Dynamical Simulation, Sensitivity, and Productivity Analysis of a Light-Photoacclimation Model for Microalgae-Based Carbohydrate Production in Continuous Photobioreactors
Abstract
1. Introduction
1.1. Dynamic Photoacclimation Model
1.1.1. Specific Growth Rate
1.1.2. Nutrient Uptake
1.1.3. Average Photon Flux Density throughout the Culture
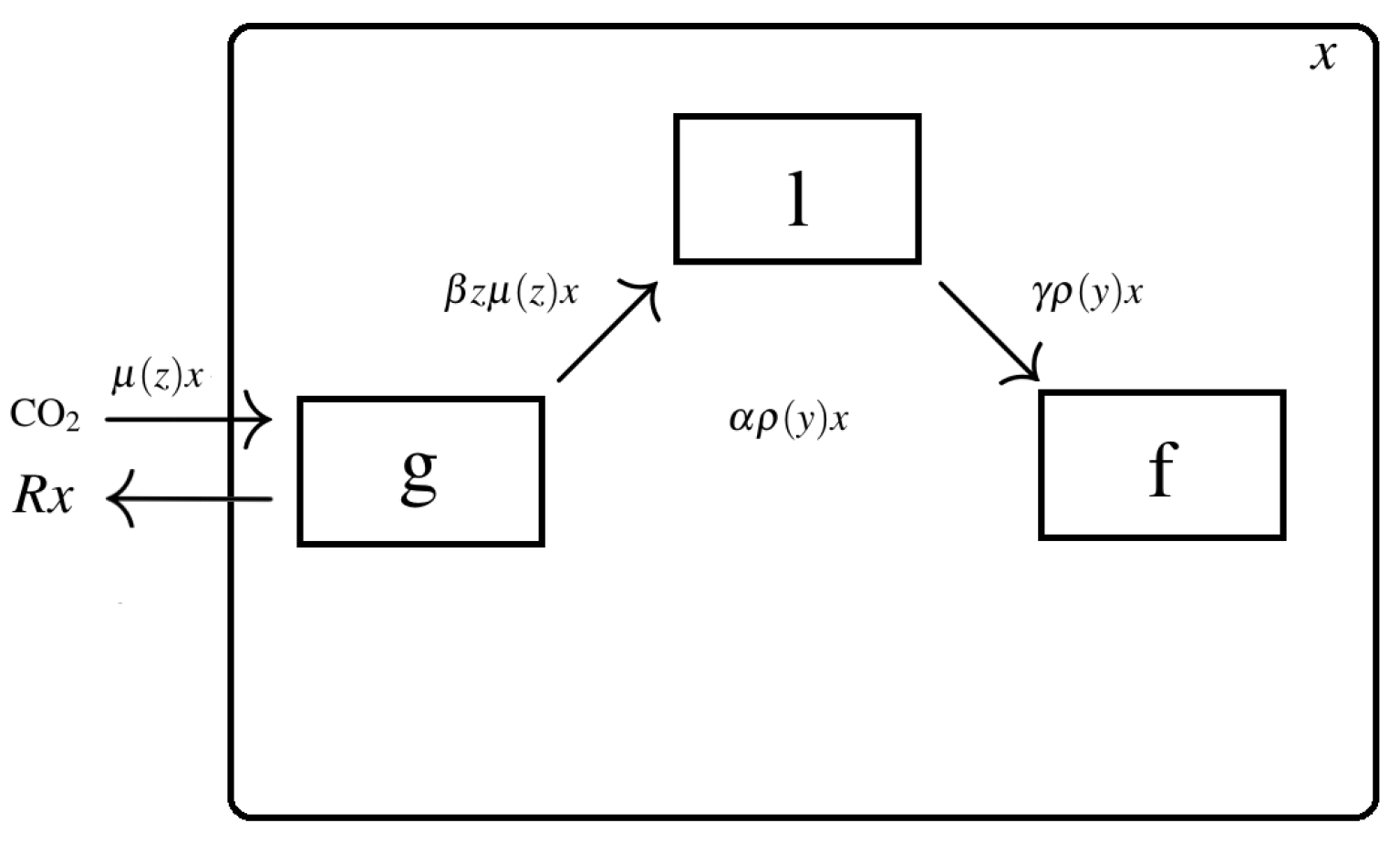
1.2. Carbon Flux and Carbohydrates Dynamics
Carbohydrate Dynamics Deduction
2. Materials and Methods
2.1. The Fermentation Process Simulation
2.1.1. Methodology for Dynamic Simulation
2.1.2. Methodology for Equilibrium and Productivity Analysis
2.1.3. Methodology for Sensitivity Analysis
3. Results and Discussion
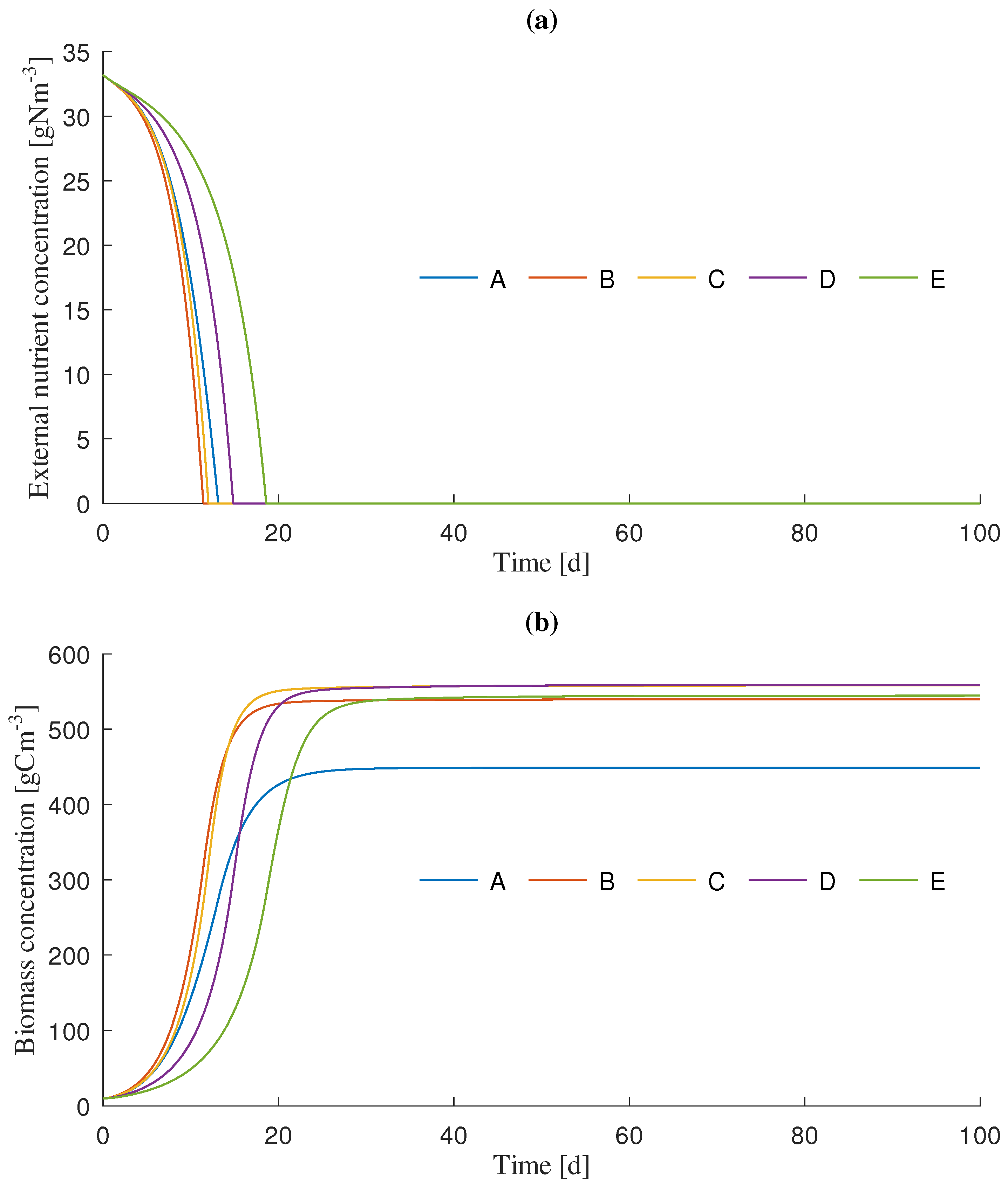
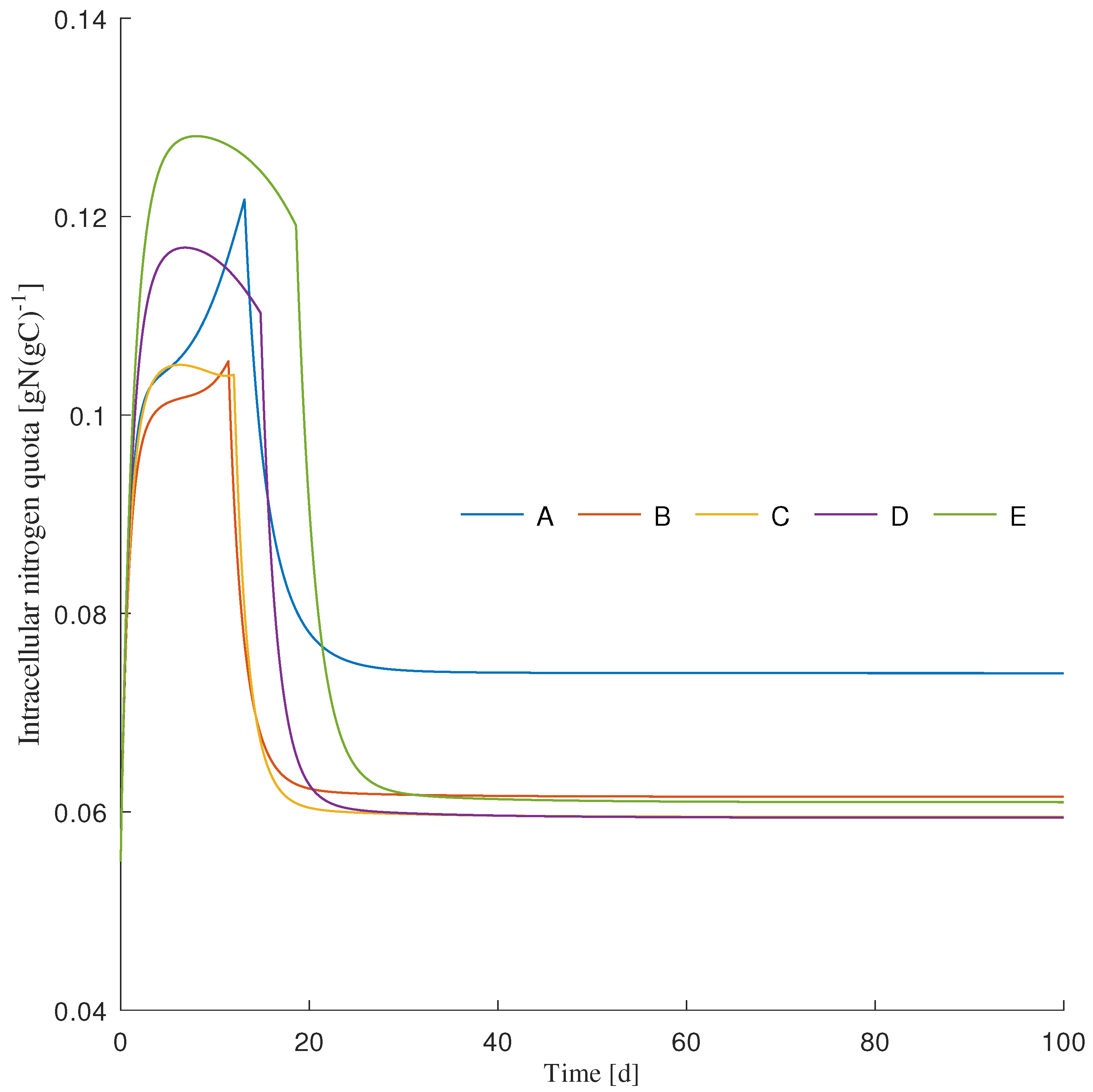
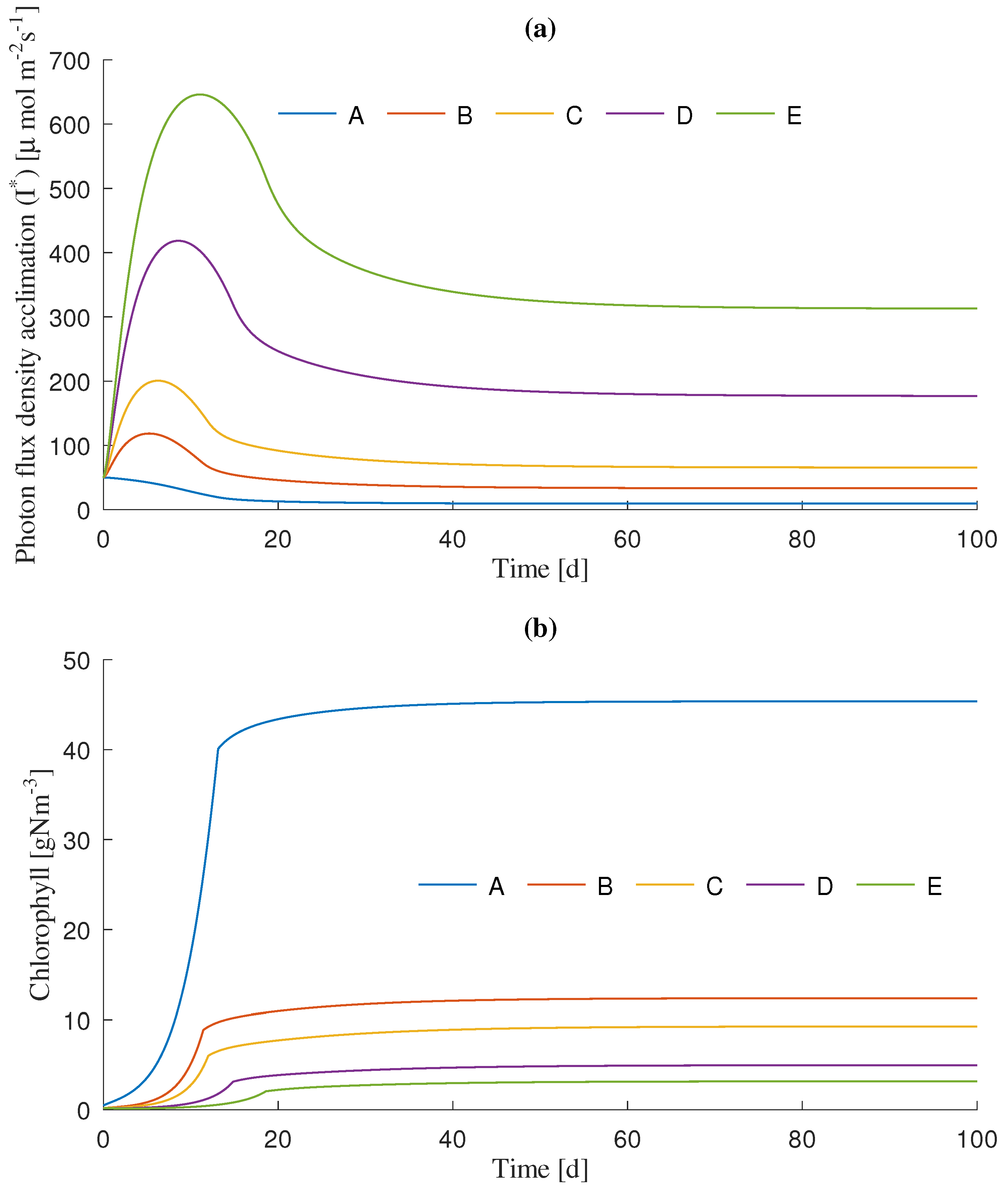
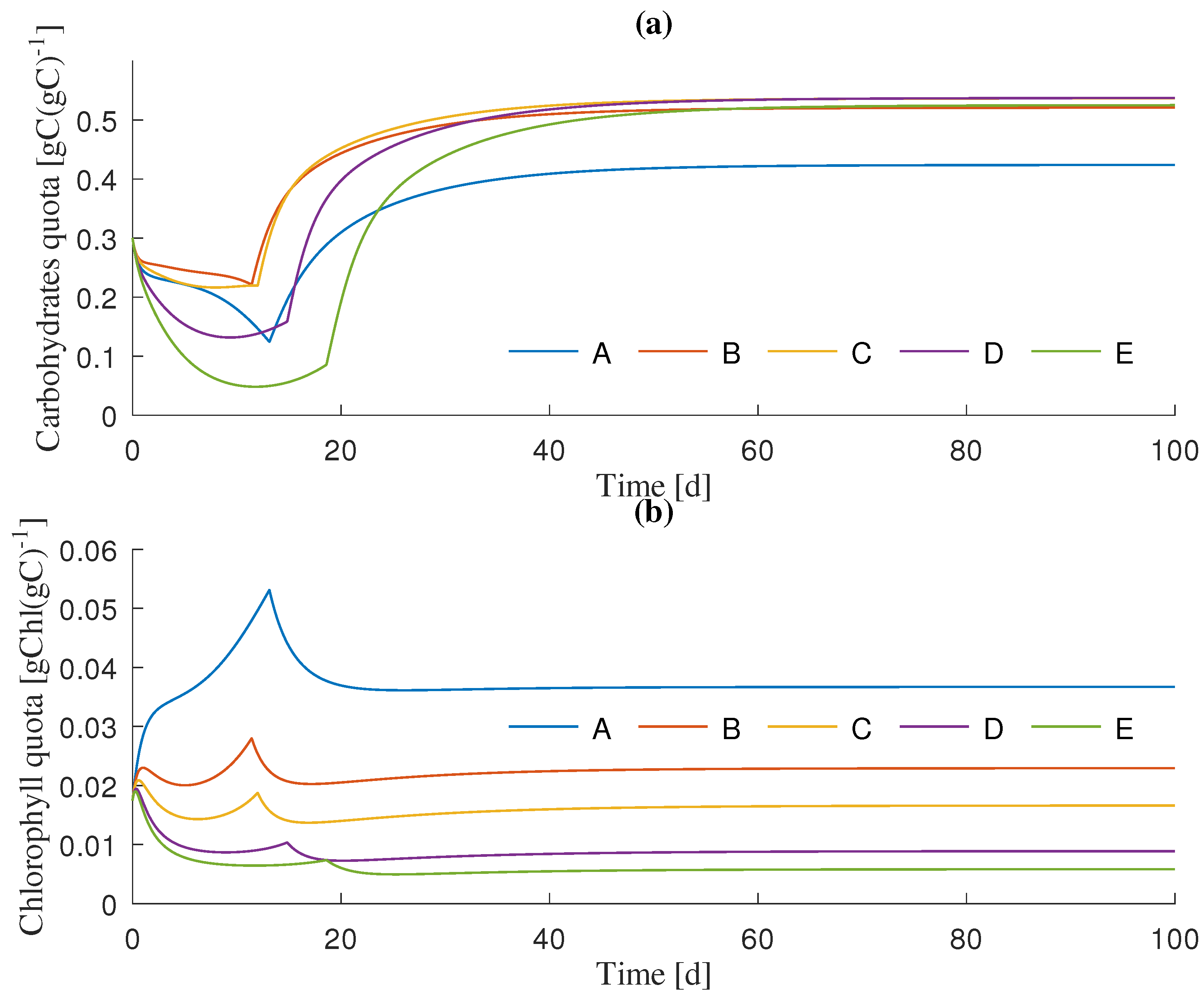
3.1. Dynamics Simulation
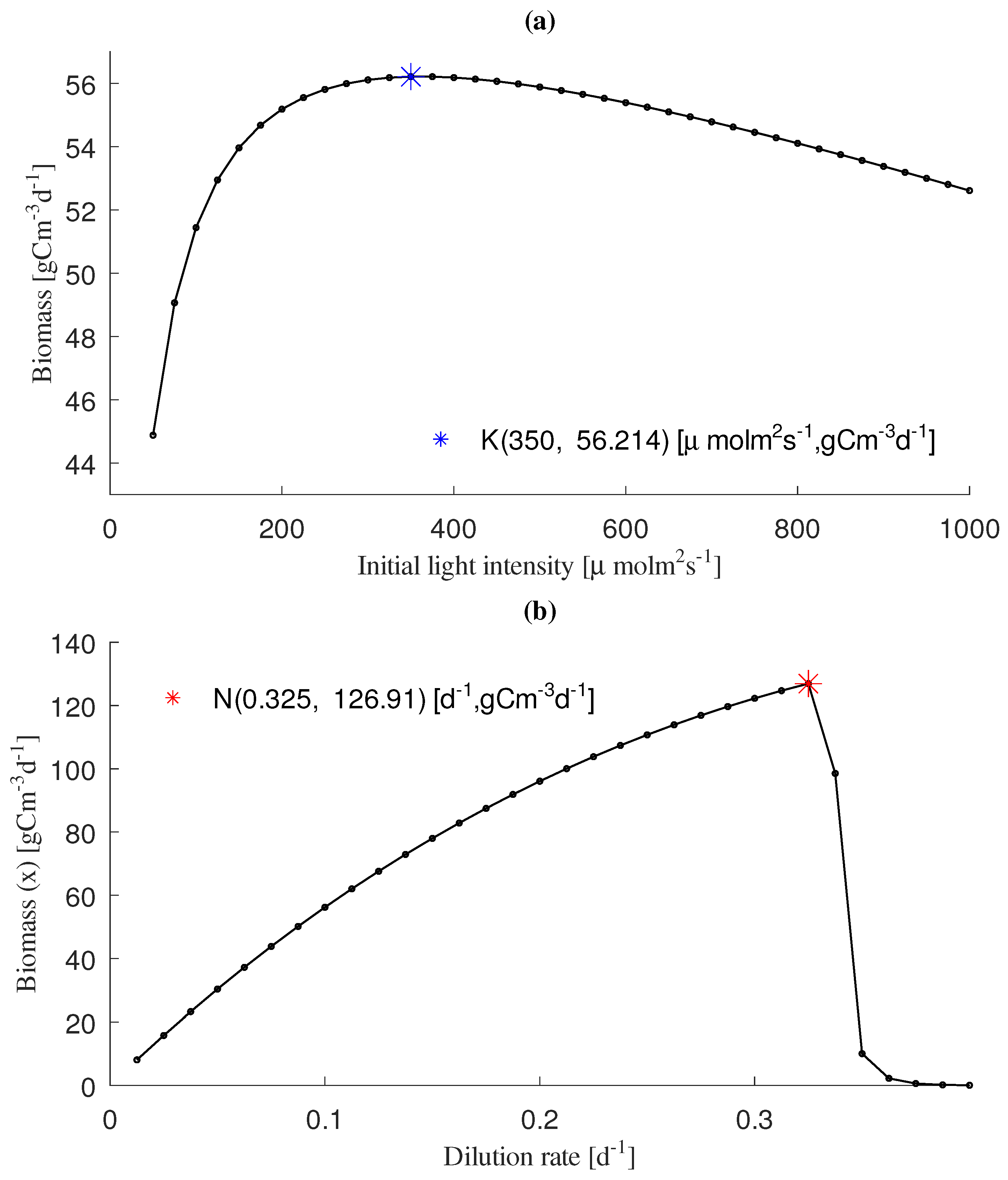
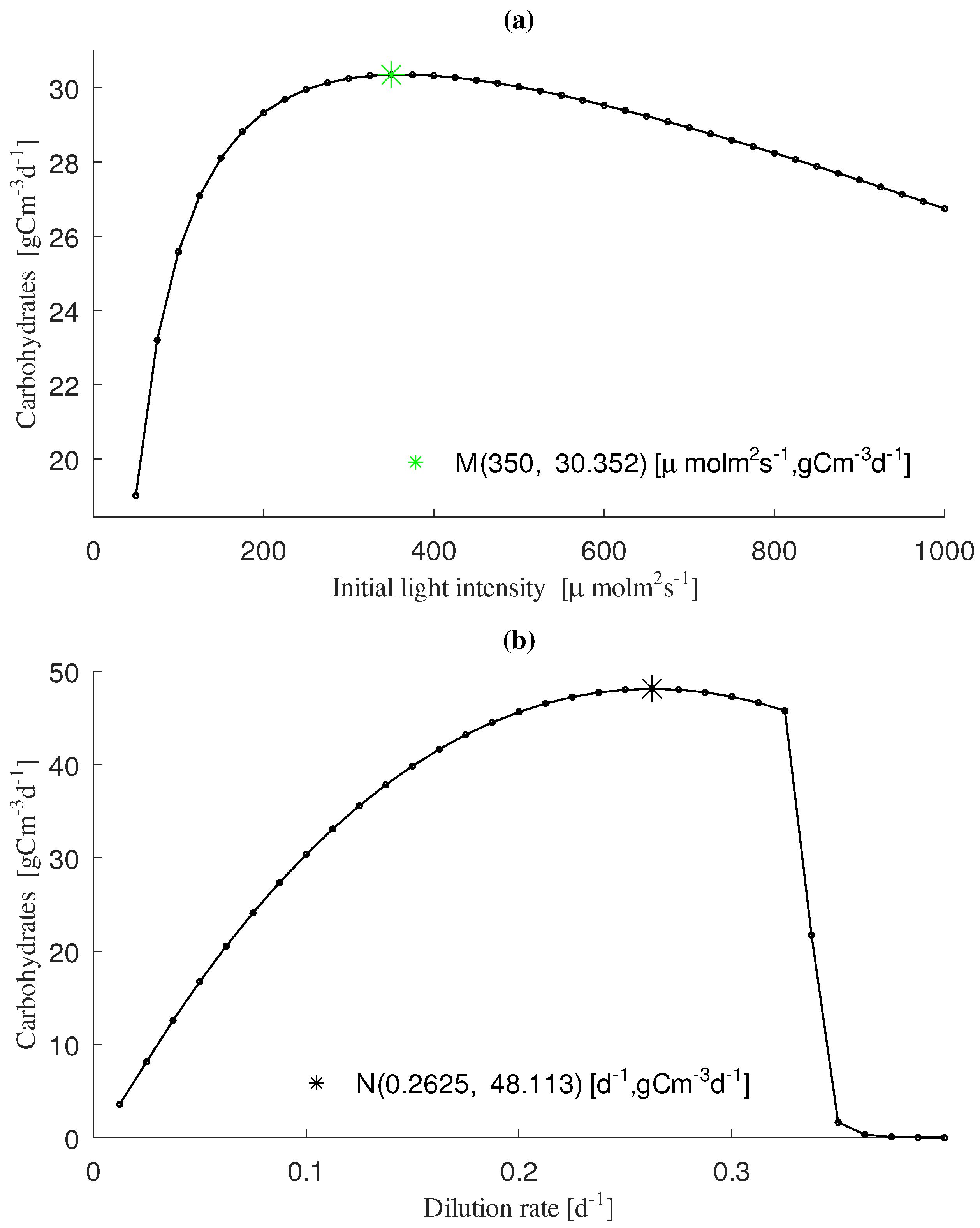
3.2. Analysis of Productivity
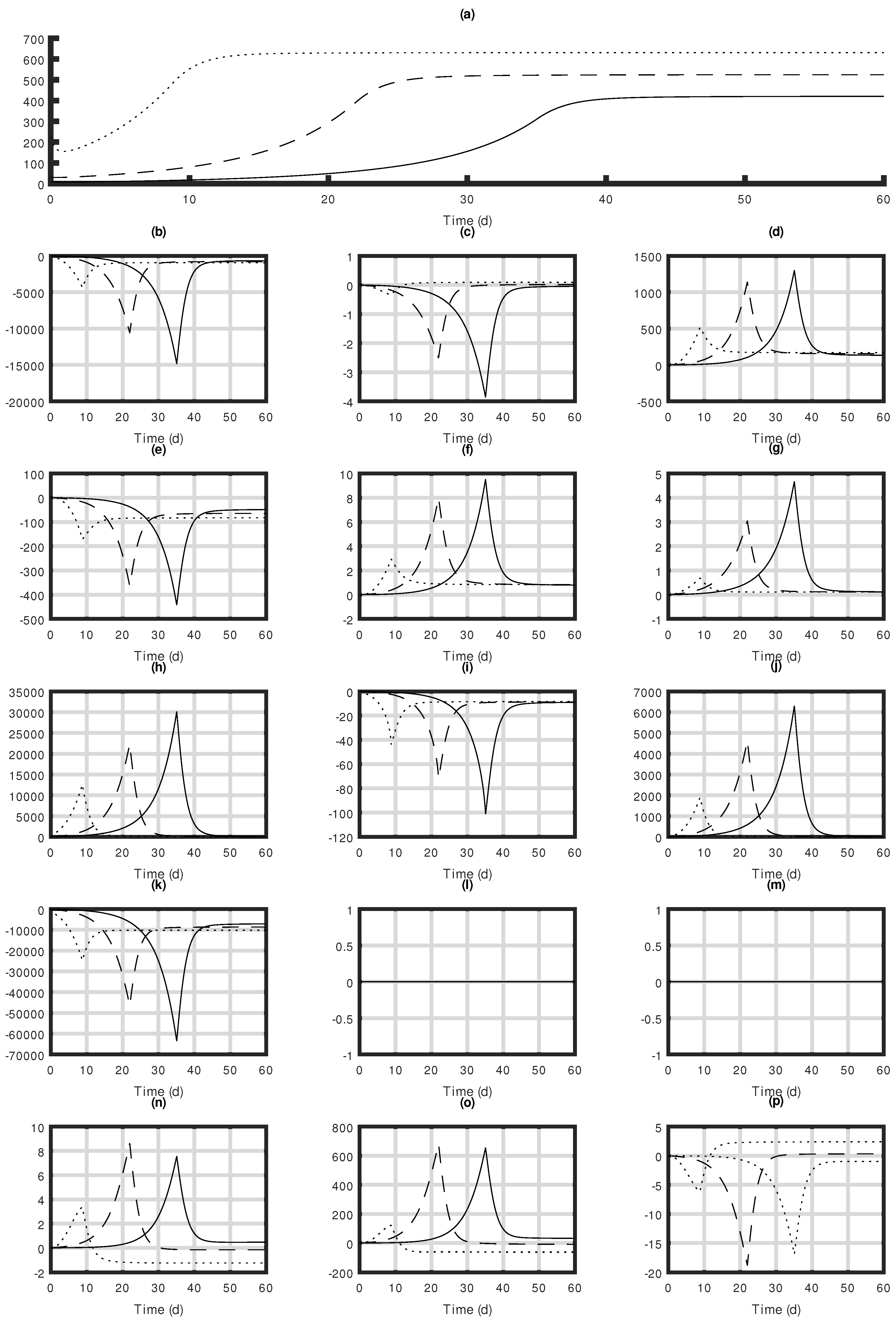
3.3. Sensitivity Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| NADPH | Nicotinamide Adenine Dinucleotide Phosphate |
| PBR | photobioreactor |
| FFA | free fatty acid |
| ODE | ordinary differential equation |
| LSODE | Livermore solver for ordinary differential equations |
| ATP | adenosine triphosphate |
References
- Chia, S.R.; Ong, H.C.; Chew, K.W.; Show, P.L.; Phang, S.M.; Ling, T.C.; Nagarajan, D.; Lee, D.J.; Chang, J.S. Sustainable approaches for algae utilisation in bioenergy production. Renew. Energy 2018, 129, 838–852. [Google Scholar] [CrossRef]
- Vadiveloo, A.; Moheimani, N. Effect of continuous and daytime mixing on Nannochloropsis growth in raceway ponds. Algal Res. 2018, 33, 190–196. [Google Scholar] [CrossRef]
- Li, Y.; Horsman, M.; Wu, N.; Lan, C.Q.; Dubois-Calero, N. Biofuels from Microalgae. Biotechnol. Prog. 2008, 24, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Mohan, D.; Pittman, C.U.; Steele, P.H. Pyrolysis of wood/biomass for bio-oil: A critical review. Energy Fuels 2006, 24, 848–889. [Google Scholar] [CrossRef]
- Babu, S.S.; Gondi, R.; Vincent, G.S.; JohnSamuel, G.C.; Jeyakumar, R.B. Microalgae Biomass and Lipids as Feedstock for Biofuels: Sustainable Biotechnology Strategies. Sustainability 2022, 14, 15070. [Google Scholar] [CrossRef]
- Jagadevan, S.; Banerjee, A.; Banerjee, C.; Guria, C.; Tiwari, R.; Baweja, M.; Shukla, P. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnol. Biofuels 2018, 11, 185. [Google Scholar] [CrossRef]
- Culaba, A.B.; Ubando, A.T.; Ching, P.M.L.; Chen, W.-H.; Chang, J.-S. Biofuel from Microalgae: Sustainable Pathways. Sustainability 2020, 12, 8009. [Google Scholar] [CrossRef]
- Kings, A.J.; Raj, R.E.; Miriam, L.R.M.; Visvanathan, M.A. Cultivation, extraction and optimization of biodiesel production from potential microalgae Euglena sanguinea using eco-friendly natural catalyst. Energy Convers. Manag. 2017, 141, 224–235. [Google Scholar] [CrossRef]
- Arora, P.; Chance, R.R.; Hendrix, H.; Realff, M.J.; Thomas, V.M.; Yuan, Y. Greenhouse Gas Impact of Algal Bio-Crude Production for a Range of CO2 Supply Scenarios. Appl. Sci. 2021, 11, 11931. [Google Scholar] [CrossRef]
- Liu, C.M.; Sandhu, N.K.; McCoy, S.T.; Bergerson, J.A. A life cycle assessment of greenhouse gas emissions from direct air capture and Fischer-Tropsch fuel production. Sustain. Energy Fuels 2020, 4, 3129–3142. [Google Scholar] [CrossRef]
