Expression, Characterization, and Immobilization of a Novel D-Lactate Dehydrogenase from Salinispirillum sp. LH 10-3-1
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Strains
2.2. Primary and Secondary Structure Analysis
2.3. Gene Cloning
2.4. Expression and Purification of SaLDH
2.5. Molecular Mass Determination
2.6. SaLDH Activity Assay and Protein Quantification
2.7. Characterization of SaLDH
2.8. Synthesis and Characterization of SaLDH/Cu3(PO4)2 Hybrid Nanoflowers
2.9. Catalytic Properties of SaLDH/Cu3(PO4)2 Hybrid Nanoflowers
2.10. Biotransformation of D-Lactic Acid by Free and Immobilized SaLDH
3. Results
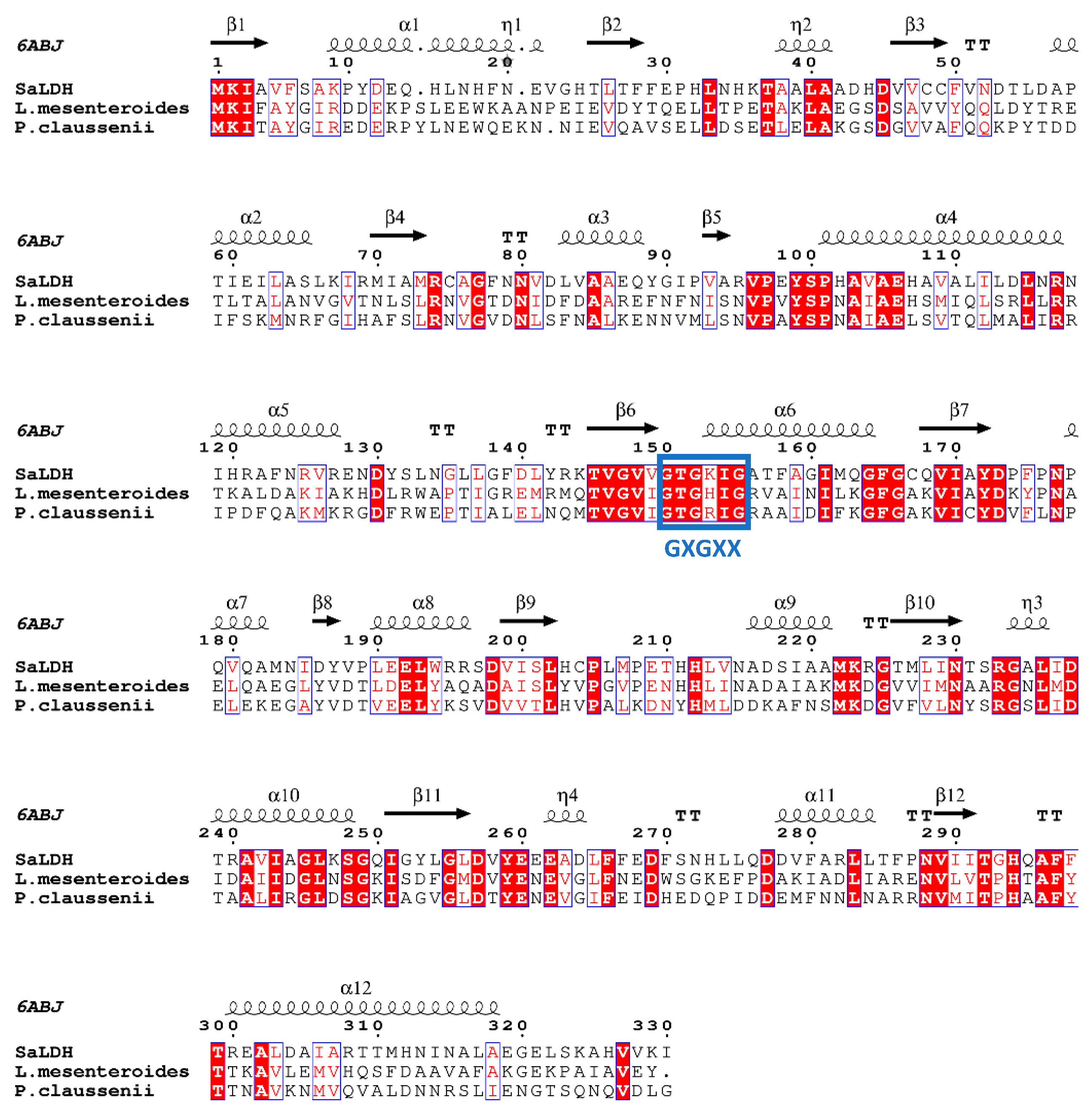
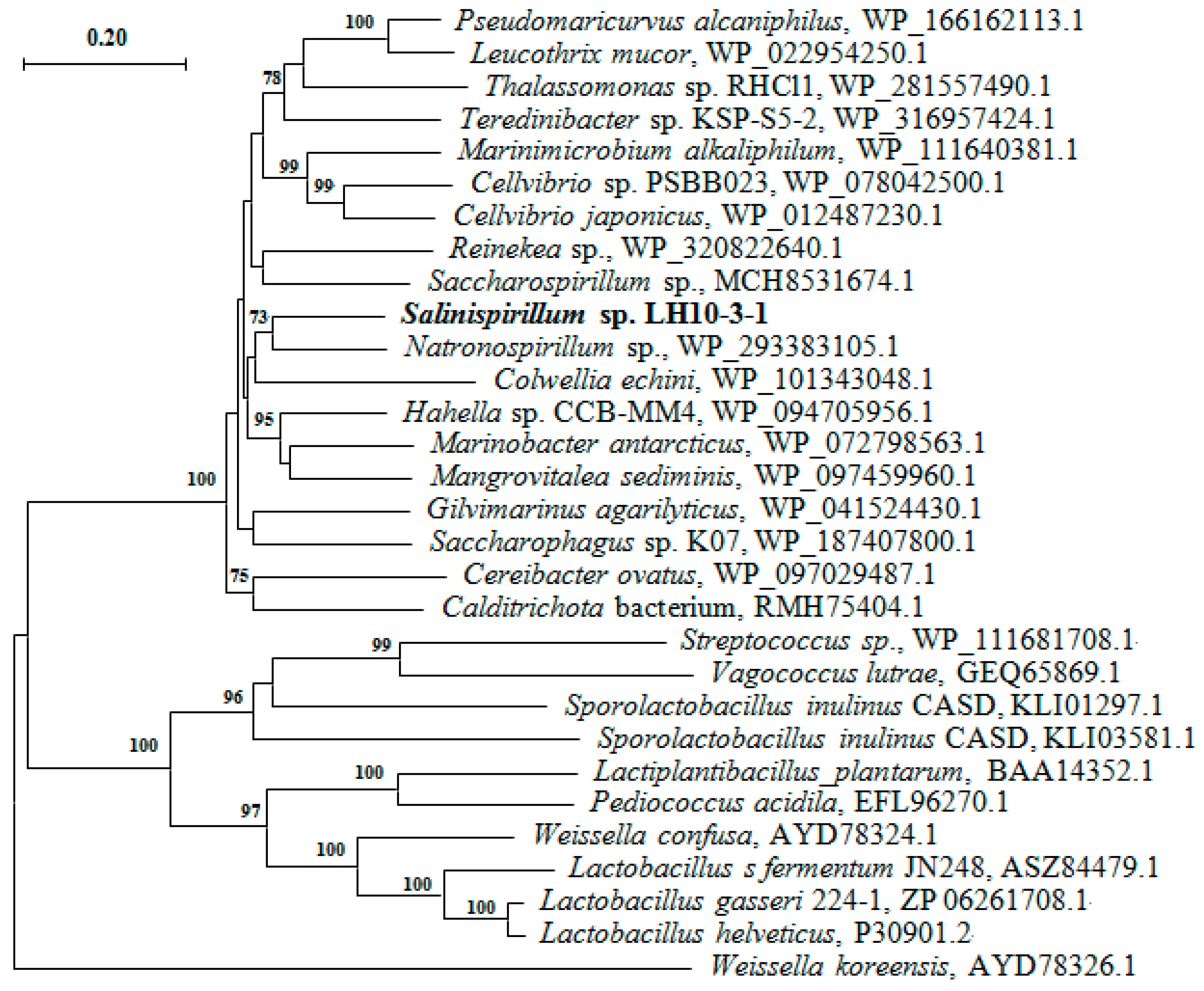
3.1. Sequence Analysis of SaLDH
3.2. Enzyme Purification and Molecular Mass Determination
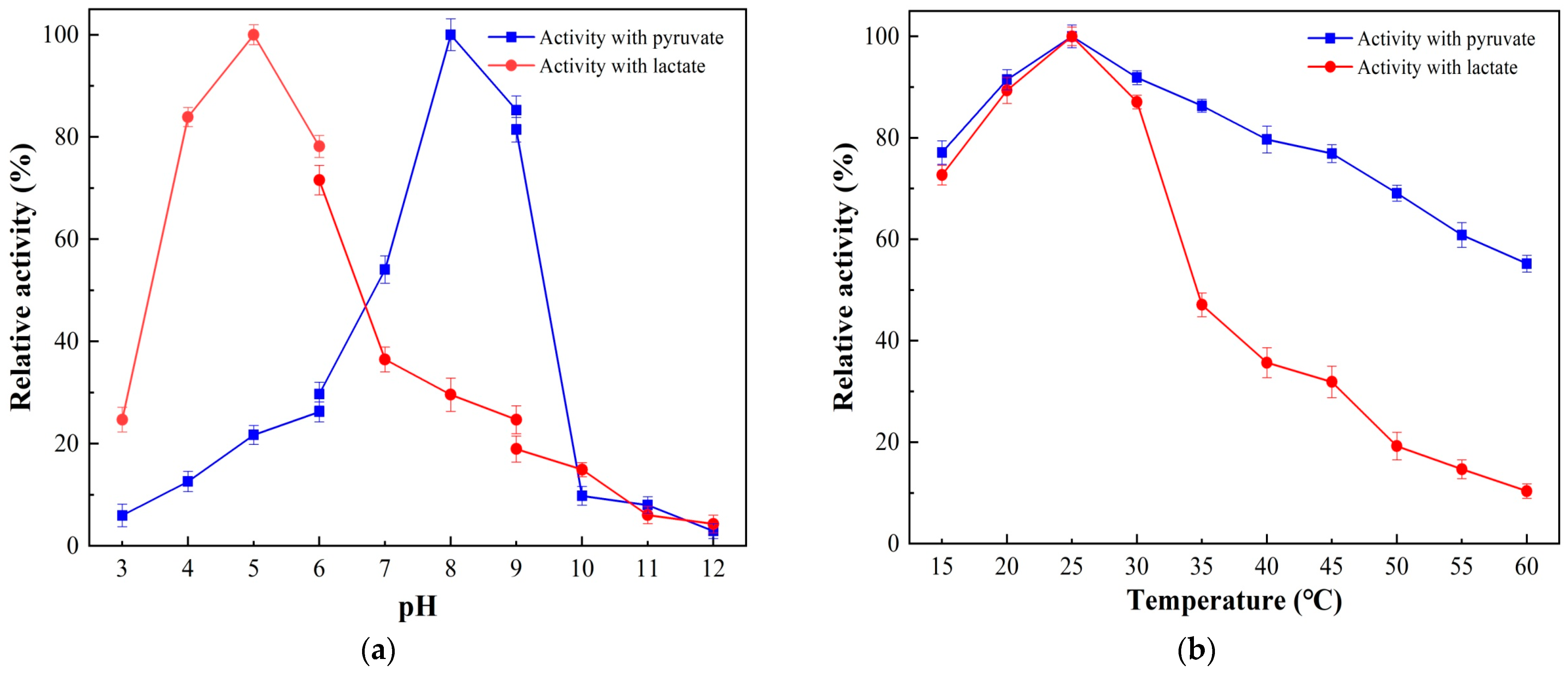
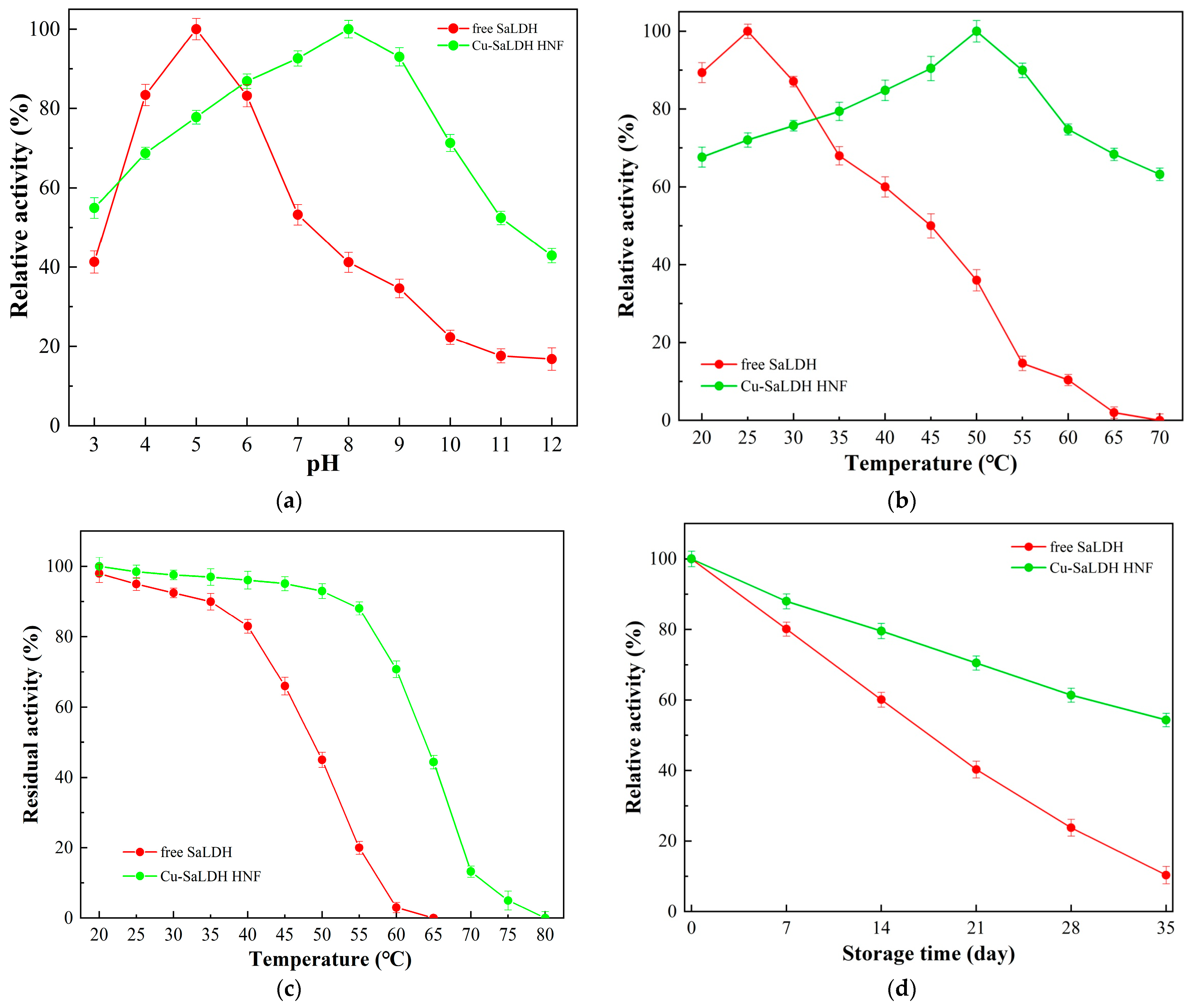
3.3. Effects of pH and Temperature on SaLDH Activity
3.4. Effects of Metal Ions, Organic Solvents, and NaCl Concentration on SaLDH Activity
3.5. Substrate Specificity and Kinetic Parameters of SaLDH
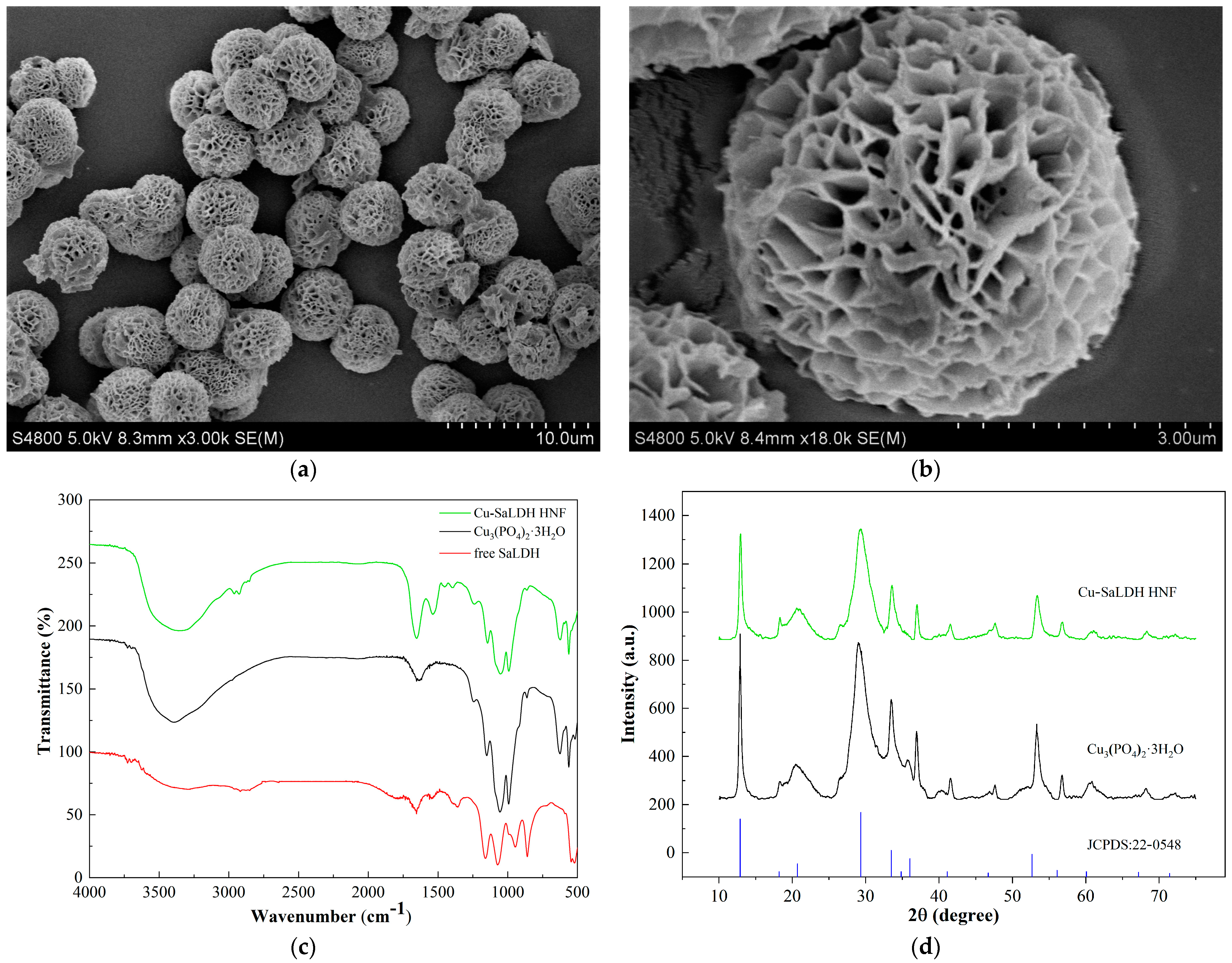
3.6. Synthesis and Characterization of SaLDH/Cu3(PO4)2 Hybrid Nanoflowers
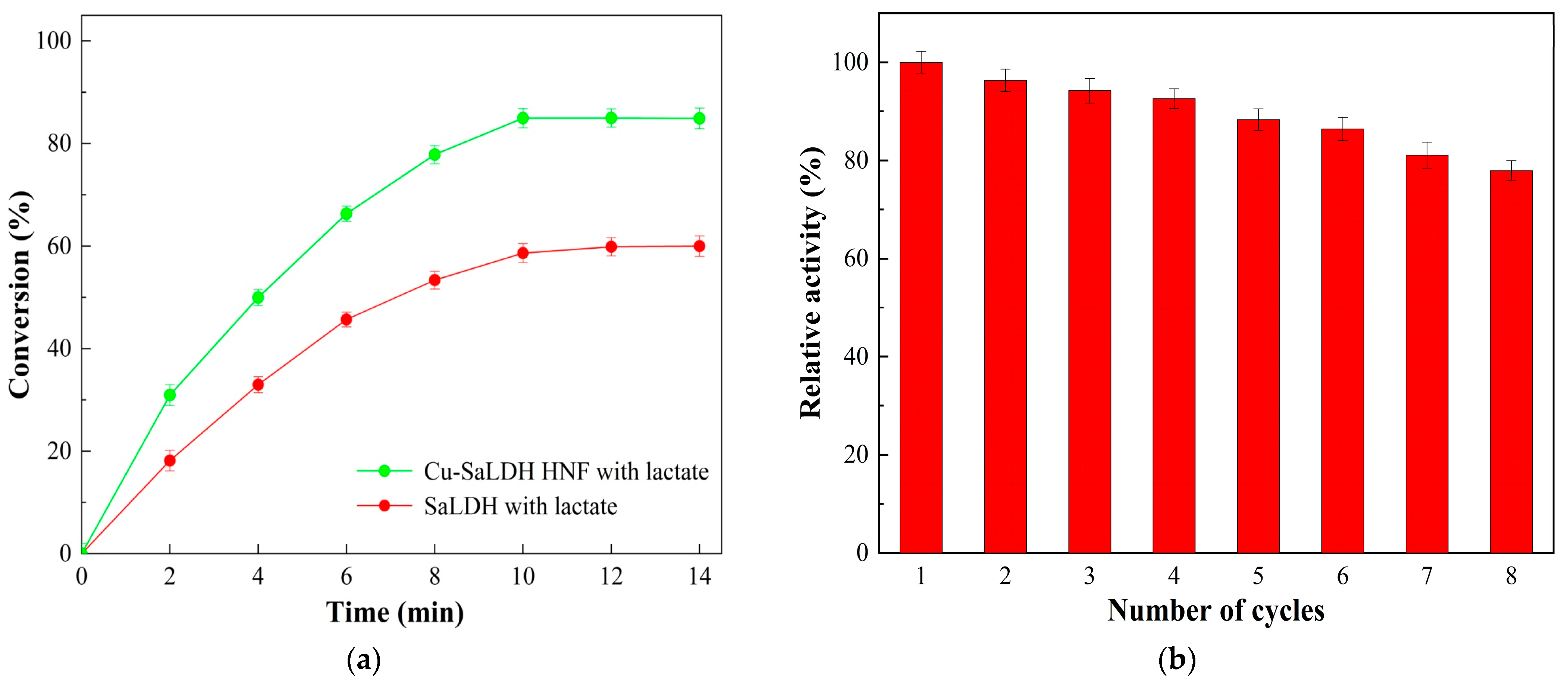
3.7. Catalytic Properties of SaLDH/Cu3(PO4)2 Hybrid Nanoflowers
3.8. Biotransformation of D-Lactic Acid by Free and Immobilized SaLDH
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, L.; Cai, Y.; Zhu, L.; Guo, H.; Yu, B. Major role of NAD-dependent lactate dehydrogenases in the production of L-lactic acid with high optical purifity by the thermophile Bacillus coagulans. Appl. Environ. Microb. 2014, 80, 7134–7141. