Abstract
Gene expression data are usually known for having a large number of features. Usually, some of these features are irrelevant and redundant. However, in some cases, all features, despite being numerous, show high importance and contribute to the data analysis. In a similar fashion, gene expression data sometimes have limited instances with a high rate of imbalance among the classes. This can limit the exposure of a classification model to instances of different categories, thereby influencing the performance of the model. In this study, we proposed a cancer detection approach that utilized data preprocessing techniques such as oversampling, feature selection, and classification models. The study used SVMSMOTE for the oversampling of the six examined datasets. Further, we examined different techniques for feature selection using dimension reduction methods and classifier-based feature ranking and selection. We trained six machine learning algorithms, using repeated 5-fold cross-validation on different microarray datasets. The performance of the algorithms differed based on the data and feature reduction technique used.
1. Introduction
Gene expression involves the translation of information encoded in a gene into gene products, including proteins, tRNA, rRNA, mRNA, or snRNA. With the increase in technological standards, gene expression continues to have a sporadic increase in importance for health and life science applications [1]. Prognostic risk scores from gene expression have shown prominent clinical values as they are promising biomarkers. They can be used for the prediction of prognosis, including the identification of mortality and metastasis risks in patients. They can also be used to determine the response of patients to treatment [2]. Identifying the risk of cancer recurrence or metastasis in patients can help clinicians strategically recommend effective treatment. Furthermore, the determination of response to treatment can identify the overall survival of the patients and intuitively develop novel drugs or appropriate treatment based on each patient’s classification. In the majority of cancer types, HLA gene expression has been shown to prolong overall survival [3]. On the other hand, an increase in the expression of Human Endogenous Retrovirus K mRNA in the blood is linked to the presence of breast cancer, which shows it is a biomarker [4]. BRCA2 is another gene whose expression is associated with highly proliferative and aggressive breast cancer. The higher the expression of BRCA2, the more aggressiveness the breast cancer [5]. This indicates its potential as a biomarker for breast cancer. In essence, biological determinants such as predictive gene expression signatures can now be used for the effective classification of tumors according to their subgroup [6]. The profiling of gene expression for breast cancer or other cancer types can be further improved using clinicopathological and microenvironmental features [7,8,9].
The generation of data in the biomedical and health fields has increased sporadically while yielding samples with a high number of features [10,11]. The challenge of high-dimensional data is the difficulty associated with manual analysis and the redundancy that comes along with some of the features. Over the years, several studies have been carried out based on feature selection. In [12], the authors utilized a hybrid filter and wrapper method for the selection of features in gene expression data. The authors also used LASSO, an embedded technique, and reported that the performance of the machine learning algorithms was better with the implementation of LASSO on the examined high dimensional datasets. Townes et al. [13] implemented a simple multinomial method using generalized principal component analysis and carried out feature selection using deviance. Different combinations of methods were further compared with current methods to show their performance. Evolutionary algorithms have also been implemented in some studies for the improvement of feature selection. Jain et al. [14] implemented a correlation-based feature selection method improved with binary particle swarm optimization for the selection of genes before classifying the cancer types using Naïve Bayes algorithm. This method improved the classifier’s performance. In the same vein, Kabir et al. [15] compared two different dimension reduction techniques—PCA, and autoencoders for the selection of features in a prostate cancer classification analysis. Two machine learning methods—neural networks and SVM—were further used for classification. The study showed that the classifiers performed better on the reduced dataset.