- Guest, G.; Cherubini, F.; Strømman, A.H. Global Warming Potential of Carbon Dioxide Emissions from Biomass Stored in the Anthroposphere and Used for Bioenergy at End of Life. J. Ind. Ecol. 2013, 17, 20–30. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef]
- Park, S.; Ahn, Y.; Pandi, K.; Ji, M.-K.; Yun, H.-S.; Choi, J.-Y. Microalgae Cultivation in Pilot Scale for Biomass Production Using Exhaust Gas from Thermal Power Plants. Energies 2019, 12, 3497. [Google Scholar] [CrossRef]
- Rehman, M.; Kesharvani, S.; Dwivedi, G.; Gidwani, K. Impact of cultivation conditions on microalgae biomass productivity and lipid content. Mater. Today Proc. 2022, 56, 282–290. [Google Scholar] [CrossRef]
- Bekirogullari, M.; Figueroa-Torres, G.M.; Pittman, J.K.; Theodoropoulos, C. Models of microalgal cultivation for added-value products—A review. Biotechnol. Adv. 2020, 44, 107609. [Google Scholar] [CrossRef]
- Pignolet, O.; Jubeau, S.; Vaca-Garcia, C.; Michaud, P. Highly valuable microalgae: Biochemical and topological aspects. J. Ind. Microbiol. Biotechnol. 2016, 28, 1453–1457. [Google Scholar] [CrossRef]
- Mooij, P.R.; de Jongh, L.D.; van Loosdrecht, M.C.; Kleerebezem, R. Influence of silicate on enrichment of highly productive microalgae from a mixed culture. J. Appl. Phycol. 2013, 40, 781–796. [Google Scholar] [CrossRef] [PubMed]
- Markou, G.; Depraetere, O.; Vandamme, D.; Muylaert, K. Cultivation of Chlorella vulgaris and Arthrospira platensis with recovered phosphorus from wastewater by means of zeolite sorption. Int. J. Mol. Sci. 2015, 16, 4250–4264. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, J.; Ma, X.; Guo, B.; Liu, B.; Wu, T.; Jiang, Y.; Chen, F. Genetic engineering of the Calvin cycle toward enhanced photosynthetic CO2 fixation in microalgae. Biotechnol. Biofuels 2017, 10, 229. [Google Scholar] [CrossRef] [PubMed]
- Silvello, M.A.D.C.; Gonçalves, I.S.; Azambuja, S.P.H.; Costa, S.S.; Silva, P.G.P.; Santos, L.O.; Goldbeck, R. Microalgae-based carbohydrates: A green innovative source of bioenergy. Bioresour. Technol. 2022, 344, 126304. [Google Scholar] [CrossRef]
- Lam, G.P.; Giraldo, J.B.; Vermue, M.H.; Olivieri, G.; Eppink, M.H.; Wijffels, R.H. Understanding the salinity effect on cationic polymers in inducing flocculation of the microalga Neochloris oleoabundans. J. Biotechnol. 2016, 225, 10–17. [Google Scholar] [CrossRef]
- Lakatos, G.E.; Ranglová, K.; Manoel, J.C.; Grivalský, T.; Kopecký, J.; Masojídek, J. Bioethanol production from microalgae polysaccharides. Folia Microbiol. 2019, 64, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Tan, L.; Jin, X.; Wu, H.; Houbo, W.; Tao, L.; Wenzhou, X. Evaluating the potential of carbohydrate-rich microalga Rhodosorus sp. SCSIO-45730 as a feedstock for biofuel and β-glucans using strategies of phosphate optimization and low-cost harvest. J. Appl. Phycol. 2020, 32, 3051–3061. [Google Scholar] [CrossRef]
- Gupta, S.K.; Kumar, N.M.; Guldhe, A.; Ansari, F.A.; Rawat, I.; Nasr, M.; Bux, F. Wastewater to biofuels: Comprehensive evaluation of various flocculants on biochemical composition and yield of microalgae. Ecol. Eng. 2018, 117, 62–68. [Google Scholar] [CrossRef]