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.A.; Jun, C.; Joo, J.C.; Kim, S.; Lee, S.H.; Kim, Y.H. Higher thermostability of L-lactate dehydrogenases is a key factor in decreasing the optical purity of D-lactic acid produced from Lactobacillus coryniformis. Enzym. Microb. Tech. 2014, 58–59, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Yu, S.; Jiang, B.; Li, X. Characterization of D-lactate dehydrogenase from Pediococcus acidilactici that converts phenylpyruvic acid into phenyllactic acid. Biotechnol. Lett. 2012, 34, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.C.; Yuan, S.F.; Wang, C.A.; Huang, Y.J.; Guo, G.L.; Hwang, W.S. Production of optically pure L-lactic acid from lignocellulosic hydrolysate by using a newly isolated and D-lactate dehydrogenase gene-deficient Lactobacillus paracasei strain. Bioresour. Technol. 2015, 198, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Leonida, M.D.; Starczynowski, D.T.; Waldman, R.; Aurian-Blajeni, B. Polymeric FAD used as enzyme-friendly mediator in lactate detection. Anal. Bioanal. Chem. 2003, 376, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Richter, N.; Zienert, A.; Hummel, W. A single-point mutation enables lactate dehydrogenase from Bacillus subtilis to utilize NAD+ and NADP+ as cofactor. Eng. Life Sci. 2011, 11, 26–36. [Google Scholar] [CrossRef]
- Jun, C.; Sa, Y.S.; Gu, S.A.; Joo, J.C.; Kim, S.; Kim, K.J.; Kim, Y.H. Discovery and characterization of a thermostable D-lactate dehydrogenase from Lactobacillus jensenii through genome mining. Process Biochem. 2013, 48, 109–117. [Google Scholar]
- Zhou, W.; Zhang, W.; Cai, Y. Enzyme-enhanced adsorption of laccase immobilized graphene oxide for micro-pollutant removal. Sep. Purif. Technol. 2022, 294, 121178. [Google Scholar] [CrossRef]
- Singh, T.A.; Jajoo, A.; Bhasin, S. Optimization of various encapsulation systems for efficient immobilization of actinobacterial glucose isomerase. Biocatal. Agric. Biotechnol. 2020, 29, 101766. [Google Scholar] [CrossRef]
