Physiological and Metabolic Challenges of Flocculating Saccharomyces cerevisiae in D-Lactic Acid Fermentation Under High-Glucose and Inhibitory Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Medium
2.2. Batch Fermentation
2.3. Batch Fermentation with Calcium Carbonate Supplementation
2.4. Measurements of Fermentation Products
2.5. Measurement of Cell Growth
2.6. Cell Morphological Observation
2.7. Gene Expression Analysis by Quantitative PCR
2.8. Extraction of Internal Metabolites
2.9. GC/MS Analysis for Untargeted Metabolites
2.10. LC/MS Analysis for Carbon Central Metabolites
2.11. Multivariate Analysis
3. Results
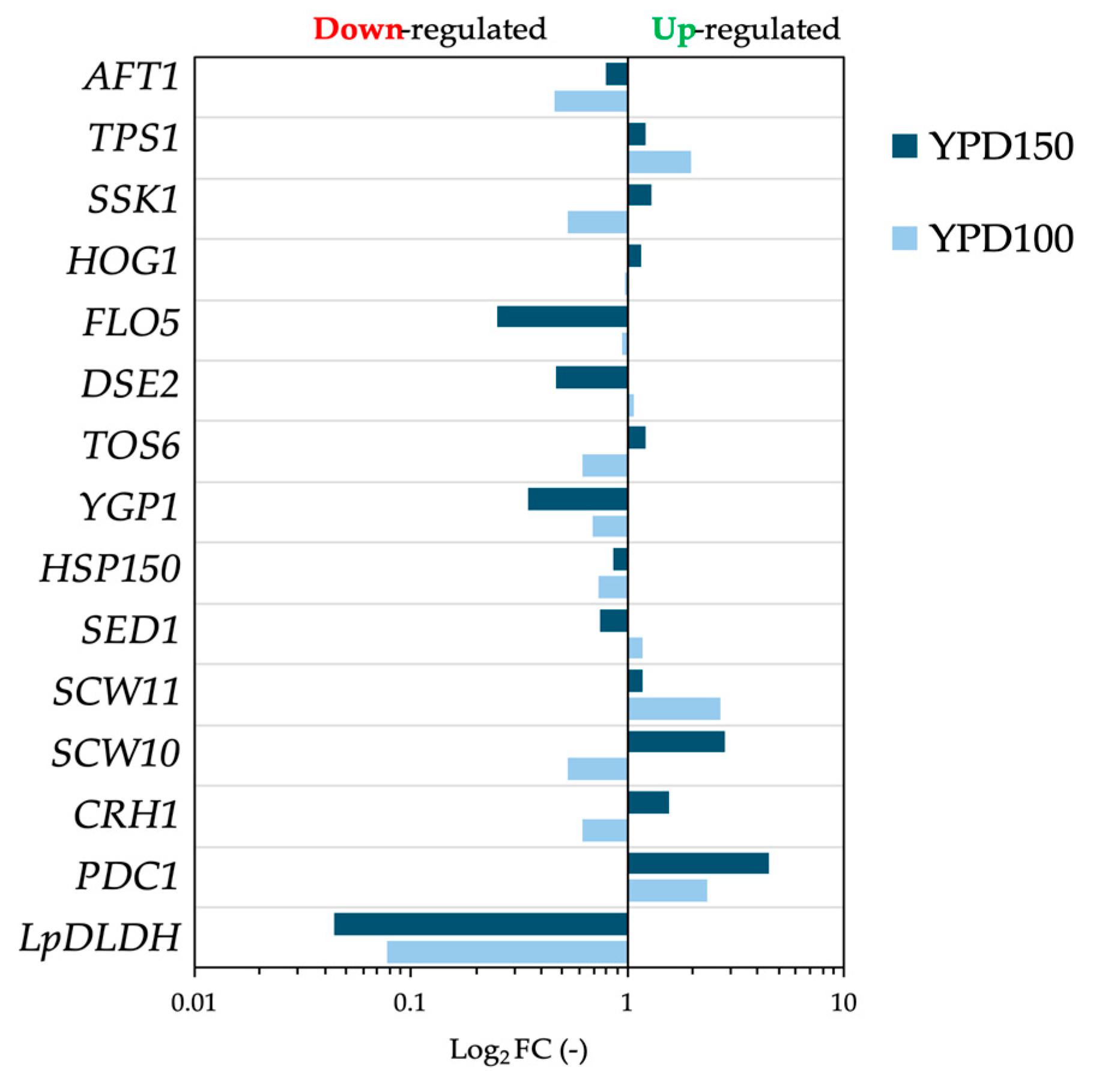
3.1. D-LA Production at Different Glucose Levels
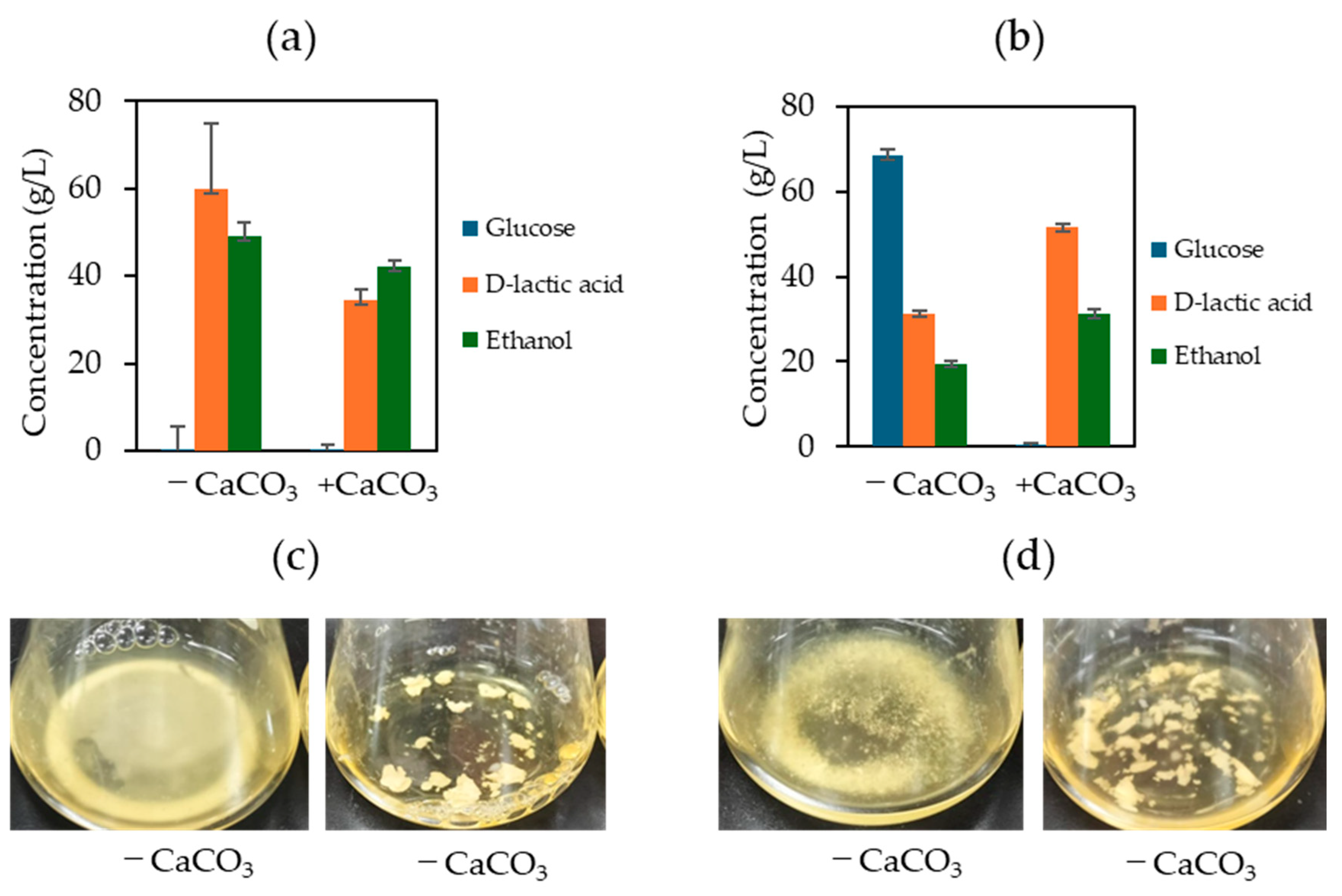
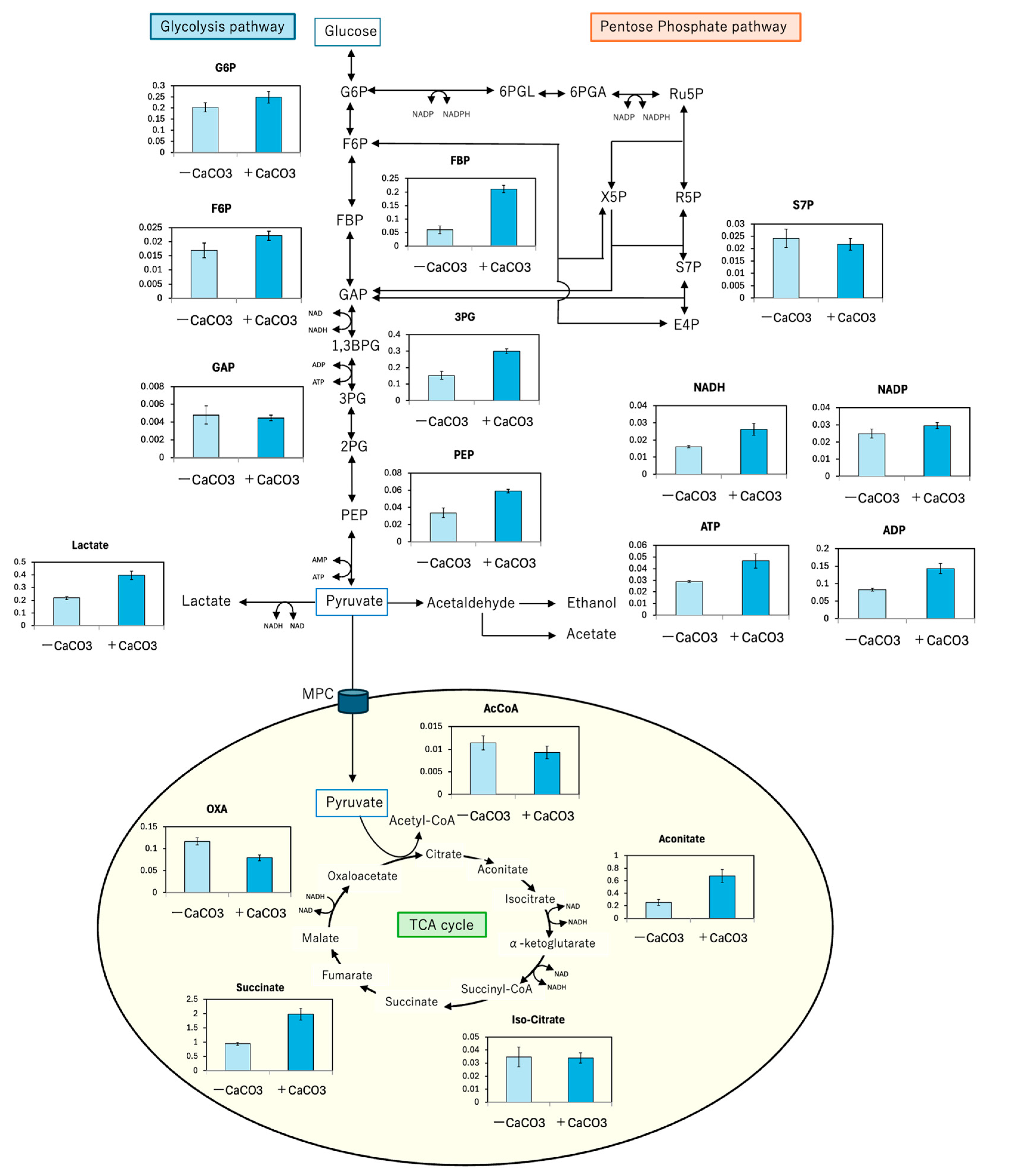
3.2. Calcium Carbonate Supplementation Effect on D-LA Production
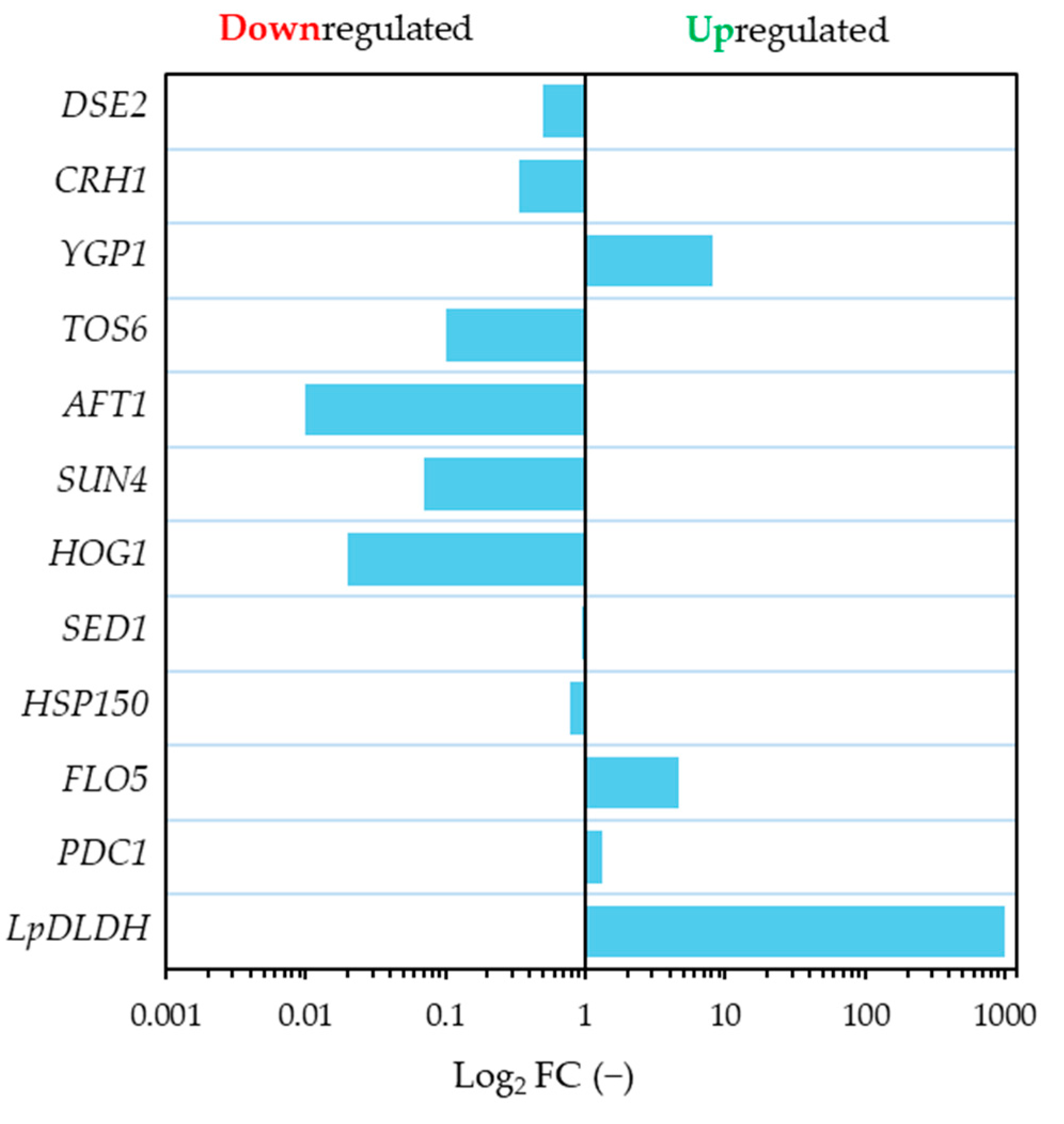
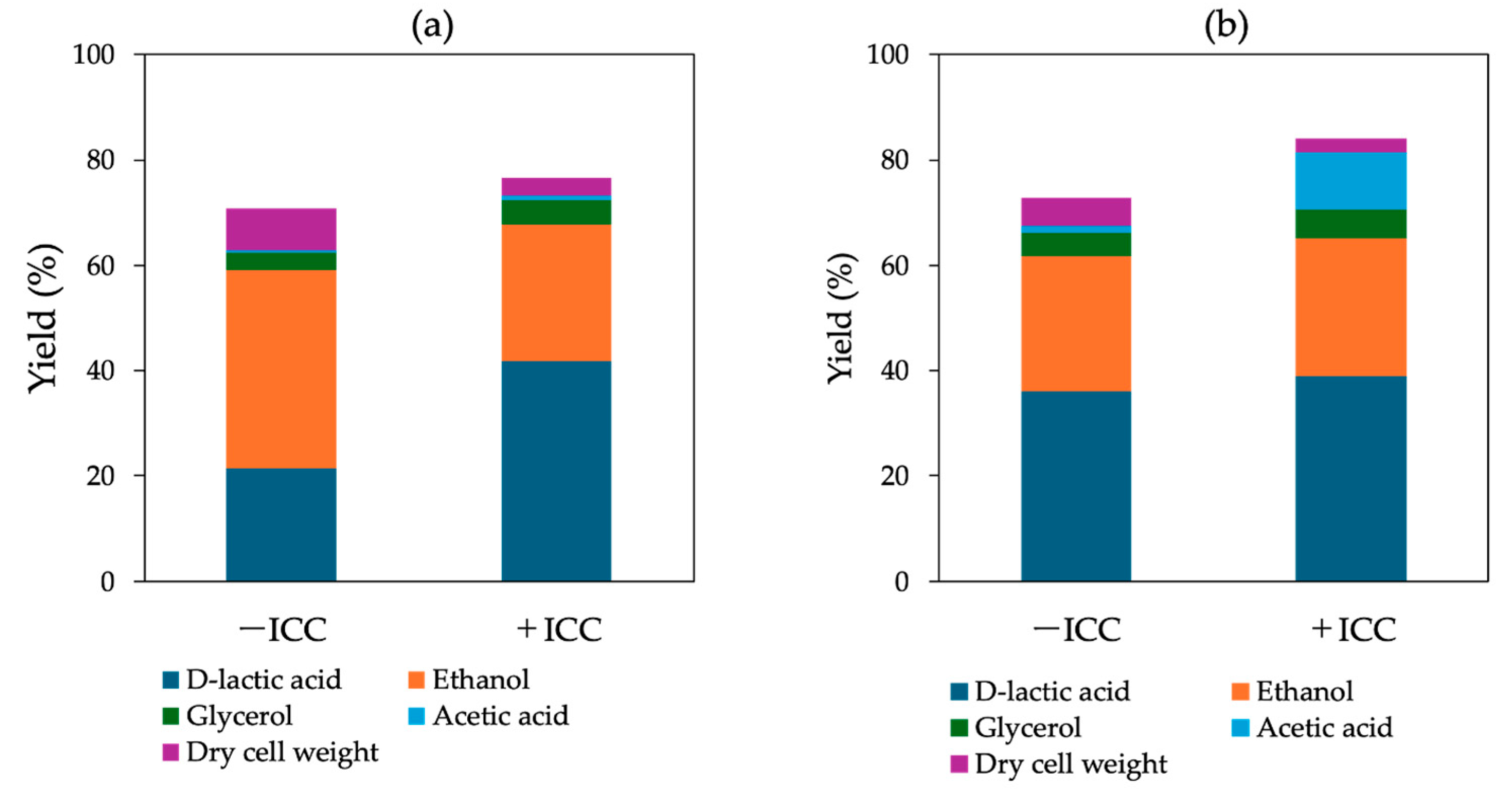
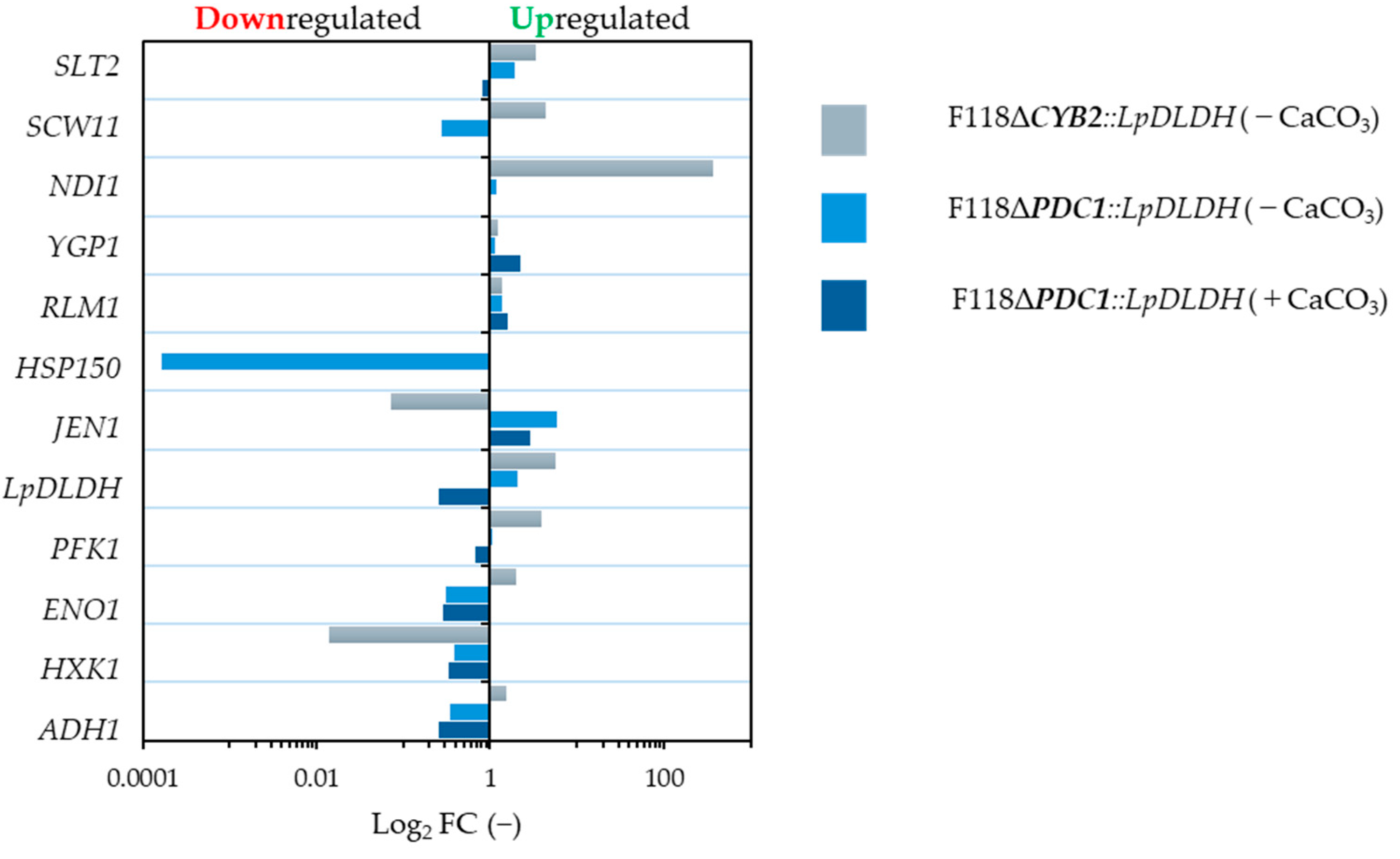
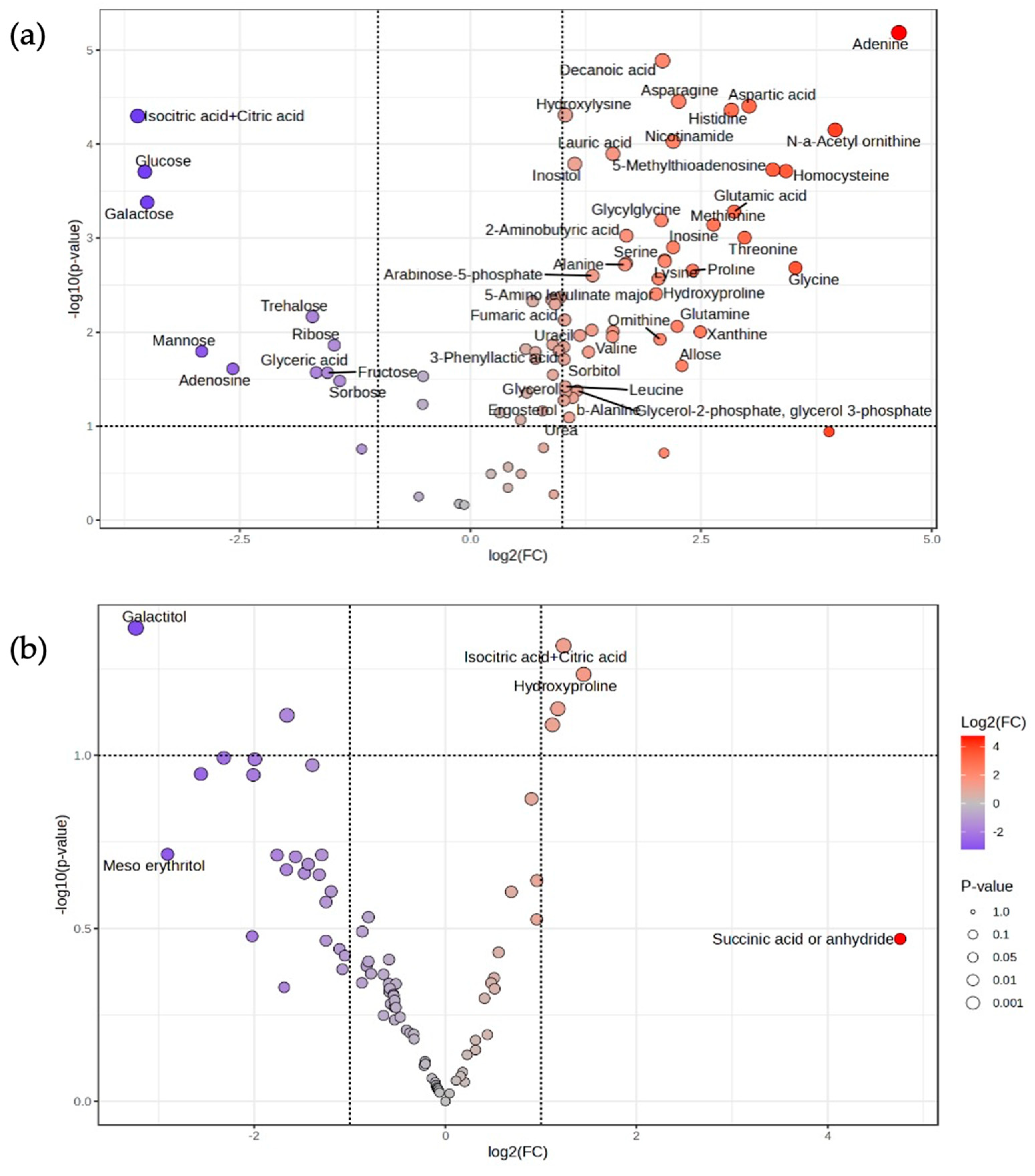
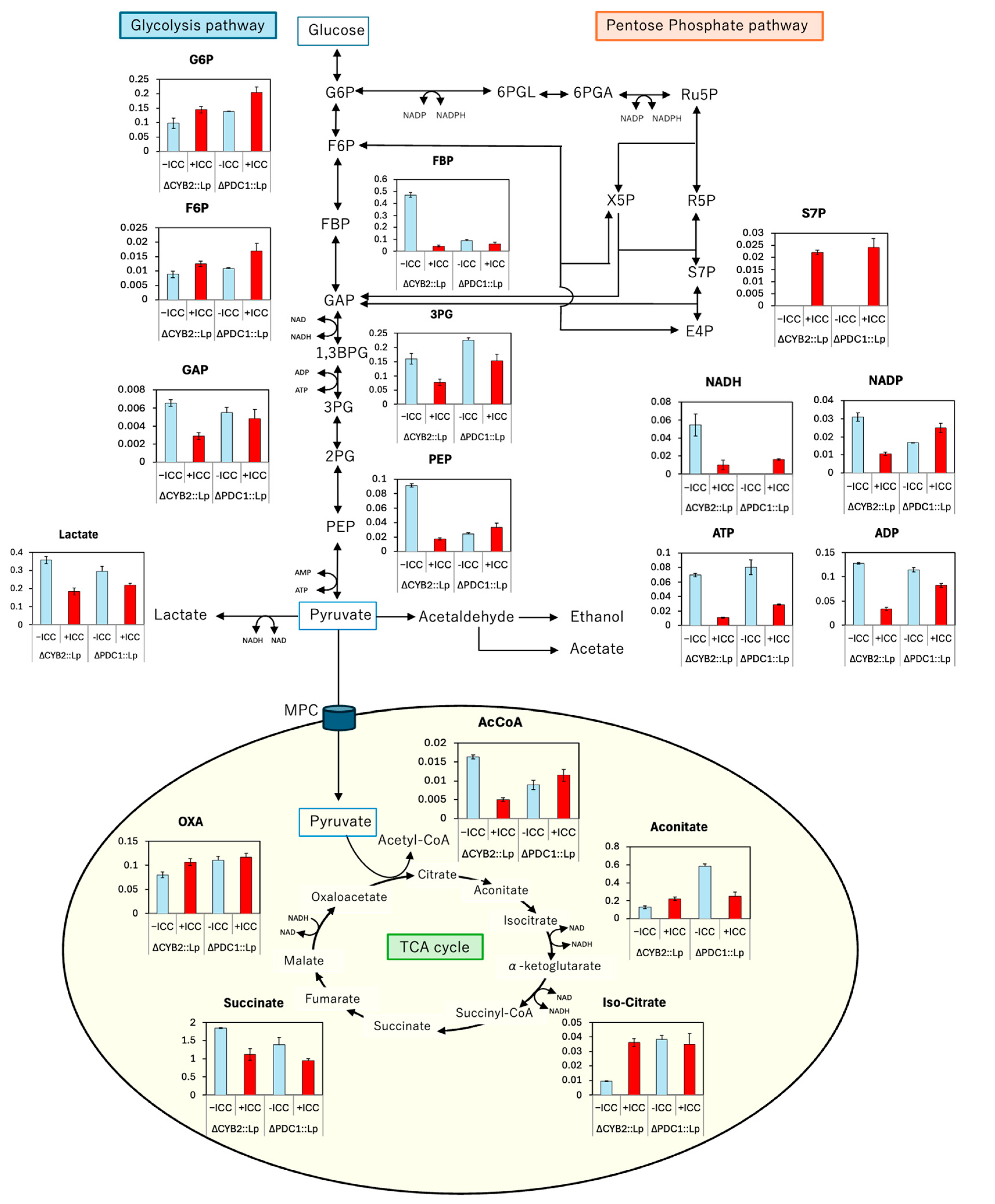
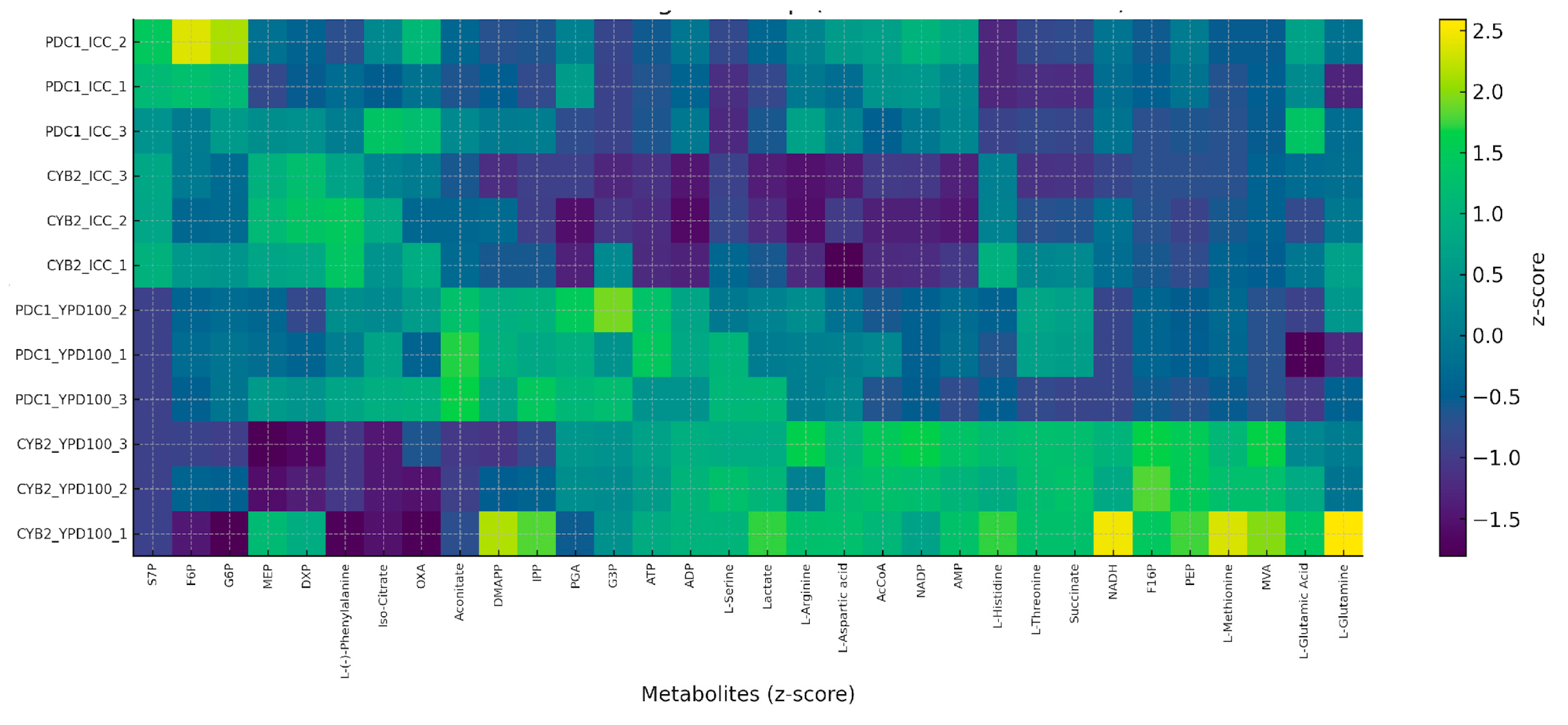
3.3. Effect of Inhibitory Chemical Compounds on D-LA Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADP | Adenosine diphosphate |
| AMP | Adenosine monophosphate |
| ATP | Adenosine triphosphate |
| Ct | Threshold cycle (qPCR) |
| CYB2 | Cytochrome b2 (locus) |
| DCW | Dry cell weight |
| D-LA | D-lactic acid |
| D-LDH | D-lactate dehydrogenase |
| FDR | False discovery rate |