Prasad et al. [16] used a recursive particle swarm optimization technique with the integration of filter-based methods for ranking. The authors also reported improved performance based on five datasets. Another similar approach for gene selection involves the hybridization of ant colony optimization and cellular learning automata. Based on the ROC curve evaluation of three classifiers, the proposed method selected the minimum gene needed for maximum performance of the classifiers [17]. Similarly, Alhenawi et al. [18] proposed a hybrid feature selection technique using Hill Climbing, the Novel LS algorithm, and Tabu search for microarray data. This is similar to the filter-wrapper and embedded technique utilized for gene expression data in [12]. However, Keshta et al. [19] proposed a multi-stage algorithm for the extraction and selection of features in a cancer detection study. It was reported that despite the reduction in the number of features used for classification, the performance of classifiers was either enhanced or unchanged. In addition, a nested genetic algorithm consisting of an outer genetic algorithm and an inner genetic algorithm has previously been implemented for the gene selection of a colon and lung dataset using 5-fold cross-validation [20]. A significant increase in classification accuracy was also reported. Several other feature/gene selection techniques are being improved and implemented to improve the accuracy of cancer classification. These include robust linear discriminant analysis [21], adaptive principal component analysis [22], and the use of deep variational autoencoders, especially in studies that involve the use of deep learning [23].
In this study, we considered the problem of imbalanced data, which is common in health data, before using dimension reduction techniques such as principal component analysis (PCA), truncated singular value decomposition (TSVD), and T-stochastic neighbor embedding (TSNE) to address the high dimensionality issue peculiar to gene expression data. We also utilized the ability of some machine learning algorithms to rank some genes and make selections based on a specified threshold.
2. Materials and Methods
2.1. Dataset Description
The six gene expression datasets used in this study are the brain, colon, leukemia, lymphoma, prostate, and small blue round cell tumor (SBRCT) datasets, as explained in our previous work [24] and shown in Table 1. Forty-two (42) patient samples make up the brain cancer microarray dataset. The tumors include 10 medulloblastomas, 5 atypical teratoid/rhabdoid tumors of the central nervous system (CNS), 5 rhabdoid tumors of the renal and extrarenal organs, 8 supratentorial primitive neuroectodermal tumors (PNETs), 10 non-embryonal brain tumors, and 4 normal human cerebella. 6817 genes are present in the first oligonucleotide microarrays. They underwent thresholding during pre-processing by [25]. Therefore, for the entire dataset with five distinct sample classes, there are 5597 genes. Alon et al. [26] conducted the initial analysis of the colon cancer microarray dataset. The raw data from the Affymetrix oligonucleotide arrays was processed by the dataset’s original authors. Samples of both normal and tumor tissue make up the dataset. The total number of samples is 62, and the 2000 gene numbers after pre-processing reported by earlier authors [27,28] are the total gene numbers. Acute lymphoblastic leukemia and acute myeloid leukemia are the two kinds of acute leukemia studied for gene expression, whose results were used to create the leukemia cancer dataset. Affymetrix high-density oligonucleotide arrays, which had 6817 genes but were reduced to 3051 genes and further analyzed by [29], were used to determine the levels of gene expression. 47 cases of ALL (38 B-cell ALL and 9 T-cell ALL) and 25 instances of AML make up the dataset. Dudoit et al. [30] performed more pre-processing on the dataset. The dataset can be acquired from [27,28].