- Ansari, S.A.; Husain, Q. Potential applications of enzymes immobilized on/in nano materials: A review. Biotechnol. Adv. 2012, 30, 512–523. [Google Scholar] [CrossRef]
- Duan, L.; Li, H.; Zhang, Y. Synthesis of hydrid nanoflower-based carbonic anhydrase for enhanced bioactivity and stability. ACS Omega 2018, 3, 18234–18241. [Google Scholar] [CrossRef]
- Ge, J.; Lei, J.; Zare, R.N. Protein–inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef]
- Aydemir, D.; Gecili, F.; Özdemir, N.; Ulusu, N.N. Synthesis and characterization of a triple enzyme-inorganic hybrid na noflower (TrpE@ihNF) as a combination of three pancreatic digestive enzymes amylase, protease and lipase. J. Biosci. Bioeng. 2020, 129, 679–686. [Google Scholar] [CrossRef]
- Cheng, Y.Y.; Park, T.H.; Seong, H.; Kim, T.J.; Han, N.S. Biological characterization of D-lactate dehydrogenase responsible for high-yield production of D-phenyllactic acid in Sporolactobacillus inulinus. Microb. Biotechnol. 2022, 15, 2717–2729. [Google Scholar] [CrossRef]
- Liu, D.; Xi, L.; Han, D.; Dou, K.; Su, S.; Liu, J. Cloning, expression, and characterization of a novel nitrilase, PaCNit, from Pannonibacter carbonis Q4.6. Biotechnol. Lett. 2019, 41, 583–589. [Google Scholar] [CrossRef]
- Li, J.; Li, Z.; Gong, H.; Ma, M.; Li, S.; Yang, H.; Zhang, H.; Liu, J. Identification and characterization of a novel high-activity amylosucrase from Salinispirillum sp. LH10-3-1. Appl. Microb. Biot. 2023, 107, 1725–1736. [Google Scholar] [CrossRef]
- Sedmak, J.J.; Grossberg, S.E. A rapid, sensitive and vertile assay for protein using Coomassia Brilliant Blue G250. Anal. Biochem. 1977, 79, 544–552. [Google Scholar] [CrossRef]