| FBP | Fructose-1,6-bisphosphate |
| G6P | Glucose-6-phosphate |
| GC/MS | Gas chromatography–mass spectrometry |
| GRAS | Generally Recognized as Safe |
| HMF | 5-Hydroxymethylfurfural |
| HOG | High-osmolarity glycerol (pathway) |
| HPLC | High-performance liquid chromatography |
| ICC; ICCs | Inhibitory chemical compound(s) |
| LC/MS | Liquid chromatography–mass spectrometry |
| LC/QqQ/MS | Liquid chromatography–triple quadrupole mass spectrometry |
| Log2 FC | Log2 fold change |
| Lp | Leuconostoc pseudomesenteroides (source of D-LDH) |
| MRMPROBS | Multiple Reaction Monitoring PROfiling Browser Software |
| MSDIAL | Mass Spectrometry–based Data Independent AnaLysis (software) |
| MSTFA | N-Methyl-N-(trimethylsilyl)trifluoroacetamide |
| NADH | Nicotinamide adenine dinucleotide (reduced form) |
| NADPH | Nicotinamide adenine dinucleotide phosphate (reduced form) |
| NBRC | NITE Biological Resource Center |
| OD600 | Optical density at 600 nm |
| PCA | Principal component analysis |
| PDC1 | Pyruvate decarboxylase 1 |
| PEP | Phosphoenolpyruvate |
| PLA | Poly(lactic acid) |
| PPP | Pentose phosphate pathway |
| PTFE | Polytetrafluoroethylene |
| qPCR | Quantitative polymerase chain reaction |
| RID | Refractive index detector |
| S7P | Sedoheptulose-7-phosphate |
| SD | Standard deviation |
| TCA | Tricarboxylic acid (cycle) |
| YPD | Yeast extract–peptone–dextrose medium |
References