Table 1.
Detailed Information about the datasets.
The dataset for the lymphoma microarray is found in [25]. It comprises 62 samples and 4026 genes. The majority of the data samples are from three distinct adult lymphoid malignancies: 42 samples represent diffuse large B-cell lymphoma (DLBCL), 9 samples come from follicular lymphoma (FL), and 11 samples come from chronic lymphocytic leukemia (CLL). The dataset is also available can be found in the literature [27,28]. 50 of the samples in the prostate cancer dataset are normal prostate specimens, while the remaining 52 are tumors. The collection contains 102 patterns of gene expression. About 12,600 genes make up this microarray dataset, which is based on an oligonucleotide microarray. The dataset still contains 6033 genes after pre-processing [31]. There are four distinct classifications in the Small Round Blue-Cell Tumor (SRBCT) microarray dataset, which initially had 6567 genes and 63 samples. Whereas, 8 samples come from NHL, 12 from NB, 20 from the RMS, and 23 from EWS. The number of genes decreased to 2308 after pre-processing. This dataset was produced using [32] and is available in [27,28].
2.2. Methods
From the information about the datasets contained in Table 1, there is a clear indication of the high dimensionality of the datasets. This high dimensionality informs the basics of the feature selection and dimension reduction methods used in the study. Before analysis, we normalized the data using the min-max normalization method. The formula for min-max normalization is given as , where is the vector in a feature column, is the minimum value in the column and is the maximum value in the column. Because the analysis deals with the selection of features, it is crucial to balance the features so that a feature does not have more contributing capacity to the analysis than another column. For further analysis, we note the imbalance of the data.
For health-related data, it is always essential to deal with the imbalance in order to present a false representation of the evaluation, especially using accuracy. Subsequently, oversampling was carried out on each of the datasets using the SVMSMOTE technique [24]. This technique uses the support vector machine to predict new and unknown samples around the borderline. In a support vector machine, the borderline or margin is crucial for the establishment of the decision boundary. This is why SVMSMOTE focuses on instances of the minority class that are found along the borderline and therefore generates more samples in such a way that new instances of the minority class will appear where there are fewer instances of the majority class. In this study, the nearest neighbor parameter was set to 3 and the random state to 42.
Two categories of feature reduction methods were considered in this study. The first one entails the use of dimension reduction methods, and the second category entails the use of classifiers for feature ranking and selection. The dimension reduction methods used are principal component analysis (PCA), truncated singular value decomposition (TSVD), and T-distributed stochastic neighbor embedding (TSNE). On the other hand, the classifiers used for ranking are random forest (RF) and logistic regression (LR). PCA tries to maintain the data variance as much as possible. Basis vectors, also known as principal components, along with the maximum variance of the data are chosen with the goal of minimizing the reconstruction error over all the data points. To do this, the eigenvectors with the largest eigenvalues are selected first. TSVD uses a matrix factorization technique that is similar to PCA. However, TSVD is performed on the data matrix, while PCA is performed on the covariance matrix. The name “truncated” comes from the number of columns being equal to the truncation. That is, matrices with a specified number of columns are produced. The primary goal of TSNE is to preserve pairwise similarities as much as possible. However, like PCA, it does not maintain inputs. This makes it useful for visualization and exploration. It treats similarities in the original space as probabilities and finds the embedding that preserves probability structure. TSNE uses Kullback-Leibler (KL) divergence as a measure of the similarity between two probability distributions.
All examined models went through a 5-fold cross-validation repeated for a total of five replicates and were evaluated based on four different metrics: accuracy, precision, recall, and F1-score. These evaluation criteria are calculated based on Equations (1)–(4) respectively.
where TP, FP, TN, and FN are true positive, false positive, true negative, and false negative, respectively.
We implemented all analyses in this study on a Windows 10 64-bit operating system on a x64-based processor computer with 64 GB of RAM. The processor specification is an Intel (R) Core (TM) i7-9700 K CPU @ 3.6 GHz. Python 3.6 and libraries, including scikit-learn, were utilized for the analyses. Figure 1 further shows the procedure for study analysis.
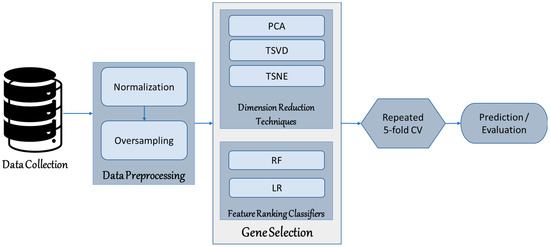
Figure 1.
Process Diagram of the Study.
3. Results and Discussions
For all analyses, we used repeated 5-fold cross-validation. The 5-fold cross-validation had the parameter shuffle set to true and was repeated five times to generate 25 results per evaluation metric. The mean of each analysis is reported as the result, while a full report of the result with the standard deviation is given in the Supplementary Material. For this study, we examined the different feature reduction techniques using six different classifiers, logistic regression (LR), random forest (RF), support vector machine (SVM), gradient boosting classifier (GBC), Gaussian Naïve Bayes (GNB), and k-nearest neighbor (KNN). These are all commonly used classifiers that behave differently, which has thus influenced their choice for the analyses. For the logistic regression, the parameter for maximum iteration was set to 500, and liblinear was selected as the solver. In a similar manner, for the random forest and gradient boosting classifiers, the number of estimators was 500. For the support vector machine, Gaussian naïve bayes, and k-nearest neighbor, the default sklearn parameters were used.
3.1. Performance of Classification Methods after Oversampling
Firstly, we carried out the analysis on all the datasets without any prior oversampling or reduction. The only preprocessing technique that was applied was normalization. We further compared the result with the result generated after the SVMSMOTE oversampling technique was used. The results from the two analyses are shown in Table 2, Table 3, Table 4 and Table 5. The better result is highlighted. From the tables, we deduce that the majority of the analysis with oversampling has better performance than the one without oversampling. A peculiar benefit of oversampling is that it allows the model to be exposed and trained with a balanced number of both the majority and minority samples. For the lymphoma dataset, there is consistency between the original dataset and the oversampled dataset using random forest and support vector machines.

