Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview
Abstract
1. Introduction
2. Spectroscopic Methods for the Identification and Characterization of Microbial Pathogens
2.1. Wavelength-Based Microbial Growth Using Spectroscopic Analysis
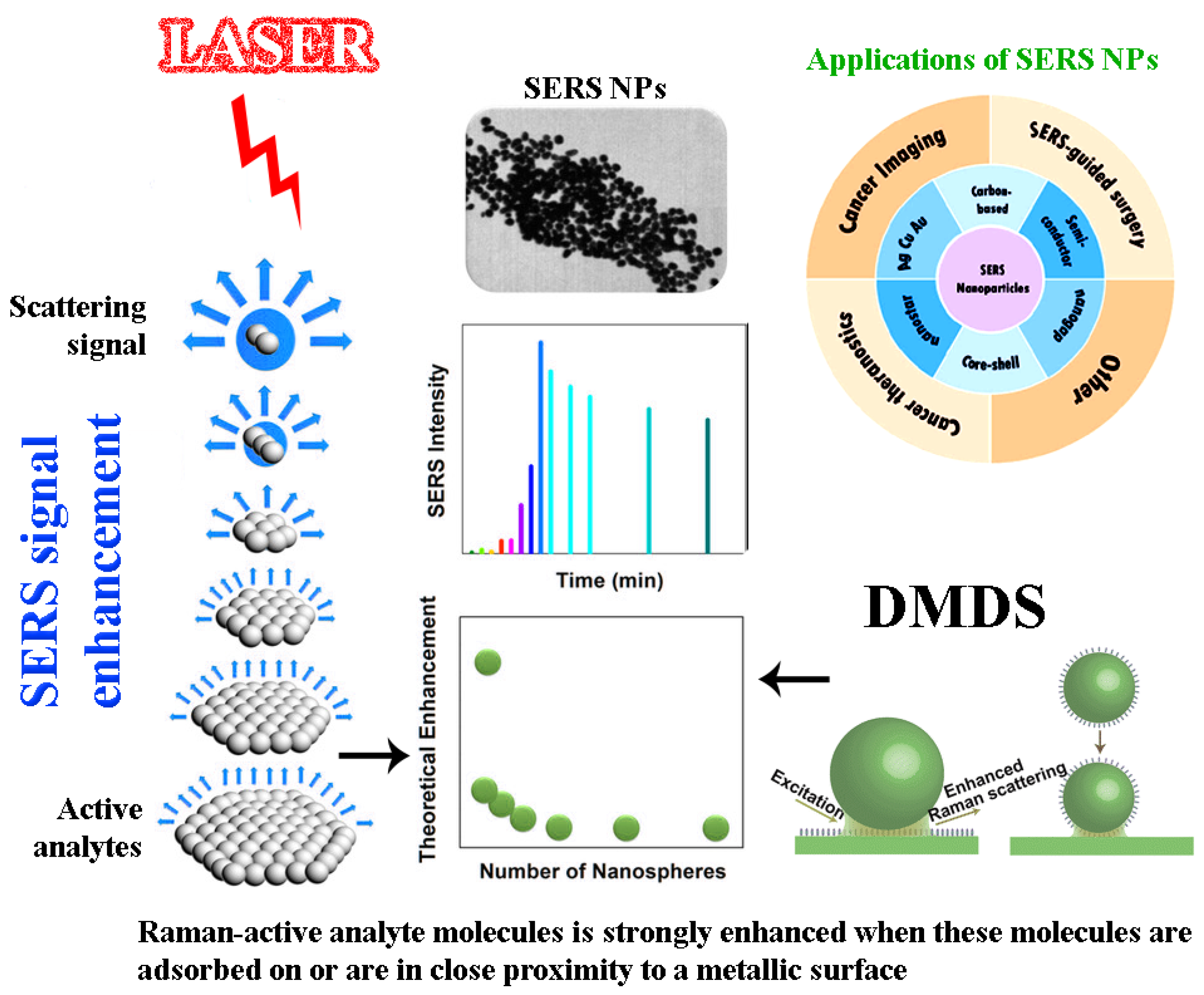
2.2. Surface-Enhanced Raman Spectroscopy (SERS)
2.3. Fourier Transform Infrared Spectroscopy (FTIR)
2.4. Electrochemical Impedance Spectroscopy (EIS)