- Kavšček, M.; Stražar, M.; Curk, T.; Natter, K.; Petrovič, U. Yeast as a Cell Factory: Current State and Perspectives. Microb. Cell Factories 2015, 14, 94. [Google Scholar] [CrossRef]
- Aiello, E.; Arena, M.P.; De Vero, L.; Montanini, C.; Bianchi, M.; Mescola, A.; Alessandrini, A.; Pulvirenti, A.; Gullo, M. Wine Yeast Strains Under Ethanol-Induced Stress: Morphological and Physiological Responses. Fermentation 2024, 10, 631. [Google Scholar] [CrossRef]
- Walker, G.M.; Basso, T.O. Mitigating Stress in Industrial Yeasts. Fungal Biol. 2020, 124, 387–397. [Google Scholar] [CrossRef]
- Baig, K.S.; Wu, J.; Turcotte, G. Future Prospects of Delignification Pretreatments for the Lignocellulosic Materials to Produce Second Generation Bioethanol. Int. J. Energy Res. 2019, 43, 1411–1427. [Google Scholar] [CrossRef]
- Allen, S.A.; Clark, W.; McCaffery, J.M.; Cai, Z.; Lanctot, A.; Slininger, P.J.; Liu, Z.L.; Gorsich, S.W. Furfural Induces Reactive Oxygen Species Accumulation and Cellular Damage in Saccharomyces cerevisiae. Biotechnol. Biofuels 2010, 3, 2. [Google Scholar] [CrossRef]
- Deparis, Q.; Claes, A.; Foulquié-Moreno, M.R.; Thevelein, J.M. Engineering Tolerance to Industrially Relevant Stress Factors in Yeast Cell Factories. FEMS Yeast Res. 2017, 17, fox036. [Google Scholar] [CrossRef]
- He, M.; Wu, B.; Shui, Z.; Hu, Q.; Wang, W.; Tan, F.; Tang, X.; Zhu, Q.; Pan, K.; Li, Q.; et al. Transcriptome Profiling of Zymomonas mobilis under Furfural Stress. Appl. Microbiol. Biotechnol. 