Table 2.
Comparison between the accuracy of models before and after oversampling.

Table 3.
Comparison between the precision of models before and after oversampling.

Table 4.
Comparison between the recall of models before and after oversampling.

Table 5.
Comparison between F1 scores of models before and after oversampling.
3.2. Performance of Classification Methods Based on Dimension Reduction Techniques
Due to the better performance of the models using the oversampled data in Table 2, Table 3, Table 4 and Table 5, the same data was used for the rest of the analyses. In Table 6, Table 7, Table 8 and Table 9, we compared the performance of the trained models based on three-dimensional reduction methods, namely principal component analysis (PCA), truncated singular value decomposition (TSVD), and t-distributed stochastic neighbor embedding (TSNE). The parameters of PCA and TSVD were set to make the cumulative explained variance of the chosen principal components 0.99. For TSNE, the number of components was set at 3, and perplexity was set at 50. From the results shown in Table 6, Table 7, Table 8 and Table 9, the performance of PCA and TSVD is relatively similar, although in many cases, PCA had better performance. In the majority of the analyses, the performance of the PCA or TSVD dimension-reduced analyses was better than the performance of the classifiers before reduction. TSNE, on the other hand, had generally poor performance for all the datasets and classifiers examined. We suppose that this can be attributed to the fact that TSNE has random probability and attempts to retain the variance of neighboring points, that is, local variance. In many cases, TSNE is used just for data visualization, especially for 2D or 3D visualization of images while retaining the local variance.

Table 6.
Comparison between model accuracy for PCA, TSVD, and TSNE using an oversampled dataset.

Table 7.
Comparison between model precision for PCA, TSVD, and TSNE using an oversampled dataset.

Table 8.
Comparison between model recall for PCA, TSVD, and TSNE using an oversampled dataset.

Table 9.
Comparison between model F1 scores for PCA, TSVD, and TSNE using an oversampled dataset.
3.3. Performance of Classification Methods Based on Classifier-Based Gene Ranking and Selection
Furthermore, we used two classifiers (random forest and logistic regression) for feature ranking and selection. The oversampled datasets were also used for analyses. For the feature selection based on random forest, we used 500 estimators as the parameters and only selected features that were above the mean threshold. On the other hand, for feature selection based on logistic regression, only features above the median threshold for each were selected. We have employed different thresholds for the two techniques to discover if there would be a significant difference in the performance of the classifiers based on the threshold of the feature selection. For the Lymphoma dataset, both random forest and logistic regression had the same performance across evaluations. This was similarly noticed with the small blue round cell tumor (SBRCT) dataset, except with the use of gradient boosting classifiers, and k-nearest neighbor. Overall, Table 10, Table 11, Table 12 and Table 13 show that both classifiers had good performance in the ranking and selection of features, although they utilize different strategies for their threshold.

