoms and oral antibiotic use in children with autism spectrum disorder: Retrospective analysis of a privately insured U.S. population. J. Autism Dev. Disord. 2019, 49, 647–659. [Google Scholar] [CrossRef]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef]
- Krajmalnik-Brown, R.; Lozupone, C.; Kang, D.-W.; Adams, J.B. Gut bacteria in children with autism spectrum disorders: Challenges and promise of studying how a complex community influences a complex disease. Microb. Ecol. Health Dis. 2015, 26, 26914. [Google Scholar] [CrossRef]
- Vuong, H.E.; Hsiao, E.Y. Emerging roles for the gut microbiome in autism spectrum disorder. Biol. Psychiatry 2017, 81, 411–423. [Google Scholar] [CrossRef]
- Frye, R.E.; Slattery, J.; MacFabe, D.F.; Allen-Vercoe, E.; Parker, W.; Rodakis, J.; Adams, J.B.; Krajmalnik-Brown, R.; Bolte, E.; Kahler, S.; et al. Approaches to studying and manipulating the enteric microbiome to improve autism symptoms. Microb. Ecol. Health Dis. 2015, 26, 26878. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhou, J.-M. The microbiota–gut–brain axis and its potential therapeutic role in autism spectrum disorder. Neuroscience 2016, 324, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, M.J.; Weingarden, A.R.; Sadowsky, M.J.; Khoruts, A. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile Infection. Am. J. Gastroenterol. 2012, 107, 761–767. [Google Scholar] [CrossRef]
- Revicki, D.A.; Wood, M.; Wiklund, I.; Crawley, J. Reliability and validity of the Gastrointestinal Symptom Rating Scale in patients with gastroesophageal reflux disease. Qual. Life Res. 1998, 7, 75–83. [Google Scholar] [CrossRef]
- Long, T.; Hicks, M.; Yu, H.-C.; Biggs, W.H.; Kirkness, E.F.; Menni, C.; Zierer, J.; Small, K.S.; Mangino, M.; Messier, H.; et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017, 49, 568–578. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Adams, J.B.; Howsmon, D.P.; Kruger, U.; Geis, E.; Gehn, E.; Fimbres, V.; Pollard, E.; Mitchell, J.; Ingram, J.; Hellmers, R.; et al. Significant association of urinary toxic metals and autism-related symptoms—A nonlinear statistical analysis with cross validation. PLoS ONE 2017, 12, e0169526. [Google Scholar] [CrossRef]
- Howsmon, D.P.; Vargason, T.; Rubin, R.A.; Delhey, L.; Tippett, M.; Rose, S.; Bennuri, S.C.; Slattery, J.C.; Melnyk, S.; James, S.J.; et al. Multivariate techniques enable a biochemical classification of children with autism spectrum disorder versus typically-developing peers: A comparison and validation study. Bioeng. Transl. Med. 2018, 3, 156–165. [Google Scholar] [CrossRef]
- Hicks, S.D.; Rajan, A.T.; Wagner, K.E.; Barns, S.; Carpenter, R.L.; Middleton, F.A. Validation of a salivary RNA test for childhood autism spectrum disorder. Front. Genet. 2018, 9, 534. [Google Scholar] [CrossRef]
- Banjanovic, E.S.; Osborne, J.W. Confidence intervals for effect sizes: Applying bootstrap resampling. Pract. Assess. Res. Eval. 2016, 21, 5. [Google Scholar]
- Kang, D.-W.; Krajmalnik-Brown, R.; Adams, J.B. Distinct fecal and plasma metabolites in children with autism spectrum disorder and their modulation after Microbiota Transfer Therapy. In preparation.