- Zhong, W.; Yang, M.; Mu, T.; Wu, F.; Hao, X.; Chen, R.; Sharshar, M.M.; Thygesen, A.; Wang, Q.; Xing, J. Systematically redesigning and optimizing the expression of D-lactate dehydrogenase efficiently produces high-optical-purity D-lactic acid in Saccharomyces cerevisiae. Biochem. Eng. J. 2019, 144, 217–226. [Google Scholar] [CrossRef]
- Sam, K.K.; Lau, N.S.; Furusawa, G.; Amirul, A.A. Draft genome sequence of halophilic Hahella sp. strain CCB-MM4, isolated from Matang Mangrove Forest in Perak, Malaysia. Genome Announc. 2017, 5, e01147-17. [Google Scholar] [CrossRef]
- Liao, H.; Li, Y.Q.; Guo, X.T.; Lin, X.L.; Lai, Q.L.; Xu, H.; Zheng, T.L.; Tian, Y. Mangrovitalea sediminis gen. nov. sp. nov. a member of the family Alteromonadaceae isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 2017, 67, 5172–5178. [Google Scholar] [CrossRef]
- Christiansen, L.; Bech, P.K.; Schultz-Johansen, M.; Martens, H.J.; Stougaard, P. Colwellia echini sp. nov. an agar- and carrageenan-solubilizing bacterium isolated from sea urchin. Int. J. Syst. Evol. Microbiol. 2018, 68, 687–691. [Google Scholar] [CrossRef]
- Liu, C.; Chen, C.X.; Zhang, X.Y.; Yu, Y.; Liu, A.; Li, G.W.; Chen, X.L.; Chen, B.; Zhou, B.C.; Zhang, Y.Z. Marinobacter antarcticus sp. nov. a halotolerant bacterium isolated from Antarctic intertidal sandy sediment. Int. J. Syst. Evol. Microbiol. 2012, 62, 1838–1844. [Google Scholar] [CrossRef]
- Zaboli, M.; Saeidnia, F.; Zaboli, M.; Torkzadeh-Mahani, M. Stabilization of recombinant D-Lactate dehydrogenase enzyme with trehalose: Response surface methodology and molecular dynamics simulation study. Process Biochem. 2021, 101, 26–35. [Google Scholar] [CrossRef]
- Kochhar, S.; Hunziker, P.E.; Leong-Morgenthaler, P.; Hottinger, H. Primary structure, physicochemical properties, and chemical modification of NAD+-dependent D-lactate dehydrogenase. J. Biol. Chem. 1992, 267, 8499–8513. [Google Scholar] [CrossRef]