Table 10.
Comparison between model accuracy for RF and LR using an oversampled dataset.

Table 11.
Comparison between model precision for RF and LR using an oversampled dataset.

Table 12.
Comparison between model recall for RF and LR using an oversampled dataset.

Table 13.
Comparison between model F1 scores for RF and LR using an oversampled dataset.
4. Conclusions
Data generated in the medical and bioinformatics fields are known to have a large number of features. Analyzing these features manually is exhausting, and this is where the utilization of machine learning tools comes in handy. Another major issue with health data is the high rate of class imbalance. This sometimes affects the integrity and robustness of models built using such data. Models might be unable to accurately predict the class of unseen instances in the minority class. In this study, we have considered SVMSMOTE for oversampling the data and thereby increasing the number of instances of each data point. We discover that, generally, the performance of the examined models was better after oversampling. The application of the dimension reduction techniques and feature ranking and selection further improved the performance in many instances. Principal component analysis and truncated singular value decomposition performed better than t-distributed stochastic neighbor embedding. TSNE in fact had a generally bad performance, as shown in Table 6, Table 7, Table 8 and Table 9. It is advised to use TSNE primarily for dimension reduction for data visualization. In the same vein, both random forest and logistic regression classifiers were effective in the selection of features despite utilizing different threshold criteria. Although a low number of analyses with the original dataset had good performance, the majority of the analyses with SVMSMOTE and feature reduction had better performance, saved time, and enhanced the interpretability of models. Future works will investigate the analysis of gene expression data and cancer classification using different oversampling and dimension reduction methods on different microarray datasets.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pr11071940/s1, Table S1: Comparison between performance with original dataset and performance after oversampling. Table S2: Comparison between performance of PCA, TSVD, and TSNE using oversampled dataset. Table S3: Comparison between performance of RF and LR using oversampled dataset.
Author Contributions
Conceptualization, O.O.P. and F.S.; methodology, O.O.P., F.S. and N.S.; software, O.O.P.; validation, M.T., Z.L. and I.O.M.; formal analysis, O.O.P., F.S., M.T. and Z.L.; investigation, M.T., Z.L. and I.O.M.; resources, N.S. and I.O.M.; data curation, O.O.P.; writing—original draft preparation, O.O.P., M.T., Z.L. and I.O.M.; writing—review and editing, O.O.P., F.S. and N.S.; visualization, O.O.P.; supervision, F.S. and N.S.; project administration, F.S.; funding acquisition, N.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research Management Center at Universiti Teknologi Malaysia (Vot No: Q.J130000.21A6.00P48) and the Ministry of Higher Education, Malaysia (JPT(BKPI)1000/016/018/25(58)) through the Malaysia Big Data Research Excellence Consortium (BiDaREC) (Vot No: R.J130000.7851.4L933), (Vot No: R.J130000.7851.5F568), (Vot No: R.J130000.7851.4L942), (Vot No: R.J130000.7851.4L938), and (Vot No: R.J130000.7851.4L936). We are also grateful to (Project No: KHAS-KKP/2021/FTMK/C00003) and (Project No: KKP002-2021) for their financial support of this research.
Data Availability Statement
The datasets are available and can be downloaded form Microarray Datasets: https://csse.szu.edu.cn/staff/zhuzx/Datasets.html.