- Vargason, T.; Howsmon, D.P.; McGuinness, D.L.; Hahn, J. On the use of multivariate methods for analysis of data from biological networks. Processes 2017, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Cruz, N.J.; Kang, D.-W.; Gandal, M.J.; Wang, B.; Kim, Y.-M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell 2019, 177, 1600–1618.e17. [Google Scholar] [CrossRef] [PubMed]
- Howsmon, D.P.; Kruger, U.; Melnyk, S.; James, S.J.; Hahn, J. Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and DNA methylation. PLoS Comput. Biol. 2017, 13, e1005385. [Google Scholar] [CrossRef]
- Anwar, A.; Abruzzo, P.M.; Pasha, S.; Rajpoot, K.; Bolotta, A.; Ghezzo, A.; Marini, M.; Posar, A.; Visconti, P.; Thornalley, P.J.; et al. Advanced glycation endproducts, dityrosine and arginine transporter dysfunction in autism-a source of biomarkers for clinical diagnosis. Mol. Autism 2018, 9, 3. [Google Scholar] [CrossRef]
- Barone, R.; Alaimo, S.; Messina, M.; Pulvirenti, A.; Bastin, J.; Group, M.-A.; Ferro, A.; Frye, R.E.; Rizzo, R. A subset of patients with autism spectrum disorders show a distinctive metabolic profile by dried blood spot analyses. Front. Psychiatry 2018, 9, 636. [Google Scholar] [CrossRef]
- Mizejewski, G.J.; Lindau-Shepard, B.; Pass, K.A. Newborn screening for autism: In search of candidate biomarkers. Biomark. Med. 2013, 7, 247–260. [Google Scholar] [CrossRef]
- Momeni, N.; Bergquist, J.; Brudin, L.; Behnia, F.; Sivberg, B.; Joghataei, M.T.; Persson, B.L. A novel blood-based biomarker for detection of autism spectrum disorders. Transl. Psychiatry 2012, 2, e91. [Google Scholar] [CrossRef]
- Rangel-Huerta, O.D.; Gomez-Fernández, A.; de la Torre-Aguilar, M.J.; Gil, A.; Perez-Navero, J.L.; Flores-Rojas, K.; Martín-Borreguero, P.; Gil-Campos, M. Metabolic profiling in children with autism spectrum disorder with and without mental regression: Preliminary results from a cross-sectional case–control study. Metabolomics 2019, 15, 99. [Google Scholar] [CrossRef]
- Smith, A.M.; King, J.J.; West, P.R.; Ludwig, M.A.; Donley, E.L.R.; Burrier, R.E.; Amaral, D.G. Amino acid dysregulation metabotypes: Potential biomarkers for diagnosis and individualized treatment for subtypes of autism spectrum disorder. Biol. Psychiatry 2019, 85, 345–354. [Google Scholar] [CrossRef]
- Wang, H.; Liang, S.; Wang, M.; Gao, J.; Sun, C.; Wang, J.; Xia, W.; Wu, S.; Sumner, S.J.; Zhang, F.; et al. Potential serum biomarkers from a metabolomics study of autism. J. Psychiatry Neurosci. 2016, 41, 27–37. [Google Scholar] [CrossRef] [PubMed]
- West, P.R.; Amaral, D.G.; Bais, P.; Smith, A.M.; Egnash, L.A.; Ross, M.E.; Palmer, J.A.; Fontaine, B.R.; Conard, K.R.; Corbett, B.A.; et al. Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS ONE 2014, 9, e112445. [Google Scholar] [CrossRef] [PubMed]
- Wiest, M.M.; German, J.B.; Harvey, D.J.; Watkins, S.M.; Hertz-Picciotto, I. Plasma fatty acid profiles in autism: A case-control study. Prostaglandins Leukot. Essent. Fat. Acids 2009, 80, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Sun, C.; Liang, S.; Sun, Y.; Li, D.; Li, L.; Fan, L.; Wu, L.; Xia, W. Fisher discriminant analysis for classification of autism spectrum disorders based on folate-related metabolism markers. J. Nutr. Biochem. 2019, 64, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Stein, T.P.; Barnes, V.; Rhodes, N.; Guo, L. Metabolic perturbance in autism spectrum disorders: A metabolomics study. J. Proteome Res. 2012, 11, 5856–5862. [Google Scholar] [CrossRef] [PubMed]
- Abu-Akel, A.; Bousman, C.; Skafidas, E.; Pantelis, C. Mind the prevalence rate: Overestimating the clinical utility of psychiatric diagnostic classifiers. Psychol. Med. 2018, 48, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Ousley, O.; Cermak, T. Autism spectrum disorder: Defining dimensions and subgroups. Curr. Dev. Disord. Rep. 2014, 1, 20–28. [Google Scholar] [CrossRef]
- Sacco, R.; Lenti, C.; Saccani, M.; Curatolo, P.; Manzi, B.; Bravaccio, C.; Persico, A.M. Cluster analysis of autistic patients based on principal pathogenetic components. Autism Res. 2012, 5, 137–147. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).