- Andreeva, A.; Budenkova, E.; Babich, O.; Sukhikh, S.; Dolganyuk, V.; Michaud, P.; Ivanova, S. Influence of Carbohydrate Additives on the Growth Rate of Microalgae Biomass with an Increased Carbohydrate Content. Mar. Drugs 2021, 19, 381. [Google Scholar] [CrossRef] [PubMed]
- Yen, H.W.; Hu, I.C.; Chen, C.Y.; Ho, S.H.; Lee, D.J.; Chang, J.S. Microalgae-based biorefinery—From biofuels to natural products. Bioresour. Technol 2012, 135, 166–174. [Google Scholar] [CrossRef]
- Huang, Q.; Jiang, F.; Wang, L.; Yang, C. Design of Photobioreactors for Mass Cultivation of Photosynthetic Organisms. Engineering 2017, 3, 318–329. [Google Scholar] [CrossRef]
- Gojkovic, Ž.; Garbayo-Nores, I.; Gómez-Jacinto, V.; García-Barrera, T.; Gómez-Ariza, J.L.; Márová, I.; Vilchez-Lobato, C. Continuous production of selenomethionine-enriched Chlorella sorokiniana biomass in a photobioreactor. Process Biochem. 2013, 48, 1235–1241. [Google Scholar] [CrossRef]
- Sero, E.T.; Siziba, N.; Bunhu, T.; Shoko, R.; Jonathan, E. Biophotonics for improving algal photobioreactor performance: A review. Int. J. Energy Res. 2020, 44, 5071–5092. [Google Scholar] [CrossRef]
- Albarello, A.; Simionato, D.; Morosinotto, T.; Bezzo, F. Model-Based Optimization of Microalgae Growth in a Batch Plant. Ind. Eng. Chem. Res. 2019, 58, 5121–5130. [Google Scholar] [CrossRef]
- Guzmán, J.L.; Acién, F.G.; Berenguel, M. Modelling and control of the microalgae production in industrial photobioreactors. Rev. Iberoam. Autom. Inf. 2021, 18, 1–18. [Google Scholar]
- Rizwan, M.; Mujtaba, G.; Memon, S.A.; Lee, K.; Rashid, N. Exploring the potential of microalgae for new biotechnology applications and beyond: A review. Renew. Sustain. Energy Rev. 2018, 92, 394–404. [Google Scholar] [CrossRef]
- Guterman, H.; Vonshak, A.; Ben-Yaakov, S. A macromodel for outdoor algal mass production. Biotechnol. Bioeng. 1990, 35, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.H.; Wang, F.S.; Chang, M.S. Dynamic sensitivity analysis of biological systems. BMC Bioinform. 2008, 9, S17. [Google Scholar] [CrossRef] [PubMed]
- Formighieri, C.; Franck, F.; Bassi, R. Regulation of the pigment optical density of an algal cell: Filling the gap between photosynthetic productivity in the laboratory and in mass culture. J. Biotechnol. 2012, 162, 115–123. [Google Scholar] [CrossRef]
- Briones-Baez, M.F.; Aguilera-Vazquez, L.; Rangel-Valdez, N.; Martinez-Salazar, A.L.; Zuñiga, C. Multi-Objective Optimization of Microalgae Metabolism: An Evolutive Algorithm Based on FBA. Metabolites 2022, 12, 603. [Google Scholar] [CrossRef]
- Lee, E.; Jalalizadeh, M.; Zhang, Q. Growth kinetic models for microalgae cultivation: A review. Algal Res. 2015, 12, 497–512. [Google Scholar] [CrossRef]
- Solimeno, A.; Samsó, R.; García, J. Parameter sensitivity analysis of a mechanistic model to simulate microalgae growth. Algal Res. 2016, 15, 217–223. [Google Scholar] [CrossRef]
- Grognard, F.; Akhmetzhanov, A.R.; Bernard, O. Optimal strategies for biomass productivity maximization in a photobioreactor using natural light. Automatica 2014, 50, 359–368. [Google Scholar] [CrossRef]
- San Pedro, A.; González-López, C.V.; Acién, F.G.; Molina-Grima, E. Marine microalgae selection and culture conditions optimization for biodiesel production. Bioresour. Technolo. 2013, 134, 353–361. [Google Scholar] [CrossRef]
