Modelling Approach for the Continuous Biocatalytic Synthesis of N-Acetylneuraminic Acid in Packed Bed Reactors
Abstract
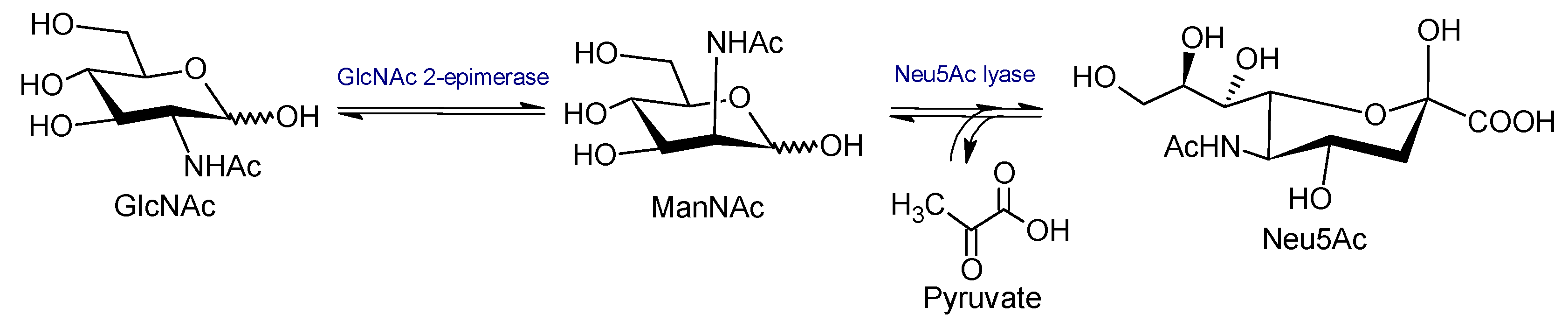
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Enzyme Expression and Purification
2.3. Immobilisation
2.4. Activity Assays in Batch Experiment
2.5. Impact of the Reaction Conditions
2.5.1. Impact of pH
2.5.2. Impact of Activators
2.6. Characterisation of the Immobilised Enzymes in a Packed Bed Reactor
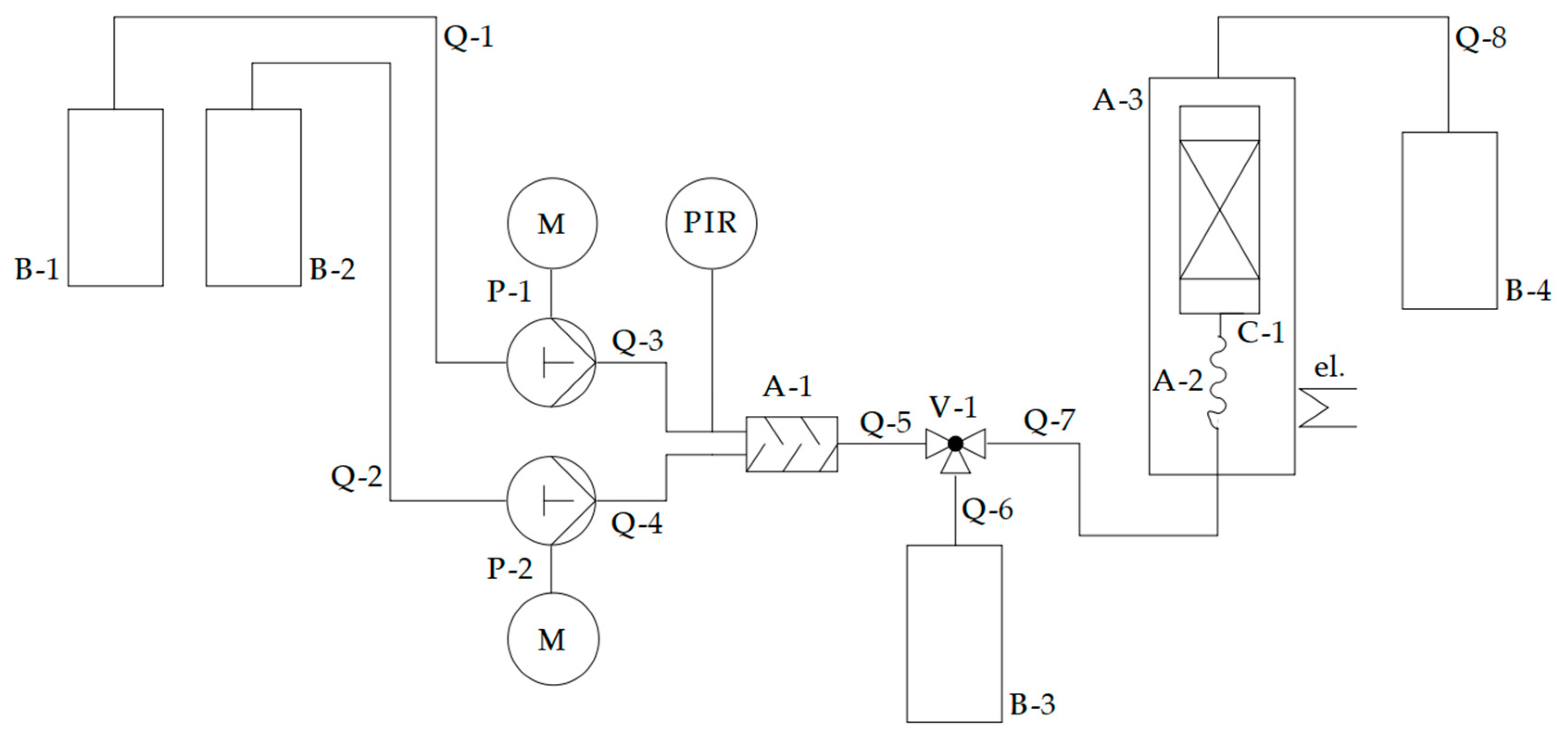
2.6.1. Experimental Setup
2.6.2. Packing of the Reactor
2.6.3. Enzyme Activity in Packed Bed Reactors
2.6.4. Stability of Immobilised Enzymes in Packed Bed Reactors Under Continuous Flow
2.6.5. Substrate Adsorption on Carrier
2.7. Kinetics
2.8. Modelling of Progress Curves
2.8.1. Residence Time Determination
2.8.2. Determination of Reactor Volume
2.9. Model Validation
3. Results and Discussion
3.1. Enzyme Expression and Purification
3.2. Immobilisation
3.3. Impact of the Reaction Conditions
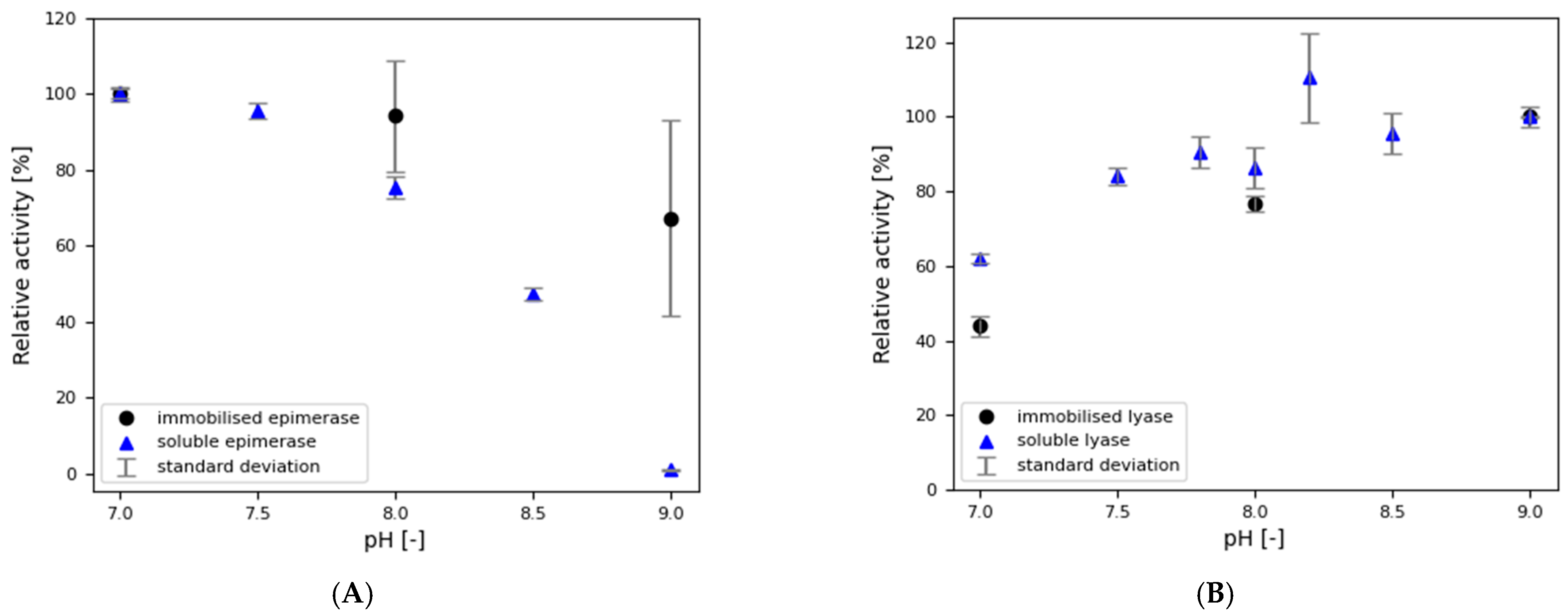
3.3.1. Impact of pH
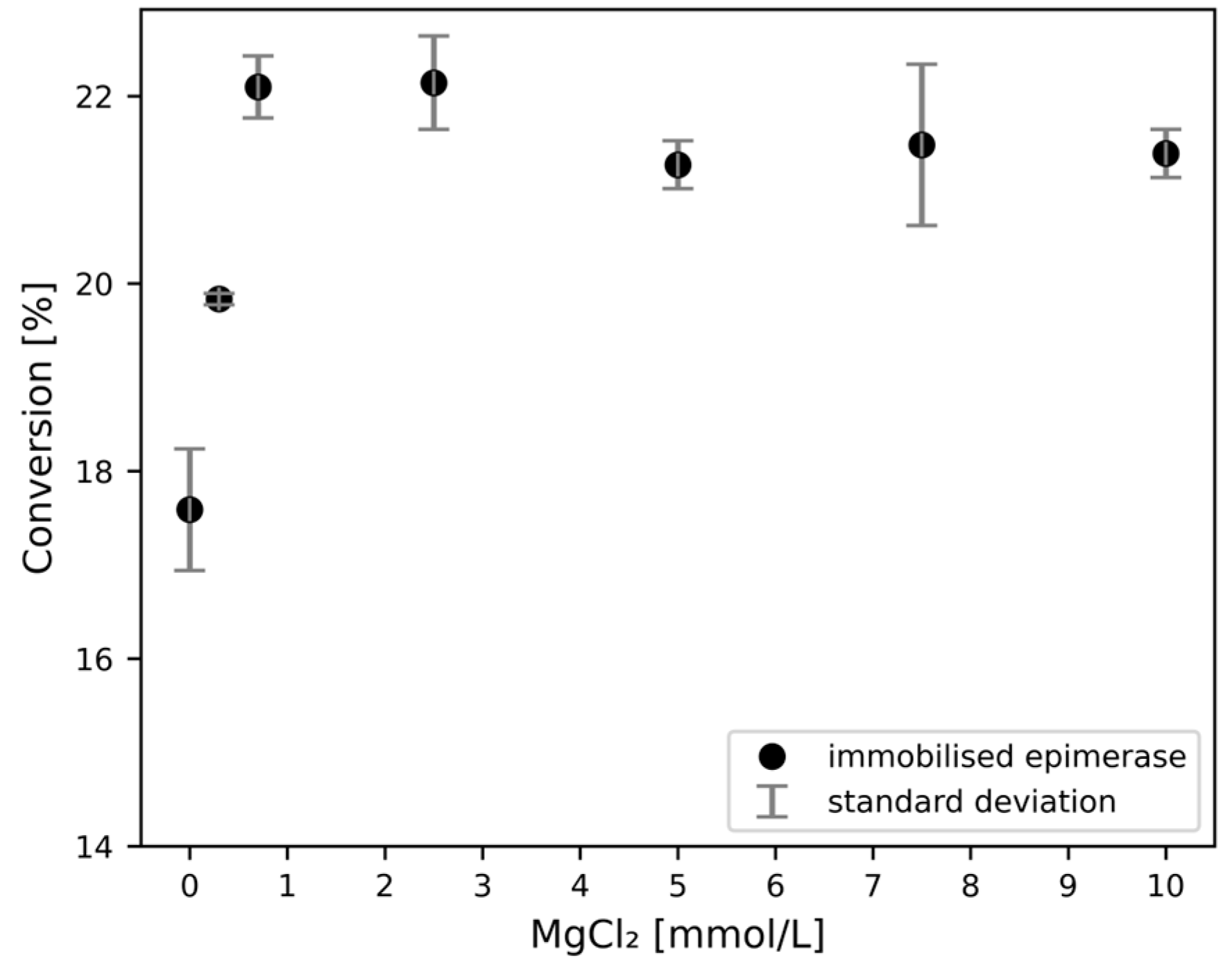
3.3.2. Impact of Activators
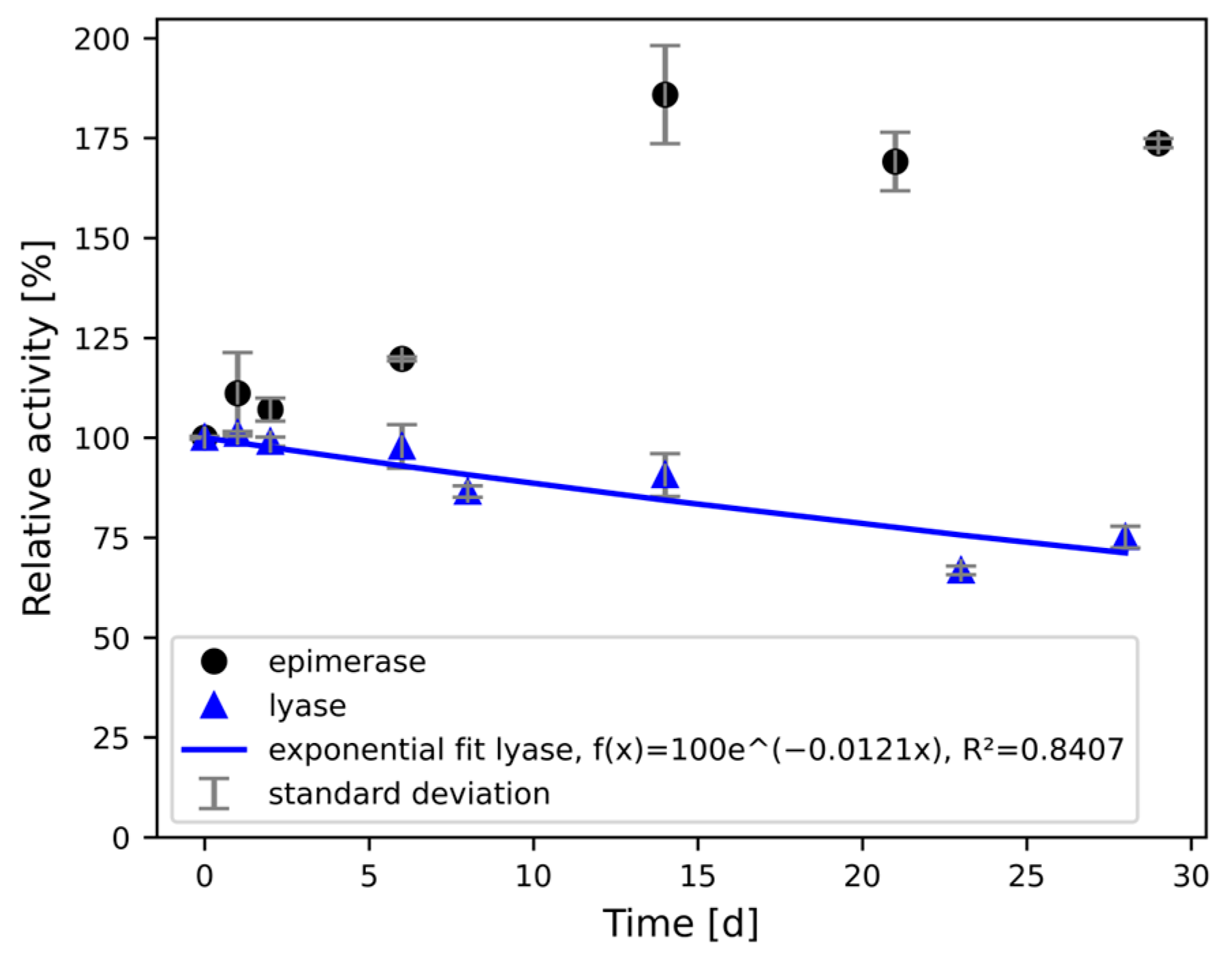
3.4. Stability of Immobilised Enzymes in Packed Bed Reactors Under Continuous Flow
3.5. Kinetics
3.6. Modelling of Progress Curves
3.6.1. Residence Time and Reactor Volume Determination