2012, 95, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Soares, E.V. Flocculation in Saccharomyces cerevisiae: A Review: Yeast Flocculation: A Review. J. Appl. Microbiol. 2011, 110, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ye, P.-L.; Wang, X.-Q.; Yuan, B.; Liu, C.-G.; Zhao, X.-Q. Manipulating Cell Flocculation-Associated Protein Kinases in Saccharomyces cerevisiae Enables Improved Stress Tolerance and Efficient Cellulosic Ethanol Production. Bioresour. Technol. 2022, 348, 126758. [Google Scholar] [CrossRef]
- Westman, J.O.; Mapelli, V.; Taherzadeh, M.J.; Franzén, C.J. Flocculation Causes Inhibitor Tolerance in Saccharomyces cerevisiae for Second-Generation Bioethanol Production. Appl. Environ. Microbiol. 2014, 80, 6908–6918. [Google Scholar] [CrossRef] [PubMed]
- Kahar, P.; Itomi, A.; Tsuboi, H.; Ishizaki, M.; Yasuda, M.; Kihira, C.; Otsuka, H.; Azmi, N.B.; Matsumoto, H.; Ogino, C.; et al. The Flocculant Saccharomyces cerevisiae Strain Gains Robustness via Alteration of the Cell Wall Hydrophobicity. Metab. Eng. 2022, 72, 82–96. [Google Scholar] [CrossRef]
- Wijaya, H.; Sasaki, K.; Kahar, P.; Yopi; Kawaguchi, H.; Sazuka, T.; Ogino, C.; Prasetya, B.; Kondo, A. Repeated Ethanol Fermentation from Membrane-Concentrated Sweet Sorghum Juice Using the Flocculating Yeast Saccharomyces cerevisiae F118 Strain. Bioresour. Technol. 2018, 265, 542–547. [Google Scholar] [CrossRef]
- Swetha, T.A.; Ananthi, V.; Bora, A.; Sengottuvelan, N.; Ponnuchamy, K.; Muthusamy, G.; Arun, A. A Review on Biodegradable Polylactic Acid (PLA) Production from Fermentative Food Waste—Its Applications and Degradation. Int. J. Biol. Macromol. 2023, 234, 123703. [Google Scholar] [CrossRef]