Acknowledgments
The authors would like to thank the Research Management Center at Universiti Teknologi Malaysia for funding this research using (Vot No: Q.J130000.21A6.00P48) and the Ministry of Higher Education, Malaysia (JPT(BKPI)1000/016/018/25(58)) through the Malaysia Big Data Research Excellence Consortium (BiDaREC) (Vot No: R.J130000.7851.4L933), (Vot No: R.J130000.7851.5F568), (Vot No: R.J130000.7851.4L942), (Vot No: R.J130000.7851.4L938), and (Vot No: R.J130000.7851.4L936). We are also grateful to (Project No: KHAS-KKP/2021/FTMK/C00003) and (Project No: KKP002-2021) for their financial support of this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Thakur, T.; Batra, I.; Luthra, M.; Vimal, S.; Dhiman, G.; Malik, A.; Shabaz, M. Gene expression-assisted cancer prediction techniques. J. Healthc. Eng. 2021, 2021, 4242646. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, P.; Kolhe, R.; Gahlay, G.K. The clinical relevance of gene expression based prognostic signatures in colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188513. [Google Scholar] [CrossRef] [PubMed]
- Schaafsma, E.; Fugle, C.M.; Wang, X.; Cheng, C. Pan-cancer association of HLA gene expression with cancer prognosis and immunotherapy efficacy. Br. J. Cancer 2021, 125, 422–432. [Google Scholar] [CrossRef]
- Tourang, M.; Fang, L.; Zhong, Y.; Suthar, R.C. Association between Human Endogenous Retrovirus K gene expression and breast cancer. Cell. Mol. Biomed. Rep. 2021, 1, 7–13. [Google Scholar] [CrossRef]
- Satyananda, V.; Oshi, M.; Endo, I.; Takabe, K. High BRCA2 gene expression is associated with aggressive and highly proliferative breast cancer. Ann. Surg. Oncol. 2021, 28, 7356–7365. [Google Scholar] [CrossRef]
- Qian, Y.; Daza, J.; Itzel, T.; Betge, J.; Zhan, T.; Marmé, F.; Teufel, A. Prognostic cancer gene expression signatures: Current status and challenges. Cells 2021, 10, 648. [Google Scholar] [CrossRef] [PubMed]
- Munkácsy, G.; Santarpia, L.; Győrffy, B. Gene Expression Profiling in Early Breast Cancer—Patient Stratification Based on Molecular and Tumor Microenvironment Features. Biomedicines 2022, 10, 248. [Google Scholar] [CrossRef]
- Oliveira, L.J.C.; Amorim, L.C.; Megid, T.B.C.; De Resende, C.A.A.; Mano, M.S. Gene expression signatures in early Breast Cancer: Better together with clinicopathological features. Crit. Rev. Oncol. Hematol. 2022, 175, 103708. [Google Scholar] [CrossRef]
- Schettini, F.; Chic, N.; Brasó-Maristany, F.; Paré, L.; Pascual, T.; Conte, B.; Martínez-Sáez, O.; Adamo, B.; Vidal, M.; Barnadas, E.; et al. Clinical, pathological, and PAM50 gene expression features of HER2-low breast cancer. NPJ Breast Cancer 2021, 7, 1. [Google Scholar] [CrossRef]
- Zhong, Y.; Chalise, P.; He, J. Nested cross-validation with ensemble feature selection and classification model for high-dimensional biological data. Commun. Stat. Simul. Comput. 2023, 52, 110–125. [Google Scholar] [CrossRef]
- Petinrin, O.O.; Saeed, F.; Li, X.; Ghabban, F.; Wong, K.C. Reactions’ descriptors selection and yield estimation using metaheuristic algorithms and voting ensemble. Comput. Mater. Contin. 2022, 70, 4745–4762. [Google Scholar]
- Hameed, S.S.; Petinrin, O.O.; Hashi, A.O.; Saeed, F. Filter-wrapper combination and embedded feature selection for gene expression data. Int. J. Adv. Soft Compu. Appl. 2018, 10, 90–105. [Google Scholar]
- Townes, F.W.; Hicks, S.C.; Aryee, M.J.; Irizarry, R.A. Feature selection and dimension reduction for single-cell RNA-Seq based on a multinomial model. Genome Biol. 2019, 20, 295. [Google Scholar] [CrossRef] [PubMed]