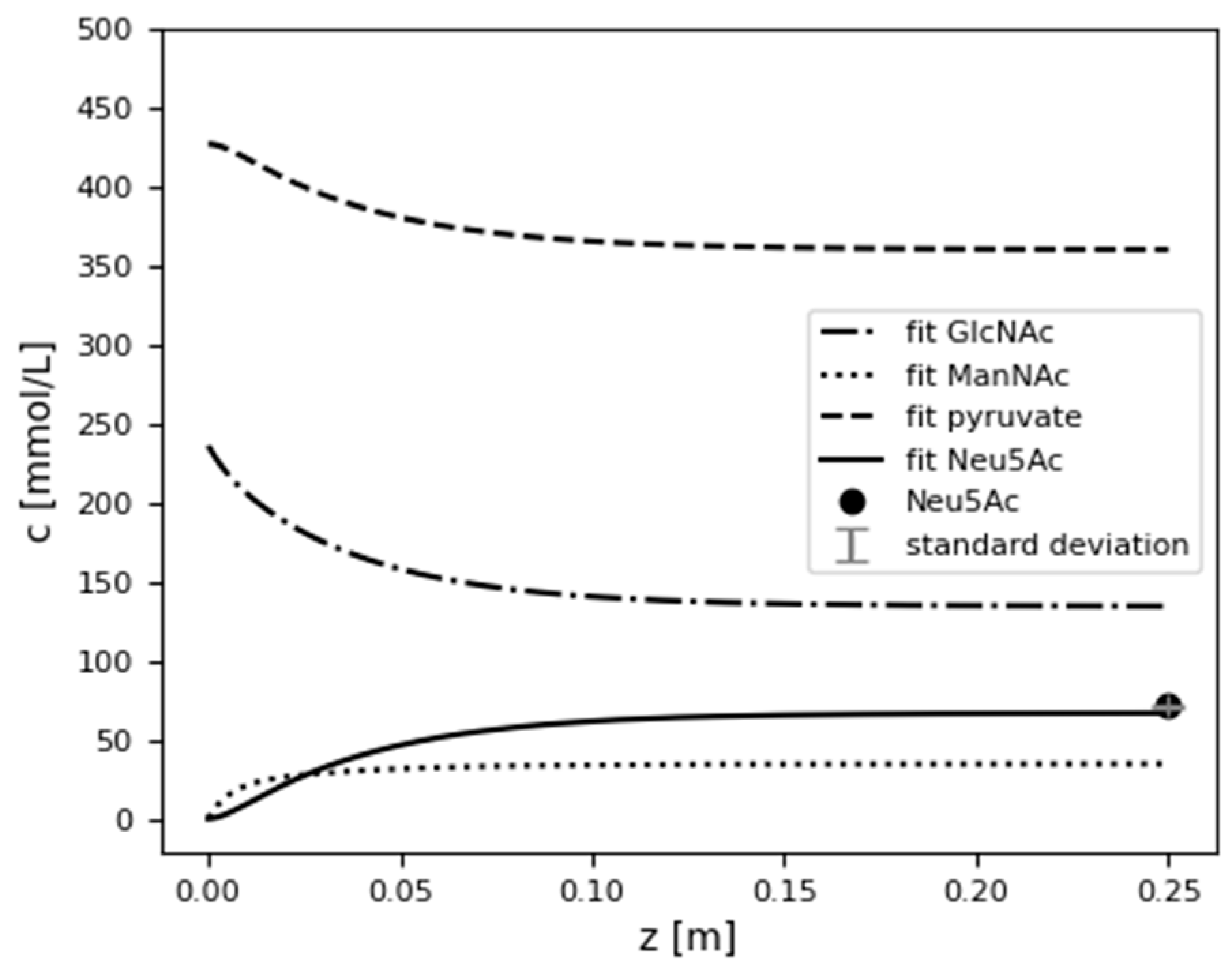
3.6.2. Model Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Böhm, P.; Dauber, S.; Baumeister, L. Über Neuraminsäure, Ihr Vorkommen Und Ihre Bestimmung Im Serum. Klin. Wochenschr. 1954, 32, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, X. Sialic Acid Metabolism and Sialyltransferases: Natural Functions and Applications. Appl. Microbiol. Biotechnol. 2012, 94, 887–905. [Google Scholar] [CrossRef]
- Wang, B.; Brand-Miller, J. The Role and Potential of Sialic Acid in Human Nutrition. Eur. J. Clin. Nutr. 2003, 57, 1351–1369. [Google Scholar] [CrossRef] [PubMed]
- Vimr, E.R.; Kalivoda, K.A.; Deszo, E.L.; Steenbergen, S.M. Diversity of Microbial Sialic Acid Metabolism. Microbiol. Mol. Biol. Rev. 2004, 68, 132–153. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhu, Y.; Wang, H.; Zhang, W.; Mu, W. Recent Advances on N-Acetylneuraminic Acid: Physiological Roles, Applications, and Biosynthesis. Synth. Syst. Biotechnol. 2023, 8, 2405–2805. [Google Scholar] [CrossRef] [PubMed]
- Kragl, U.; Gygax, D.; Ghisalba, O.; Wandrey, C. Enzymatic Two-Step Synthesis of N-Acetylneuraminic Acid in the Enzyme Membrane Reactor. Angew. Chem. Int. 1991, 30, 827–828. [Google Scholar] [CrossRef]