- Ishida, N.; Saitoh, S.; Ohnishi, T.; Tokuhiro, K.; Nagamori, E.; Kitamoto, K.; Takahashi, H. Metabolic Engineering of Saccharomyces cerevisiae for Efficient Production of Pure L-(+)-Lactic Acid. Appl. Biochem. Biotechnol. 2006, 131, 795–807. [Google Scholar] [CrossRef]
- Baek, S.-H.; Kwon, E.Y.; Kim, Y.H.; Hahn, J.-S. Metabolic Engineering and Adaptive Evolution for Efficient Production of D-Lactic Acid in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2016, 100, 2737–2748. [Google Scholar] [CrossRef]
- Liu, T.; Sun, L.; Zhang, C.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Combinatorial Metabolic Engineering and Process Optimization Enables Highly Efficient Production of L-Lactic Acid by Acid-Tolerant Saccharomyces cerevisiae. Bioresour. Technol. 2023, 379, 129023. [Google Scholar] [CrossRef]
- Rahmasari, D.; Kahar, P.; De Oliveira, A.V.; Putra, F.J.N.; Kondo, A.; Ogino, C. Factors Affecting D-Lactic Acid Production by Flocculant Saccharomyces cerevisiae Under Non-Neutralizing Conditions. Microorganisms 2025, 13, 618. [Google Scholar] [CrossRef]
- Long, Y.; Han, X.; Meng, X.; Xu, P.; Tao, F. A Robust Yeast Chassis: Comprehensive Characterization of a Fast-Growing Saccharomyces cerevisiae. mBio 2024, 15, e03196-23. [Google Scholar] [CrossRef] [PubMed]