- Taguchi, H.; Ohta, T. D-lactate dehydrogenase is a member of D-isomer-specific 2-hydroxy acid dehydrogenase family. Cloning, sequencing, and expression in Escherichia coli of the D-lactate dehydrogenase gene of Lactobacillus plantarum. J. Biol. Chem. 1991, 266, 12588–12594. [Google Scholar] [CrossRef]
- Bhowmik, T.; Lueck, M.; Steele, J.L. Purification and partial characterization of D-(−)-lactate dehydrogenase from Lactobacillus helveticus CNRZ 32. J. Indust. Microbiol. 1993, 12, 35–41. [Google Scholar] [CrossRef]
- Isobe, K.; Koide, Y.; Yokoe, M.; Wakao, N. Crystallization and some properties of D-lactate dehydrogenase from Staphylococcus sp. LDH-1. J. Biosci. Bioeng. 2002, 94, 330–335. [Google Scholar] [CrossRef]
- Gulmez, C.; Altinkaynak, C.; Özdemir, N.; Atakisi, O. Proteinase K hybrid nanoflowers (P-hNFs) as a novel nanobiocatalytic detergent additive. Int. J. Biol. Macromol. 2018, 119, 803–810. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, Y.; Li, G.; Liu, K.; Liu, Y.; He, J.; Lei, J. Preparation and application of a chemically modified laccase and copper phosphate hybrid flower-like biocatalyst. Biochem. Eng. J. 2019, 144, 235–243. [Google Scholar] [CrossRef]
- Bernard, N.; Ferain, T.; Garmyn, D.; Hols, P.; Delcour, J. Cloning of the D-lactate dehydrogenase gene from Lactobacillus delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FEBS Lett. 1991, 290, 61–64. [Google Scholar]
- Singh, S.K.; Ahmed, S.U.; Pandey, A. Metabolic engineering approaches for lactic acid production. Process Biochem. 2006, 41, 991–1000. [Google Scholar] [CrossRef]
- Li, X.; Jiang, B.; Pan, B.; Mu, W.; Zhang, T. Purification and partial characterization of Lactobacillus species SK007 lactate dehydrogenase (LDH) catalyzing phenylpyruvic acid (PPA) conversion into phenyllactic acid (PLA). J. Argric. Food Chem. 2008, 56, 2392–2399. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, J.; Xin, F.; Ma, J.; Zhang, W.; Wu, H.; Jiang, M.; Dong, W. Heterologous expression of a novel D-lactate dehydrogenase from Lactobacillus sp. ZX1 and its application for D-phenyllactic acid production. Int. J. Biol. Macromol. 2018, 119, 1171–1178. [Google Scholar] [CrossRef]