- Guzmán-Palomino, A.; Aguilera-Vázquez, L.; Hernández-Escoto, H.; García-Vite, P.M. Sensitivity, Equilibria, and Lyapunov Stability Analysis in Droop’s Nonlinear Differential Equation System for Batch Operation Mode of Microalgae Culture Systems. Mathematics 2021, 9, 2192. [Google Scholar] [CrossRef]
- Alligood, K.T.; Sauer, T.; Yorke, J.A. Chaos: An Introduction to Dynamical Systems; Springer: Berlin/Heidelberg, Germany, 1996. [Google Scholar]
- Anning, T.; MacIntyre, H.L.; Pratt, S.M.; Sammes, P.J.; Gibb, S.; Geider, R.J. Photoacclimation in the marine diatom Skeletonema costatum. Limnol. Oceanogr. 2000, 45, 1807–1817. [Google Scholar] [CrossRef]
- Lehman, J.T.; Botkin, D.B.; Likens, G. The assumptions and rationales of a computer model of phytoplankton population dynamics. Limnol. Oceanogr. 1975, 20, 343–364. [Google Scholar] [CrossRef]
- Geider, R.; MacIntyre, H.; Kana, T. A dynamic regulatory model of phytoplanktonic acclimation to light, nutrients, and temperature. Limnol. Oceanogr. 1998, 43, 679–694. [Google Scholar] [CrossRef]
- Pahlow, M. Linking chlorophyll-nutrient dynamics to the Redfield N:C ratio with a model of optimal phytoplankton growth. Mar. Ecol. Prog. Ser. 2005, 287, 33–43. [Google Scholar] [CrossRef]
- Faugeras, B.; Bernard, O.; Sciandra, A.; Levy, M. A mechanistic modelling and data assimilation approach to estimate the carbon/chlorophyll and carbon/nitrogen ratios in a coupled hydrodynamical-biological model. Nonlinear Process. Geophys. 2004, 11, 515–533. [Google Scholar] [CrossRef]
- Flynn, K. A mechanistic model for describing dynamic multi-nutrient, light, temperature interactions in phytoplankton. J. Plankton Res. 1991, 23, 977–997. [Google Scholar] [CrossRef]
- Zonneveld, C. A cell-based model for the chlorophyll a to carbon ratio in phytoplankton. Ecol. Model. 1998, 113, 55–70. [Google Scholar] [CrossRef]
- MacIntyre, H.; Kana, T.; Anning, T.; Geider, R. Photoacclimation of photosynthesis irradiance response curves and photosynthetic pigments in microalgae and cyanobacteria. J. Phycol. 2002, 38, 17–38. [Google Scholar] [CrossRef]
- Peeters, J.C.H.; Eilers, P.H.C. The relationship between light intensity and photosynthesis—A simple mathematical model. Hydrobiol. Bull. 1978, 12, 134–136. [Google Scholar] [CrossRef]
- Eilers, P.H.C.; Peeters, J.C.H. A model for the relationship between light intensity and the rate of photosynthesis in phytoplankton. Ecol. Model. 1988, 42, 199–215. [Google Scholar] [CrossRef]
- Hartmann, P.; Nikolaou, A.; Chachuat, B.; Bernard, O. A Dynamic Model Coupling Photoacclimation and Photoinhibition in Microalgae. In Proceedings of the 12th European Control Conference, Zürich, Switzerland, 17–19 July 2013; Institute of Electrical and Electronics Engineers: Zürich, Switzerland, 2013; pp. 4178–4183. [Google Scholar]
- Droop, M.R. The kinetics of uptake, growth and inhibition in monochrysis lutheri. J. Mar. Biol. Assoc. 1968, 48, 689–733. [Google Scholar] [CrossRef]
- Benavides, M.; Hantson, A.-L.; Van Impe, J.F.M.; Vande Wouwer, A. Parameter identification of Droop model: An experimental case study. Bioprocess Biosyst. Eng. 2015, 38, 1783–1793. [Google Scholar] [CrossRef] [PubMed]
- Bernard, O. Hurdles and challenges for modelling and control of microalgae for CO2 mitigation and biofuel production. J. Process Control 2011, 21, 1378–1389. [Google Scholar] [CrossRef]