- Nusantara Putra, F.J.; Putri, S.P.; Fukusaki, E. Metabolomics-Based Profiling of Three Terminal Alkene-Producing Jeotgalicoccus spp. during Different Growth Phase. J. Biosci. Bioeng. 2019, 127, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Arita, M.; Kanazawa, M.; Ogiwara, A.; Bamba, T.; Fukusaki, E. MRMPROBS: A Data Assessment and Metabolite Identification Tool for Large-Scale Multiple Reaction Monitoring Based Widely Targeted Metabolomics. Anal. Chem. 2013, 85, 5191–5199. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Qu, T.; Smith, J.R.; Li, J.; Du, G.; Chen, J. Osmotic Tolerance in Saccharomyces cerevisiae: Implications for Food and Bioethanol Industries. Food Biosci. 2024, 60, 104451. [Google Scholar] [CrossRef]
- Tofalo, R.; Perpetuini, G.; Di Gianvito, P.; Arfelli, G.; Schirone, M.; Corsetti, A.; Suzzi, G. Characterization of Specialized Flocculent Yeasts to Improve Sparkling Wine Fermentation. J. Appl. Microbiol. 2016, 120, 1574–1584. [Google Scholar] [CrossRef]
- Zheng, Y.; Kong, S.; Luo, S.; Chen, C.; Cui, Z.; Sun, X.; Chen, T.; Wang, Z. Improving Furfural Tolerance of Escherichia coli by Integrating Adaptive Laboratory Evolution with CRISPR-Enabled Trackable Genome Engineering (CREATE). ACS Sustain. Chem. Eng. 2022, 10, 2318–2330. [Google Scholar] [CrossRef]
- Salek, S.S.; Van Turnhout, A.G.; Kleerebezem, R.; Van Loosdrecht, M.C.M. pH Control in Biological Systems Using Calcium Carbonate. Biotechnol. Bioeng. 2015, 112, 905–913. [Google Scholar] [CrossRef]