- Spivak, C.T.; Roseman, S. Preparation of N-Acetyl-D-Mannosamine (2-Acetamido-2-Deoxy-D-Mannose) and D-Mannosamine Hydrochloride (2-Amino-Deoxy-D-Mannose). J. Am. Chem. Soc. 1959, 81, 2403–2404. [Google Scholar] [CrossRef]
- Ferrero, M.Á.; Aparicio, L.R. Biosynthesis and Production of Polysialic Acids in Bacteria. Appl. Microbiol. Biotechnol. 2010, 86, 1621–1635. [Google Scholar] [CrossRef]
- Maru, I.; Ohnishi, J.; Ohta, Y.; Tsukada, Y. Simple and Large-Scale Production of N-Acetylneuraminic Acid from N-Acetyl-D-Glucosamine and Pyruvate Using N-Acyl-D-Glucosamine 2-Epimerase and N-Acetylneuraminate Lyase. Carbohydr. Res. 1998, 306, 575–578. [Google Scholar] [CrossRef]
- Mahmoudian, M.; Noble, D.; Drake, C.S.; Middleton, R.F.; Montgomery, D.S.; Piercey, J.E.; Ramlakhan, D.; Todd, M.; Dawson, M.J. An Efficient Process for Production of N-Acetylneuraminic Acid Using N-Acetylneuraminic Acid Aldolase. Enzyme Microb. Technol. 1997, 20, 393–400. [Google Scholar] [CrossRef]
- Hu, S.; Chen, J.; Yang, Z.; Shao, L.; Bai, H.; Luo, J.; Jiang, W.; Yang, Y. Coupled Bioconversion for Preparation of N-Acetyl-D-Neuraminic Acid Using Immobilized N-Acetyl-D-Glucosamine-2-Epimerase and N-Acetyl-D-Neuraminic Acid Lyase. Appl. Microbiol. Biotechnol. 