- Kochhar, S.; Lamzin, V.S.; Razeto, A.; Delley, M.; Hottinger, H.; Germond, J.E. Roles of His205, His296, His303 and Asp259 in catalysis by NAD+-specific D-lactate dehydrogenase. Eur. J. Biochem. 2000, 267, 1633–1639. [Google Scholar] [CrossRef]
- Razeto, A.; Kochhar, S.; Hottinger, H.; Dauter, M.; Wilson, K.S.; Lamzin, V.S. Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus. J. Mol. Biol. 2002, 318, 109–119. [Google Scholar] [CrossRef]
- Satomura, T.; Kawakami, R.; Sakuraba, H.; Ohshima, T. A novel flavin adenine dincleotide (FAD) containing D-lactate dehydrogenase from the thermoacidophilic crenarchaeota Sulfolobus tokodaii strain 7: Purification, characterization and expression in Escherichia coli. J. Biosci. Bioeng. 2008, 106, 16–21. [Google Scholar] [CrossRef]
- Garvie, E.I. Bacterial lacate dehydrogenases. Microbiol. Rev. 1980, 44, 106–139. [Google Scholar] [CrossRef]
- Nakano, K.; Sawada, S.; Yamada, R.; Mimitsuka, T.; Ogino, H. Enhancement of the catalytic activity of D-lactate dehydrogenase from Sporolactobacillus laevolacticus by site-directed mutagenesis. Biochem. Eng. J. 2018, 133, 214–218. [Google Scholar] [CrossRef]
- Li, J.; Li, Z.; Cao, M.; Liu, J. Expression and characterization of catechol 1,2-dioxygenase from Oceanimonas marisflavi 102-Na3. Protein Expr. Purif. 2021, 188, 105964. [Google Scholar] [CrossRef]