- Yang, P.-B.; Tian, Y.; Wang, Q.; Cong, W. Effect of Different Types of Calcium Carbonate on the Lactic Acid Fermentation Performance of Lactobacillus lactis. Biochem. Eng. J. 2015, 98, 38–46. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.-M.; Lebaka, V.R.; Wee, Y.-J. Lactic Acid for Green Chemical Industry: Recent Advances in and Future Prospects for Production Technology, Recovery, and Applications. Fermentation 2022, 8, 609. [Google Scholar] [CrossRef]
- Zara, S.; Bakalinsky, A.T.; Zara, G.; Pirino, G.; Demontis, M.A.; Budroni, M. FLO11-Based Model for Air-Liquid Interfacial Biofilm Formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2934–2939. [Google Scholar] [CrossRef] [PubMed]
- Di Gianvito, P.; Tesnière, C.; Suzzi, G.; Blondin, B.; Tofalo, R. FLO5 Gene Controls Flocculation Phenotype and Adhesive Properties in a Saccharomyces cerevisiae Sparkling Wine Strain. Sci. Rep. 2017, 7, 10786. [Google Scholar] [CrossRef]
- Pangestu, R.; Kahar, P.; Ogino, C.; Kondo, A. Comparative Responses of Flocculating and Nonflocculating Yeasts to Cell Density and Chemical Stress in Lactic Acid Fermentation. Yeast 2024, 41, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Sae-Tang, K.; Bumrungtham, P.; Mhuantong, W.; Champreda, V.; Tanapongpipat, S.; Zhao, X.-Q.; Liu, C.-G.; Runguphan, W. Engineering Flocculation for Improved Tolerance and Production of D-Lactic Acid in Pichia pastoris. J. Fungi 2023, 9, 409. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G. Yeast Flocculation—Sedimentation and Flotation. Fermentation 2018, 4, 28. [Google Scholar] [CrossRef]