2010, 85, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhuang, W.; Tang, C.; Chen, Y.; Wu, J.; Guo, T.; Ying, H. Efficient Immobilization of AGE and NAL Enzymes onto Functional Amino Resin as Recyclable and High-Performance Biocatalyst. Bioprocess. Biosyst. Eng. 2017, 40, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Bloemendal, V.R.L.J.; Moons, S.J.; Heming, J.J.A.; Chayoua, M.; Niesink, O.; van Hest, J.C.M.; Boltje, T.J.; Rutjes, F.P.J.T. Chemoenzymatic Synthesis of Sialic Acid Derivatives Using Immobilized N-Acetylneuraminate Lyase in a Continuous Flow Reactor. Adv. Synth. Catal. 2019, 361, 2443–2447. [Google Scholar] [CrossRef] [PubMed]
- Obst, F.; Mertz, M.; Mehner, P.J.; Beck, A.; Castiglione, K.; Richter, A.; Voit, B.; Appelhans, D. Enzymatic Synthesis of Sialic Acids in Microfluidics to Overcome Cross-Inhibitions and Substrate Supply Limitations. ACS Appl. Mater. Interfaces 2021, 13, 49433–49444. [Google Scholar] [CrossRef] [PubMed]
- Reich, J.A.; Aßmann, M.; Hölting, K.; Bubenheim, P.; Kuballa, J.; Liese, A. Shift of Reaction Equilibrium at High Pressure in the Continuous Synthesis of Neuraminic Acid. Beilstein 2022, 18, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Federsel, H.J.; Moody, T.S.; Taylor, S.J.C. Recent Trends in Enzyme Immobilization—Concepts for Expanding the Biocatalysis Toolbox. Molecules 2021, 26, 2822. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, J.M.; Woodley, J.M.; Fernandez-Lafuente, R. Is Enzyme Immobilization a Mature Discipline? Some Critical Considerations to Capitalize on the Benefits of Immobilization. Chem. Soc. Rev. 2022, 51, 6251–6290. [Google Scholar] [CrossRef]
- Sheldon, R.A.; van Pelt, S. Enzyme Immobilisation in Biocatalysis: Why, What and How. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef]
- De Santis, P.; Meyer, L.-E.; Kara, S. The Rise of Continuous Flow Biocatalysis—Fundamentals, Very Recent Developments and Future Perspectives. React. Chem. Eng. 2020, 5, 2155–2184. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Woodley, J.M. Role of Biocatalysis in Sustainable Chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef]
- Horiuchi, T.; Kurokawa, T. Purification from and Properties of N-Acyl-D-Mannosamine Dehydrogenase Flavobacterium Sp. 141-8. J. Biochem. 1988, 104, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Lilley, G.G.; Itzstein, M.; von Ivancic, N. High-Level Production and Purification of Escherichia Coli N-Acetylneuraminic Acid Aldolase (EC 4.1.3.3). Protein. Expr. Purif. 1992, 3, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.C.; von Hippel, P.H. Calculation of Protein Extinction Coefficients from Amino Acid Sequence Data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Swiss Institute of Bioinformatics Expasy ProtParam Tool. Available online: https://web.expasy.org/protparam/ (accessed on 9 August 2024).