- Jain, I.; Jain, V.K.; Jain, R. Correlation feature selection based improved-binary particle swarm optimization for gene selection and cancer classification. Appl. Soft Comput. 2018, 62, 203–215. [Google Scholar] [CrossRef]
- Kabir, M.F.; Chen, T.; Ludwig, S.A. A performance analysis of dimensionality reduction algorithms in machine learning models for cancer prediction. Healthc. Anal. 2023, 3, 100125. [Google Scholar] [CrossRef]
- Prasad, Y.; Biswas, K.; Hanmandlu, M. A recursive PSO scheme for gene selection in microarray data. Appl. Soft Comput. 2018, 71, 213–225. [Google Scholar] [CrossRef]
- Sharbaf, F.V.; Mosafer, S.; Moattar, M.H. A hybrid gene selection approach for microarray data classification using cellular learning automata and ant colony optimization. Genomics 2016, 107, 231–238. [Google Scholar] [CrossRef]
- Alhenawi, E.A.; Al-Sayyed, R.; Hudaib, A.; Mirjalili, S. Improved intelligent water drop-based hybrid feature selection method for microarray data processing. Comput. Biol. Chem. 2023, 103, 107809. [Google Scholar] [CrossRef]
- Keshta, I.; Deshpande, P.S.; Shabaz, M.; Soni, M.; Bhadla, M.K.; Muhammed, Y. Multi-stage biomedical feature selection extraction algorithm for cancer detection. SN Appl. Sci. 2023, 5, 131. [Google Scholar] [CrossRef]
- Sayed, S.; Nassef, M.; Badr, A.; Farag, I. A nested genetic algorithm for feature selection in high-dimensional cancer microarray datasets. Expert Syst. Appl. 2019, 121, 233–243. [Google Scholar] [CrossRef]
- Li, X.; Wang, H. On Mean-Optimal Robust Linear Discriminant Analysis. In Proceedings of the 2022 IEEE International Conference on Data Mining (ICDM), Orlando, FL, USA, 30 November–3 December 2022; pp. 1047–1052. [Google Scholar]
- Li, X.; Wang, H. Adaptive Principal Component Analysis. In Proceedings of the 2022 SIAM International Conference on Data Mining (SDM), Alexandria, VA, USA, 28–30 April 2022; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2022; pp. 486–494. [Google Scholar]
- Jiang, J.; Xu, J.; Liu, Y.; Song, B.; Guo, X.; Zeng, X.; Zou, Q. Dimensionality reduction and visualization of single-cell RNA-seq data with an improved deep variational autoencoder. Briefings Bioinform. 2023, 24, bbad152. [Google Scholar] [CrossRef] [PubMed]
- Hameed, S.S.; Muhammad, F.F.; Hassan, R.; Saeed, F. Gene Selection and Classification in Microarray Datasets using a Hybrid Approach of PCC-BPSO/GA with Multi Classifiers. J. Comput. Sci. 2018, 14, 868–880. [Google Scholar] [CrossRef]
- Dettling, M.; Bühlmann, P. Supervised clustering of genes. Genome Biol. 2002, 3, research0069.1. [Google Scholar] [CrossRef]
- Alon, U.; Barkai, N.; Notterman, D.A.; Gish, K.; Ybarra, S.; Mack, D.; Levine, A.J. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 1999, 96, 6745–6750. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Ong, Y.S.; Dash, M. Markov Blanket-Embedded Genetic Algorithm for Gene Selection. Pattern Recognit. 2007, 49, 3236–3248. [Google Scholar] [CrossRef]
- Microarray Datasets. Available online: https://csse.szu.edu.cn/staff/zhuzx/Datasets.html (accessed on 8 June 2023).
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Dudoit, S.; Fridlyand, J.; Speed, T.P. Comparison of discrimination methods for the classification of tumors using gene expression data. J. Am. Stat. Assoc. 2002, 97, 77–87. [Google Scholar] [CrossRef]
- Díaz-Uriarte, R.; De Andres, S.A. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006, 7, 3. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Cooper, E.W.; Kamei, K. Borderline over-sampling for imbalanced data classification. In Proceedings of the Fifth International Workshop on Computational Intelligence & Applications, IEEE SMC Hiroshima Chapter, Hiroshima, Japan, 10–12 November 2009. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).