- Taguchi, H.; Ohta, T. Involvement of Glu-264 and Arg-235 in the essential interaction between the catalytic imidazole and substrate for the D-lactate dehydrogenase catalysis. J. Biochem. 1997, 122, 802–809. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, Y.; Wang, C.; Wang, Z.; Chen, X.; Li, M.; Zhao, R.; Wang, L. Application of dual-enzyme nanoflower in the epoxidation of alkenes. Process Biochem. 2018, 74, 103–107. [Google Scholar] [CrossRef]
- Somturk, B.; Yilmaz, I.; Altinkaynak, C.; Karatepe, A.; Özdemir, N.; Ocsoy, I. Synthesis of urease hybrid nanoflowers and their enhanced catalytic properties. Enzym. Microb. Technol. 2016, 86, 134–142. [Google Scholar] [CrossRef] [PubMed]


| Sample | Specific Activity (U/mg) | Yield (%) | ||
|---|---|---|---|---|
| Reduction Activity | Oxidation Activity | Reduction Activity | Oxidation Activity | |
| Initial crude extract | 8.0 | 5.5 | 100.0 | 100.0 |
| Purified SaLDH | 107.5 | 40.9 | 36.1 a | 20.0 |
| Immobilized SaLDH | 0 | 66.3 | 0 | 162.1 b |
| Additive | Relative Activity (%) | |
|---|---|---|
| Activity with Pyruvate | Activity with Lactate | |
| Control | 100.0 ± 0.0 | 100.0 ± 0.0 |
| Zn2+ | 86.3 ± 1.6 | 78.9 ± 1.5 |
| Cu2+ | 24.2 ± 0.4 | 107.9 ± 0.6 |
| Mn2+ | 52.5 ± 0.3 | 64.4 ± 0.9 |
| Co2+ | 102.3 ± 1.2 | 101.2 ± 0.3 |
| Ca2+ | 105.8 ± 0.5 | 31.7 ± 0.4 |
| Mg2+ | 63.9 ± 1.1 | 65.3 ± 1.8 |
| Ba2+ | 61.6 ± 0.5 | 71.9 ± 0.6 |
| Fe3+ | 73.0 ± 0.9 | 73.6 ± 1.3 |
| EDTA | 96.5 ± 1.2 | 95.3 ± 1.4 |
| DMSO | 99.9 ± 2.1 | 98.9 ± 1.9 |
| DMF | 99.9 ± 1.8 | 99.0 ± 2.1 |
| Methanol | 99.7 ± 2.5 | 98.6 ± 1.8 |
| Ethanol | 99.3 ± 1.4 | 97.3 ± 1.7 |
| Acetone | 98.8 ± 1.7 | 97.1 ± 2.1 |
| Benzene | 90.8 ± 2.2 | 91.2 ± 1.9 |
| Substrates | Relative Activity (%) | Specific Activity (U/mg) |
|---|---|---|
| Sodium pyruvate | 100.0 | 107.5 |
| 2-Ketobutyric acid | 97.9 | 105.2 |
| Oxaloacetic acid | 74.2 | 79.8 |
| Phenylpyruvic acid | 30.3 | 32.6 |
| α-Ketoglutaric acid | 79.8 | 85.8 |
| Benzoic acid | 69.2 | 74.4 |
| D-lactic acid | 100.0 | 40.9 |
| L-lactic acid | 0.0 | 0.0 |