- Guo, B.; Yu, W.; Xu, X.; Liu, Y.; Liu, Y.; Du, G.; Liu, L.; Lv, X. Adaptively Evolved and Multiplexed Engineered Saccharomyces cerevisiae for Neutralizer-Free Production of l-Lactic Acid. J. Agric. Food Chem. 2025, 73, 9009–9018. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahmasari, D.; Kahar, P.; Putra, F.J.N.; Ogino, C. Physiological and Metabolic Challenges of Flocculating Saccharomyces cerevisiae in D-Lactic Acid Fermentation Under High-Glucose and Inhibitory Conditions. Processes 2025, 13, 3723. https://doi.org/10.3390/pr13113723
Rahmasari D, Kahar P, Putra FJN, Ogino C. Physiological and Metabolic Challenges of Flocculating Saccharomyces cerevisiae in D-Lactic Acid Fermentation Under High-Glucose and Inhibitory Conditions. Processes. 2025; 13(11):3723. https://doi.org/10.3390/pr13113723
Chicago/Turabian StyleRahmasari, Dianti, Prihardi Kahar, Filemon Jalu Nusantara Putra, and Chiaki Ogino. 2025. "Physiological and Metabolic Challenges of Flocculating Saccharomyces cerevisiae in D-Lactic Acid Fermentation Under High-Glucose and Inhibitory Conditions" Processes 13, no. 11: 3723. https://doi.org/10.3390/pr13113723
APA StyleRahmasari, D., Kahar, P., Putra, F. J. N., & Ogino, C. (2025). Physiological and Metabolic Challenges of Flocculating Saccharomyces cerevisiae in D-Lactic Acid Fermentation Under High-Glucose and Inhibitory Conditions. Processes, 13(11), 3723. https://doi.org/10.3390/pr13113723