- Mairet, F.; Bernard, O.; Masci, P.; Lacour, T.; Sciandra, A. Modelling neutral lipid production by the microalga Isochrysis aff. galbana under nitrogen limitation. Bioresour. Technol. 2011, 102, 142–149. [Google Scholar] [CrossRef]
- Gorrini, F.A.; Zamudio Lara, J.M.; Biagiola, S.I.; Figueroa, J.L.; Hernández Escoto, H.; Hantson, A.-L.; Vande Wouwer, A. Experimental Study of Substrate Limitation and Light Acclimation in Cultures of the Microalgae Scenedesmus obliquus—Parameter Identification and Model Predictive Control. Processes 2020, 8, 1551. [Google Scholar] [CrossRef]
- Eilers, P.H.C.; Peeters, J.C.H. Dynamic behaviour of a model for photosynthesis and photoinhibition. Ecol. Model. 1993, 69, 113–133. [Google Scholar] [CrossRef]
- Bernard, O.; Masci, P.; Sciandra, A. A Photobioreactor Model in Nitrogen Limited Conditions. In Proceedings of the 6th Vienna International Conference on Mathematical Modelling, Vienna, Austria, 11–13 February 2009; European Mathematical Society: Helsinki, Finland, 2009; pp. 1521–1530. [Google Scholar]
- Hindmarsh, A.C. ODEPACK, a Systematized Collection of ODE Solvers. In Scientific Computing; North-Holland: Amsterdam, The Netherlands, 1983. [Google Scholar]
- Mairet, F.; Bernard, O.; Lacour, T.; Sciandra, A. Modelling microalgae growth in nitrogen limited photobiorector for estimating biomass, carbohydrate and neutral lipid productivities. IFAC Proc. Vol. 2011, 44, 10591–10596. [Google Scholar] [CrossRef]
- Khalil, H. Nonlinear Systems, 2nd ed.; Prentice-Hall: Hoboken, NJ, USA, 1996. [Google Scholar]
- Martínez, C.; Mairet, F.; Bernard, O. Dynamics of the periodically forced light-limited Droop model. J. Differ. Equ. 2020, 269, 3890–3913. [Google Scholar] [CrossRef]
- Mishra, N.; Mohan, S.; Mishra, N. Influence of High Light Intensity and Nitrate Deprivation on Growth and Biochemical Composition of the Marine Microalgae Isochrysis galbana. Braz. Arch. Biol. Technol. 2019, 62. [Google Scholar] [CrossRef]
- Sukenik, A.; Wahnon, R. Biochemical quality of marine unicellular algae with special emphasis on lipid composition: I. Isochrysis galbana. Aquaculture 1991, 97, 61–72. [Google Scholar] [CrossRef]
- Tzovenis, I.; De Pauw, N.; Sorgeloos, P. Optimisation of T-Iso biomass production rich in essential fatty acids: I. Effect of different light regimes on growth and biomass production. Aquaculture 2003, 216, 203–222. [Google Scholar] [CrossRef]
- Dayananda, C.; Sarada, R.; Usha Rani, M.; Shamala, T.R.; Ravishankar, G.A. Autotrophic cultivation of Botryococcus braunii for the production of hydrocarbons and exopolysaccharides in various media. Biomass Bioenergy 2007, 31, 87–93. [Google Scholar] [CrossRef]
- Mandal, S.; Mallick, N. Microalga Scenedesmus obliquus as a potential source for biodiesel production. Appl. Microbiol. Biotechnol. 2009, 84, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Huppe, H.C.; Turpin, D.H. Integration of Carbon and Nitrogen Metabolism in Plant and Algal Cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1994, 45, 577–607. [Google Scholar] [CrossRef]
- Liu, N.; Li, F.; Ge, F.; Tao, N.; Zhou, Q.; Wong, M. Mechanisms of ammonium assimilation by Chlorella vulgaris F1068: Isotope fractionation and proteomic approaches. Bioresour. Technol. 2015, 190, 307–314. [Google Scholar] [CrossRef]