- Purolite Life Sciences Enzyme Immobilization Resins. 2021. Available online: https://sharevent.ams3.digitaloceanspaces.com/production/files/563/exhibitor/6449/6449_3fd71c29bd.pdf (accessed on 23 September 2024).
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Carballares, D.; Morellon-Sterling, R.; Fernandez-Lafuente, R. Stabilization of Enzymes via Immobilization: Multipoint Covalent Attachment and Other Stabilization Strategies. Biotechnol. Adv. 2021, 52, 107821. [Google Scholar] [CrossRef]
- Hölting, K.; Götz, S.; Aßmann, M.; Bubenheim, P.; Liese, A.; Kuballa, J. Resilient Enzymes through Immobilisation: Stable NDP Polyphosphate Phosphotransferase from Ruegeria Pomeroyi for Nucleotide Regeneration. Catalysts 2024, 14, 165. [Google Scholar] [CrossRef]
- Syldatk, C.; Jaeger, K.-E.; Liese, A. Einführung in Die Enzymtechnologie; Springer Spektrum: Berlin, Germany, 2018; ISBN 978-3-662-57618-2. [Google Scholar]
- Resindion, S.r.l. ReliZymeTM and SEPABEADSTM EC Enzyme Carriers. 2015. Available online: https://resindion.com/resindion/download/RESINDION_ReliZymeSepabeadsEC_PGD_050612.pdf (accessed on 23 September 2024).
- Klermund, L.; Riederer, A.; Hunger, A.; Castiglione, K. Protein Engineering of a Bacterial N-Acyl-D-Glucosamine 2-Epimerase for Improved Stability under Process Conditions. Enzyme Microb. Technol. 2016, 87, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Horecker, B.L.; Kornber, A. The Extinction Coefficients of the Reduced Band of Pyridine Nucleotides. J. Biol. Chem. 1948, 175, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Liese, A.; Hilterhaus, L. Evaluation of Immobilized Enzymes for Industrial Applications. Chem. Soc. Rev. 2013, 42, 6236–6249. [Google Scholar] [CrossRef]
- Chmiel, H.; Takors, R.; Weuster-Botz, D. Bioprozesstechnik, 4th ed.; Springer: Berlin/Heidelberg, Germany, 2018; ISBN 9783662540411. [Google Scholar]
- Müller-Erlwein, E. Chemische Reaktionstechnik; Springer Spektrum: Berlin/Heidelberg, Germany, 2015; Volume 3, ISBN 9783658093952. [Google Scholar]
- Aisaka, K.; Igarashi, A.; Yamaguchi, K.; Uwajima, T. Purification, Crystallization and Characterization of N-Acetylneuraminate Lyase from Escherichia coli. Biochem. J. 1991, 276, 541–546. [Google Scholar] [CrossRef]
- Bayne, L.; Ulijn, R.V.; Halling, P.J. Effect of Pore Size on the Performance of Immobilised Enzymes. Chem. Soc. Rev. 2013, 42, 9000–9010. [Google Scholar] [CrossRef]
- Santos, J.C.S.D.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the Support Properties for Immobilization or Purification of Enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef]
- De Santis, P.; Petrovai, N.; Meyer, L.-E.; Hobisch, M.; Kara, S. A Holistic Carrier-Bound Immobilization Approach for Unspecific Peroxygenase. Front. Chem. 2022, 10, 985997. [Google Scholar] [CrossRef] [PubMed]
- Rădoi, I.I.; Bedolla, D.E.; Vaccari, L.; Todea, A.; Zappaterra, F.; Volkov, A.; Gardossi, L. FTIR Microscopy for Direct Observation of Conformational Changes on Immobilized ω-Transaminase: Effect of Water Activity and Organic Solvent on Biocatalyst Performance. Catal. Sci. Technol. 2023, 13, 4955–4967. [Google Scholar] [CrossRef]
- Wang, S.Y.; Laborda, P.; Lu, A.M.; Duan, X.C.; Ma, H.Y.; Liu, L.; Voglmeir, J. N-Acetylglucosamine 2-Epimerase from Pedobacter Heparinus: First Experimental Evidence of a Deprotonation/Reprotonation Mechanism. Catalysts 2016, 6, 212. [Google Scholar] [CrossRef]