| D-(+)-Malic acid | 88.4 | 36.2 |
| D-(+)-3-Phenyllactic acid | 84.6 | 34.6 |
| Strains | Km (mM) | References | |||
|---|---|---|---|---|---|
| Pyruvate | NADH | D-Lactic Acid | NAD+ | ||
| Salinispirillum sp. LH 10-3-1 | 0.34 | 0.52 | 1.71 | 0.46 | This work |
| Bacillus coagulans | 5.9 | n.d. | 5.94 | n.d. | [1] |
| Leuconostoc mesenteroides | 0.09 | 0.05 | n.d. | n.d. | [23] |
| Lacobacillus bulgaricus | 1.6 | n.d. | 133 | n.d. | [24] |
| L. plantarum | 1.2 | n.d. | 7.0 | n.d. | [25] |
| L. helveticus CNRZ 32 | 0.64 | n.d. | 68 | n.d. | [26] |
| Staphylococcus sp. LDH-1 | 0.67 | n.d. | 8.5 | n.d. | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Jiang, X.; Zheng, Y.; Li, K.; Zhang, R.; Xu, J.; Wang, Z.; Zhang, Y.; Yin, H.; Li, J. Expression, Characterization, and Immobilization of a Novel D-Lactate Dehydrogenase from Salinispirillum sp. LH 10-3-1. Processes 2024, 12, 1349. https://doi.org/10.3390/pr12071349
Liu J, Jiang X, Zheng Y, Li K, Zhang R, Xu J, Wang Z, Zhang Y, Yin H, Li J. Expression, Characterization, and Immobilization of a Novel D-Lactate Dehydrogenase from Salinispirillum sp. LH 10-3-1. Processes. 2024; 12(7):1349. https://doi.org/10.3390/pr12071349
Chicago/Turabian StyleLiu, Jianguo, Xuejiao Jiang, Yaru Zheng, Kaixuan Li, Ruixin Zhang, Jingping Xu, Zhe Wang, Yuxuan Zhang, Haoran Yin, and Jing Li. 2024. "Expression, Characterization, and Immobilization of a Novel D-Lactate Dehydrogenase from Salinispirillum sp. LH 10-3-1" Processes 12, no. 7: 1349. https://doi.org/10.3390/pr12071349
APA StyleLiu, J., Jiang, X., Zheng, Y., Li, K., Zhang, R., Xu, J., Wang, Z., Zhang, Y., Yin, H., & Li, J. (2024). Expression, Characterization, and Immobilization of a Novel D-Lactate Dehydrogenase from Salinispirillum sp. LH 10-3-1. Processes, 12(7), 1349. https://doi.org/10.3390/pr12071349