- Cai, T.; Park, S.Y.; Li, Y. Nutrient recovery from wastewater streams by microalgae: Status and prospects. Renew. Sustain. Energy Rev. 2013, 19, 360–369. [Google Scholar] [CrossRef]
- Hu, Q. Environmental effects on cell composition. In Handbook of Microalgal Culture. Applied Phycology and Biotechnology, 2nd ed.; Richmond, A., Hu, Q., Eds.; Wiley Blackwell: West Sussex, UK, 2013; pp. 114–122. [Google Scholar]
- Li, Y.; Horsman, M.; Wang, B.; Wu, N.; Lan, C.Q. Effect of nitrogen source on cell growth and lipid accumulation of green alga Neochloris oleoabundans. Appl. Microbiol. Biotechnol. 2008, 81, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Simionato, D.; Block, M.A.; La Rocca, N.; Jouhet, J.; Maréchal, E.; Finazzi, G.; Morosinotto, T. The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot. Cell 2013, 12, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.K.; Uma, L.; Subramanian, G. Nitrogen stress induced changes in the marine cyanobacterium Oscillatoria willei BDU 130511. FEMS Microbiol. Ecol. 2006, 45, 263–272. [Google Scholar] [CrossRef]
- Serpa, R.; Calderón, A. Efecto de diferentes fuentes de nitrógeno en el contenido de carotenoides y clorofila de cuatro cepas peruanas de Dunaliella salina TEOD. J. Appl. Ecol. 2006, 5, 93–99. [Google Scholar] [CrossRef]
- Valenzuela-Espinoza, E.; Millán-Núñez, R.; Núñez-Cebrero, F. Protein, carbohydrate, lipid and chlorophyll a content in Isochrysis aff. galbana (clone T-Iso) cultured with a low cost alternative to the f/2 medium. Aquac. Eng. 2002, 25, 207–216. [Google Scholar] [CrossRef]
- Zarrinmehr, M.J.; Farhadian, O.; Heyrati, F.P.; Keramat, J.; Koutra, E.; Kornaros, M.; Daneshvar, E. Effect of nitrogen concentration on the growth rate and biochemical composition of the microalga, Isochrysis galbana. Egypt. J. Aquat. Res. 2020, 46, 153–158. [Google Scholar] [CrossRef]
- El-Kassas, H.Y. Growth and fatty acid profile of the marine microalga Picochlorum sp. grown under nutrient stress conditions. Egypt. J. Aquat. Res. 2013, 39, 233–239. [Google Scholar] [CrossRef]
- Courchesne, N.M.D.; Parisien, A.; Wang, B.; Lan, C.Q. Enhancement of lipid production using biochemical, genetic, and transcription factor engineering approaches. J. Biotechnol. 2009, 141, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.F.; Lourenço, S.O.; Chaloub, R.M. Effects of nitrogen starvation on the photosynthetic physiology of a tropical marine microalga Rhodomonas sp. (Cryptophyceae). Aquat. Bot. 2009, 91, 291–297. [Google Scholar] [CrossRef]
- Tan, K.W.; Lee, Y.K. The dilemma for lipid productivity in green microalgae: Importance of substrate provision in improving oil yield without sacrificing growth. Biotechnol. Biofuels 2016, 9, 255. [Google Scholar] [CrossRef]
- Siaut, M.; Cuiné, S.; Cagnon, C.; Fessler, B.; Nguyen, M.; Carrier, P.; Beyly, A. Oil accumulation in the model green alga Chlamydomonas reinhardtii: Characterization, variability between common laboratory strains, and relationship with starch reserves. BMC Biotechnol. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Praveen kumar, R.; Shameera, K.; Mahalakshmi, G.; Akbarsha, M.A.; Thajuddin, N. Influence of nutrient deprivations on lipid accumulation in a dominant indigenous microalga Chlorella sp.: Evaluation for biodiesel production. Biomass Bioenergy 2012, 37, 60–66. [Google Scholar] [CrossRef]