- Li, Y.; Yu, H.; Cao, H.; Lau, K.; Muthana, S.; Tiwari, V.K.; Son, B.; Chen, X. Pasteurella Multocida Sialic Acid Aldolase: A Promising Biocatalyst. Appl. Microbiol. Biotechnol. 2008, 79, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Maru, I.; Ohta, Y.; Murata, K.; Tsukada, Y. Molecular Cloning and Identification of N-Acyl-D-Glucosamine 2-Epimerase from Porcine Kidney as a Renin-Binding Protein. J. Biol. Chem. 1996, 271, 16294–16299. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, V.; Hennemann, H.G.; Daußmann, T.; Kragl, U. Modelling the Reaction Course of N-Acetylneuraminic Acid Synthesis from N-Acetyl-D-Glucosamine—New Strategies for the Optimisation of Neuraminic Acid Synthesis. Appl. Microbiol. Biotechnol. 2007, 76, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Izard, T.; Lawrence, M.C.; Malby, R.L.; Lilley, G.G.; Colman, P.M. The Three-Dimensional Structure of N-Acetylneuraminate Lyase from Escherichia coli. Structure 1994, 2, 361–369. [Google Scholar] [CrossRef]
- Groher, A.; Hoelsch, K. Mechanistic Model for the Synthesis of N-Acetylneuraminic Acid Using N-Acetylneuraminate Lyase from Escherichia Coli K12. J. Mol. Catal. B Enzym. 2012, 83, 1–7. [Google Scholar] [CrossRef]
- Timms, N.; Windle, C.L.; Polyakova, A.; Ault, J.R.; Trinh, C.H.; Pearson, A.R.; Nelson, A.; Berry, A. Structural Insights into the Recovery of Aldolase Activity in N -Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Γ-Thialysine by Using a Chemical Mutagenesis Strategy. ChemBioChem 2013, 14, 474–481. [Google Scholar] [CrossRef]
- Daniels, A.D.; Campeotto, I.; Van Der Kamp, M.W.; Bolt, A.H.; Trinh, C.H.; Phillips, S.E.V.; Pearson, A.R.; Nelson, A.; Mulholland, A.J.; Berry, A. Reaction Mechanism of N-Acetylneuraminic Acid Lyase Revealed by a Combination of Crystallography, QM/MM Simulation, and Mutagenesis. ACS Chem. Biol. 2014, 9, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, J.M.; López-Gallego, F. Characterization and Evaluation of Immobilized Enzymes for Applications in Flow Reactors. Curr. Opin. Green Sustain. Chem. 2020, 25, 100349. [Google Scholar] [CrossRef]



| Carrier/Order Number | Immobilisation Type | Pore Diameter */nm | Particle Diameter */µm |
|---|---|---|---|
| Epoxy methacrylate/ECR8204F (Purolite Ltd.) | covalent | 30–60 | 150–300 |
| Epoxy methacrylate/ECR8204M (Purolite Ltd.) | 30–60 | 300–710 | |
| Dimethylamino methacrylate/ECR8309F (Purolite Ltd.) | covalent via pre-activation with glutaraldehyde | 60–120 | 150–300 |
| Dimethylamino methacrylate/ECR8309M (Purolite Ltd.) | 60–120 | 300–710 | |
| Hexamethylamino methacrylate/HA403/M (Resindion S.r.l.) | 40–60 | 200–500 | |
| Dimethylamino methacrylate/EA403/M (Resindion S.r.l.) | 40–60 | 200–500 |
| PBR Number | ID/cm | Length/ cm | V */mL | Packed Bed | Packed Bed Mass/mg | Application |
|---|---|---|---|---|---|---|
| 1 | 0.3 | 3 | 0.2 | immobilised epimerase (HA403/M) blocked carrier (HA403/M) | 25 176 | Kinetic epimerase forward |
| 2 | 0.3 | 3 | 0.2 | immobilised epimerase (HA403/M) blocked carrier (HA403/M) | 119 81 | Kinetic epimerase reverse |
| 3 | 0.3 | 3 | 0.2 | immobilised epimerase (HA403/M) blocked carrier (HA403/M) | 25 110 | Stability study epimerase |
| 4 | 0.3 | 10 | 0.7 | immobilised epimerase (HA403/M) | 478 | Validation epimerase model |
| 5 | 0.3 | 3 | 0.2 | immobilised lyase (ECR8309M) | 188 | Kinetic lyase forward |
| 6 | 0.3 | 3 | 0.2 | immobilised lyase (ECR8309M) blocked carrier (ECR8309M) | 20 110 | Kinetic lyase reverse |
| 7 | 0.3 | 3 | 0.2 | immobilised lyase (ECR8309M) | 205 | Stability study lyase |
| 8 | 0.46 | 15 | 2.5 | immobilised lyase (ECR8309M) | 1969 | Validation lyase model |
| 9 | 0.46 | 15 | 2.5 | immobilised epimerase (HA403/M) immobilised lyase (ECR8309M) | 1016 1694 | Validation combined model |
| 10 | 0.46 | 5 | 0.8 | immobilised epimerase (HA403/M) immobilised lyase (ECR8309M) | 196 326 | Validation combined model |
| 11 | 2.1 | 25 | 84.6 | immobilised epimerase (ECR8204M) immobilised lyase (ECR8309M) | 38,337 38,340 | Validation combined model |