- Sánchez Saavedra, M.P.; Voltolina, D. Effect of photon fluence rates of white and blue-green light on growth efficiency and pigment content of three diatom species in batch cultures. Cienc. Mar. 2002, 28, 273–279. [Google Scholar] [CrossRef]
- Ak, I. Effects of light intensity, salinity and temperature on growth in CAMALT1 strain of Dunaliella viridis Teodoresco from Turkey. J. Biol. Sci. 2008, 8, 1356–1359. [Google Scholar] [CrossRef]
- Lacour, T.; Sciandra, A.; Talec, A.; Mayzaud, P.; Bernard, O. Neutral lipid and carbohydrate productivities as a response to nitrogen status in isochrysis sp. (t-iso; haptophyceae): Starvation versus limitation. J. Phycol. 2012, 48, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cao, Y.; Xu, H.; Liu, Y.; Sun, J.; Qiao, D.; Cao, Y. Effect of nitrogen-starvation, light intensity and iron on triacylglyceride/carbohydrate production and fatty acid profile of Neochloris oleoabundans HK-129 by a two-stage process. Bioresour. Technol. 2014, 155, 204–212. [Google Scholar] [CrossRef]
- Ho, S.H.; Huang, S.W.; Chen, C.Y.; Hasunuma, T.; Kondo, A.; Chang, J.S. Characterization and optimization of carbohydrate production from an indigenous microalga Chlorella vulgaris FSP-E. Bioresour. Technol. 2013, 135, 157–165. [Google Scholar] [CrossRef]
- Chu, F.F.; Chu, P.N.; Shen, X.F.; Lam, P.K.; Zeng, R.J. Effect of phosphorus on biodiesel production from Scenedesmus obliquus under nitrogen-deficiency stress. Bioresour. Technol. 2014, 152, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Biondi, N.; Cheloni, G.; Tatti, E.; Decorosi, F.; Rodolfi, L.; Giovannetti, L.; Viti, C.; Tredici, M.R. The bacterial community associated with Tetraselmis suecica outdoor mass cultures. J. Appl. Phycol. 2017, 29, 67–78. [Google Scholar] [CrossRef]
- Che, C.A.; Kim, S.H.; Hong, H.J.; Kityo, M.K.; Sunwoo, I.Y.; Jeong, G.T.; Kim, S.K. Optimization of light intensity and photoperiod for Isochrysis galbana culture to improve the biomass and lipid production using 14-L photobioreactors with mixed light emitting diodes (LEDs) wavelength under two-phase culture system. Bioresour. Technol. 2019, 285, 121323. [Google Scholar] [CrossRef] [PubMed]
- Ariyur, K.B.; Krstić, M. Real-Time Optimization by Extremum-Seeking Control; John Wiley and Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- Deschênes, J.S.; Wouwer, A.V. Dynamic Optimization of Biomass Productivity in Continuous Cultures of Microalgae Isochrysis galbana through Modulation of the Light Intensity. IFAC-PapersOnLine 2015, 48, 1093–1099. [Google Scholar] [CrossRef]


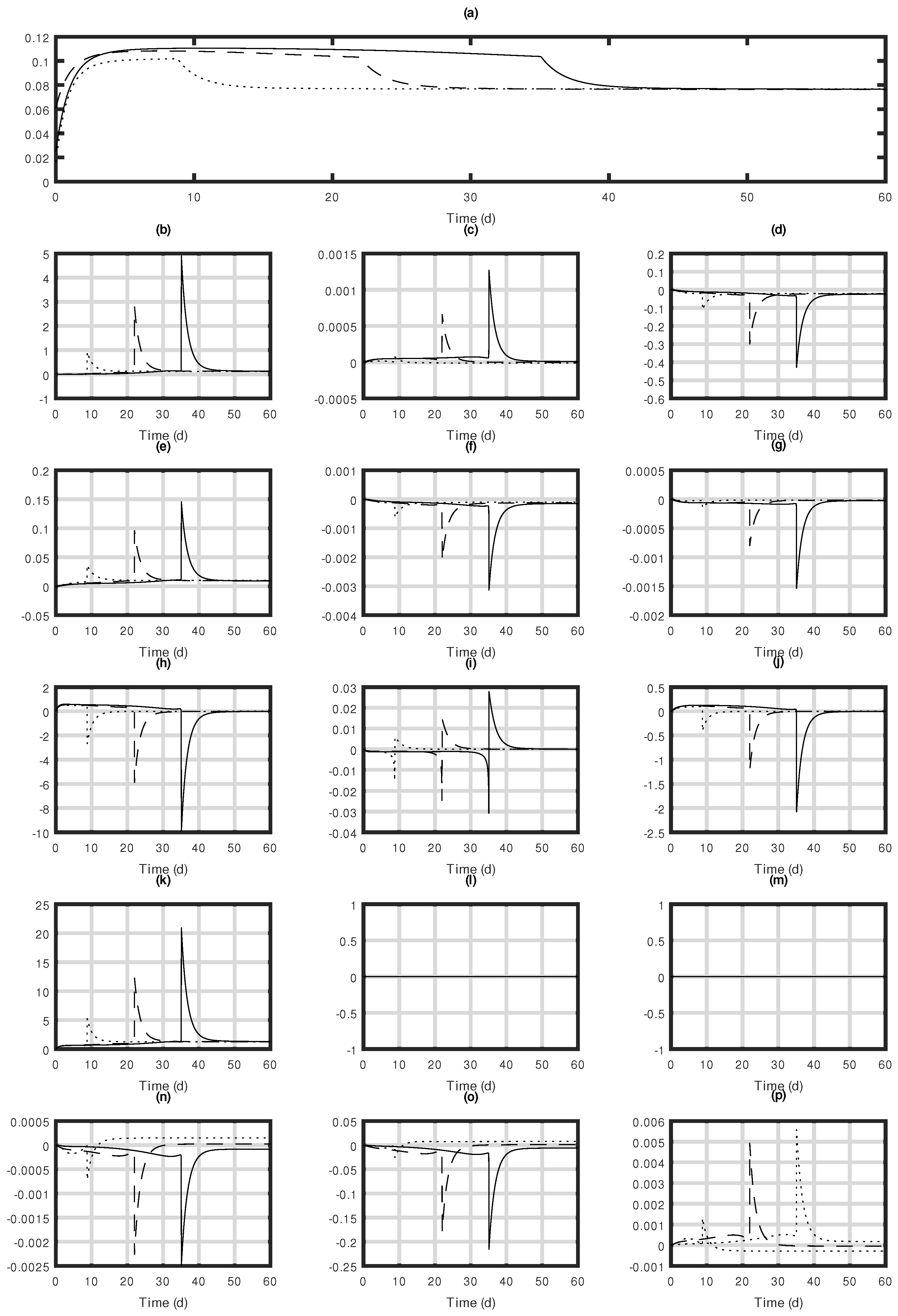

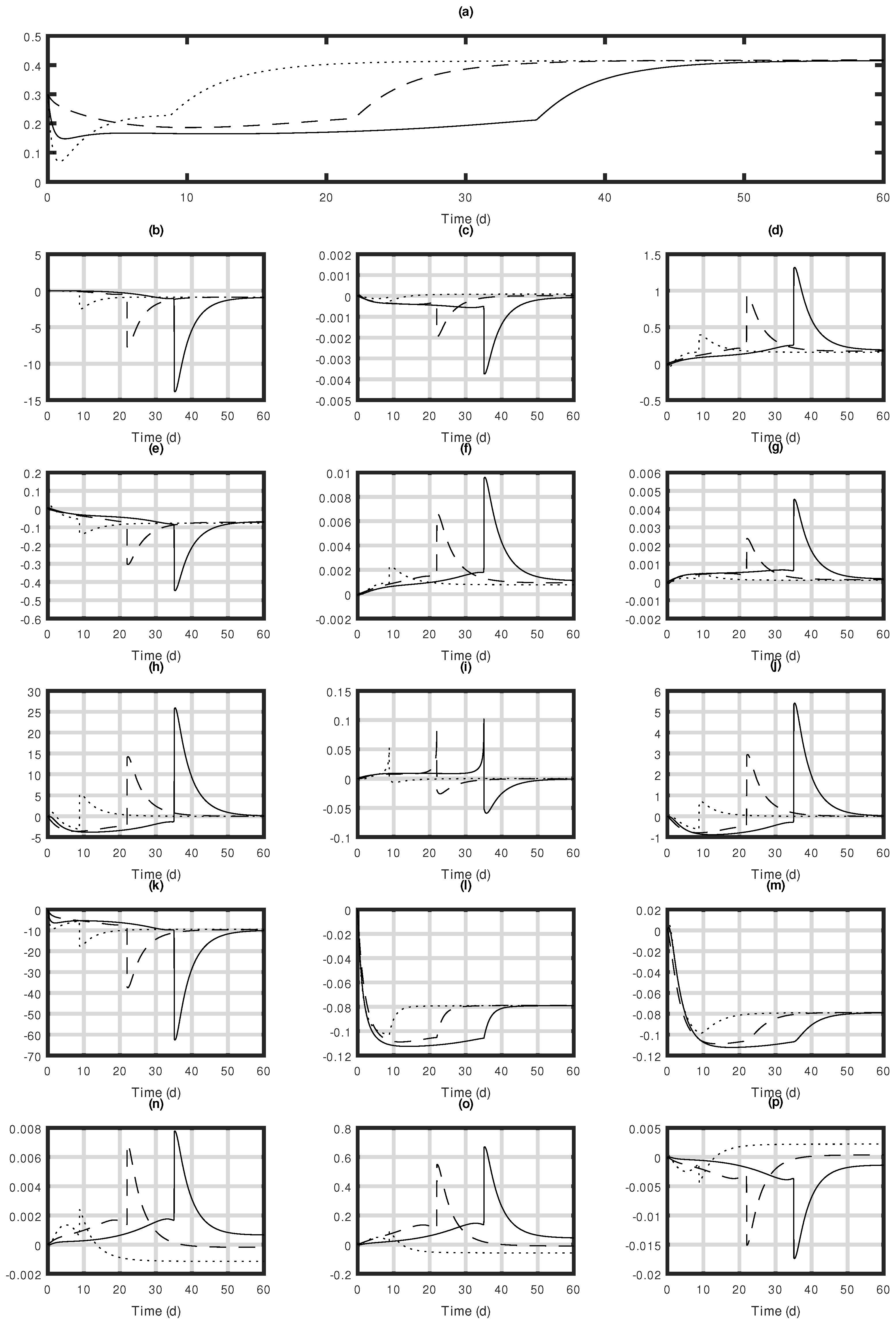
| Functions | Definition | Unit |
| Specific growth rate | gCm | |
| Maximum specific growth rate | gNm | |
| Normalized growth half saturation constant | gN(gC) | |
| Chlorophyll quota | gChl(gC) | |
| Light-attenuation rate light-attenuation coefficient | d | |
| Optical depth | m | |
| Chlorophyll concentration | gCm | |
| State variables | Definition | Unit |
| x | Biomass concentration | gCm |
| y | External nutrient concentration | gNm |
| z | Intracellular nitrogen quota | gN(gC) |
| Photon flux density acclimation | molms | |
| Carbohydrate quota | gC(gC) | |
| Parameters | Definition | Unit |
| Light intensity | molms | |
| D | Dilution rate | d |
| Nitrogen concentration in the reactor inlet | gNm | |