| Characteristic Variable | ECR8204F 1 | ECR8204M 1 | HA403/M 2 | EA403/M 3 |
|---|---|---|---|---|
| Immobilisation efficiency/% | 8.4 | 1.0 | 0.4 | 0.3 |
| Activity yield/% | 3.1 | 0.6 | 0.4 | 0.3 |
| Carrier loading/mgenzyme·gcarrier−1 | 31.6 | 33.3 | 44.0 | 42.5 |
| Carrier spec. activity 4/U·gcarrier−1 | 107.4 | 50.8 | 41.0 | 29.4 |
| Characteristic Variable | ECR8309F 1 | ECR8309M 1 |
|---|---|---|
| Immobilisation efficiency/% | 7.6 | 12.4 |
| Activity yield/% | 6.5 | 10.6 |
| Carrier loading/mgenzyme·gcarrier−1 | 84.7 | 95.1 |
| Carrier spec. activity 2/U·gcarrier−1 | 26.5 | 63.6 |
| Enzyme | Parameter | Value | Unit * |
|---|---|---|---|
| Epimerase | vmax,app,forward | 234 ± 3 | U∙gcarrier−1 |
| Km,app,forward | 266 ± 10 | mmol∙L−1 | |
| vmax,app,backward | 797 ± 46 | U∙gcarrier−1 | |
| Km,app,backward | 294 ± 45 | mmol∙L−1 | |
| KI,app,pyruvate | 940 ± 107 | mmol∙L−1 | |
| Lyase | vmax,app,forward | 294 ± 22 | U∙gcarrier−1 |
| Km,app,ManNAc,forward | 249 ± 64 | mmol∙L−1 | |
| Km,app,pyruvate,forward | 229 ± 11 | mmol∙L−1 | |
| vmax,app,backward | 151 ± 6 | U∙gcarrier−1 | |
| Km,app,backward | 205 ± 19 | mmol∙L−1 | |
| KI,app,GlcNAc | 418 ± 40 | mmol∙L−1 |
| Reactor Number a | /mL∙min−1 | b/min | /- | c/mL |
|---|---|---|---|---|
| 1 | 1.5 | 0.117 ± 0.004 | 0.82 | 0.17 |
| 4 | 1.5 | 0.367 ± 0.038 | 0.71 | 0.50 |
| 4 | 0.5 | 1.222 ± 0.008 | 0.62 | 0.43 |
| 5 | 1.5 | 0.109 ± 0.008 | 0.77 | 0.16 |
| 8 | 1.5 | 1.556 ± 0.009 | 0.69 | 1.70 |
| 8 | 0.5 | 5.131 ± 0.077 | 0.81 | 2.03 |
| 9 | 1.5 | 1.399 ± 0.045 | 0.61 | 1.53 |
| 9 | 0.5 | 4.640 ± 0.106 | 0.73 | 1.83 |
| 10 | 1.5 | 0.581 ± 0.020 | 0.79 | 0.66 |
| 10 | 0.5 | 1.527 ± 0.048 | 0.61 | 0.51 |
| 11 | 1.5 | 48 | 0.74 | 64 |
| Experiment | Reactor Number a | FlowRate/ mL∙min−1 | Measured b: ManNAc/mmol·L−1 | Model Prediction: ManNAc/mmol·L−1 | Deviation/% |
|---|---|---|---|---|---|
| 1 | 1 | 1.5 | 1.07 ± 0.01 | 1.05 | 2 |
| 2 | 4 | 1.5 | 12.96 ± 0.09 | 13.65 | 5 |
| 3 | 4 | 0.5 | 19.48 ± 0.34 | 20.72 | 6 |
| Experiment | Reactor Number a | Flow Rate/ mL∙min−1 | Measured b: Neu5Ac/mmol·L−1 | Model Prediction: Neu5Ac/mmol·L−1 | Deviation/% |
|---|---|---|---|---|---|
| 4 | 5 | 1.5 | 4.90 ± 0.11 | 5.24 | 7 |
| 5 | 8 | 1.5 | 32.52 ± 1.10 | 35.88 | 9 |
| 6 | 8 | 0.5 | 45.63 ± 0.76 | 43.08 | 6 |
| Experiment | Reactor Number a | Flow Rate/mL∙min−1 | Measured b: Neu5Ac/mmol·L−1 | Model Prediction: Neu5Ac/mmol·L−1 | Deviation/% |
|---|---|---|---|---|---|
| 7 | 9 + 10 (in series) | 1.5 | 8.43 ± 0.54 | 9.71 | 13 |
| 8 | 9 + 10 (in series) | 0.5 | 17.54 ± 0.08 | 23.11 | 24 |
| 9 | 11 | 1.5 | 71.47 ± 0.75 | 66.84 | 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hölting, K.; Aßmann, M.; Bubenheim, P.; Liese, A.; Kuballa, J. Modelling Approach for the Continuous Biocatalytic Synthesis of N-Acetylneuraminic Acid in Packed Bed Reactors. Processes 2024, 12, 2191. https://doi.org/10.3390/pr12102191
Hölting K, Aßmann M, Bubenheim P, Liese A, Kuballa J. Modelling Approach for the Continuous Biocatalytic Synthesis of N-Acetylneuraminic Acid in Packed Bed Reactors. Processes. 2024; 12(10):2191. https://doi.org/10.3390/pr12102191
Chicago/Turabian StyleHölting, Kristin, Miriam Aßmann, Paul Bubenheim, Andreas Liese, and Jürgen Kuballa. 2024. "Modelling Approach for the Continuous Biocatalytic Synthesis of N-Acetylneuraminic Acid in Packed Bed Reactors" Processes 12, no. 10: 2191. https://doi.org/10.3390/pr12102191
APA StyleHölting, K., Aßmann, M., Bubenheim, P., Liese, A., & Kuballa, J. (2024). Modelling Approach for the Continuous Biocatalytic Synthesis of N-Acetylneuraminic Acid in Packed Bed Reactors. Processes, 12(10), 2191. https://doi.org/10.3390/pr12102191