| Average photon flux density throughout the culture | molms | |
| Nitrogen intake rate | gN(gC)d | |
| Maximum specific growth rate | gN(gC)d | |
| Substrate-uptake half-saturation constant | gNm | |
| Growth half-saturation constant | molms | |
| Photon flux density saturation constant over growth | molms | |
| Chlorophyll saturation function | gChl(gN) | |
| Maximum chlorophyll saturation function | gChl(gN) | |
| Chlorophyll saturation function constant | molms | |
| Average photon flux density saturation function constant | − | |
| L | Culture depth | m |
| a | Attenuation coefficient due to chlorophyll | m(gChl) |
| b | Attenuation coefficient due to biomass | m(gC) |
| c | Attenuation coefficient due to background turbidity | m |
| Protein synthesis coefficient | gC(gN) | |
| Fatty acid synthesis coefficient | gC(gN) | |
| Maximum nitrogen quota | gN(gC) | |
| Minimum nitrogen quota | gN(gC) | |
| R | Respiration rate | d |
| Parameter | Value | Unit |
|---|---|---|
| 1.7 | d | |
| 0.0012 | gNm | |
| 0.05 | gN(gC) | |
| 0.25 | gN(gC) | |
| 0.073 | gN(gC) | |
| 1.4 | molms | |
| 295 | molms | |
| R | 0.0081 | d |
| 0.57 | gChl(gN) | |
| 63 | molms | |
| a | 16.2 | m(gChl) |
| b | 0.087 | m(gChl) |
| c | 0 | m |
| 1 | 10.6 | − |
| 2.6 | ||
| 4.8 |
| Simulation Scenario | Initial Light Intensity () 1 | Unit |
|---|---|---|
| A | 50 | molms |
| B | 150 | molms |
| C | 250 | molms |
| D | 500 | molms |
| E | 750 | molms |
| Symbol | Definition |
|---|---|
| Sensitivity function | |
| x | State variables |
| System evaluated parameters | |
| Nominal value of | |
| A | First-order partial derivatives with respect to x |
| B | First-order partial derivatives with respect to |
| Initial Conditions | |||||
|---|---|---|---|---|---|
| 10.1 | 33.2 | 0.055 | 50 | 0.3 | |
| 30.7 | 41.3 | 0.055 | 50 | 0.3 | |
| 225 | 49.9 | 0.055 | 50 | 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzmán-Palomino, A.; Aguilera-Vázquez, L.; Hernández-Escoto, H.; García-Vite, P.M.; Martínez-Salazar, A.L. Dynamical Simulation, Sensitivity, and Productivity Analysis of a Light-Photoacclimation Model for Microalgae-Based Carbohydrate Production in Continuous Photobioreactors. Processes 2023, 11, 1866. https://doi.org/10.3390/pr11071866
Guzmán-Palomino A, Aguilera-Vázquez L, Hernández-Escoto H, García-Vite PM, Martínez-Salazar AL. Dynamical Simulation, Sensitivity, and Productivity Analysis of a Light-Photoacclimation Model for Microalgae-Based Carbohydrate Production in Continuous Photobioreactors. Processes. 2023; 11(7):1866. https://doi.org/10.3390/pr11071866
Chicago/Turabian StyleGuzmán-Palomino, Abraham, Luciano Aguilera-Vázquez, Héctor Hernández-Escoto, Pedro Martin García-Vite, and Ana Lidia Martínez-Salazar. 2023. "Dynamical Simulation, Sensitivity, and Productivity Analysis of a Light-Photoacclimation Model for Microalgae-Based Carbohydrate Production in Continuous Photobioreactors" Processes 11, no. 7: 1866. https://doi.org/10.3390/pr11071866
APA StyleGuzmán-Palomino, A., Aguilera-Vázquez, L., Hernández-Escoto, H., García-Vite, P. M., & Martínez-Salazar, A. L. (2023). Dynamical Simulation, Sensitivity, and Productivity Analysis of a Light-Photoacclimation Model for Microalgae-Based Carbohydrate Production in Continuous Photobioreactors. Processes, 11(7), 1866. https://doi.org/10.3390/pr11071866