2.5. MALDI-TOF/TOF Tandem Mass Spectrometry
2.6. Near-Infrared Spectroscopy (NIRS) and Chemometrics
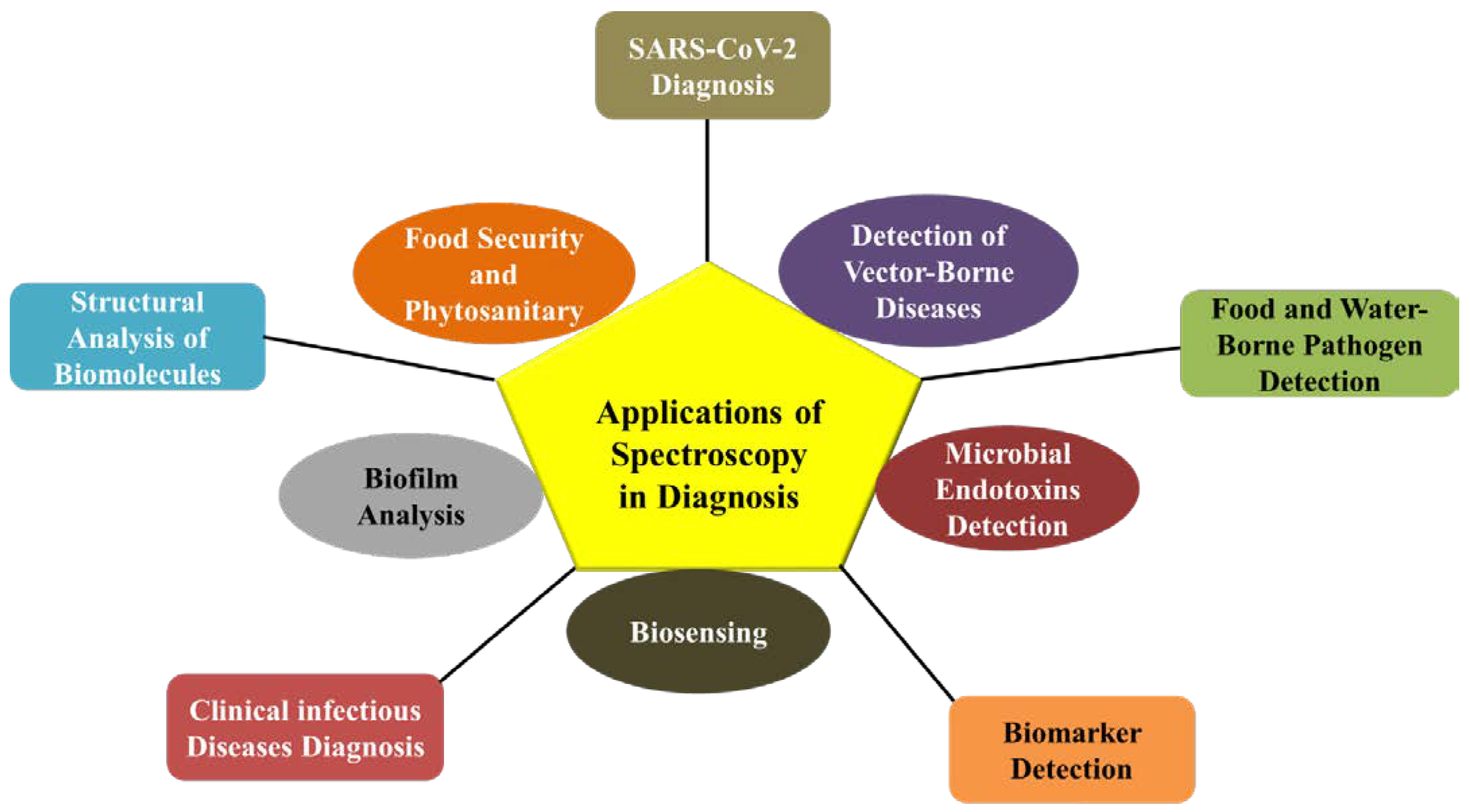
3. Applications of Spectroscopy in Diagnostics
3.1. Epidemiology
3.2. Diagnosis of Clinical Infectious and Vector-Borne Diseases
3.3. Food- and Waterborne Pathogen Detection
3.4. Antibiotic Resistance and Virulence Factors
3.5. SARS-CoV-2 Diagnosis
3.6. Microbial Endotoxins/Biomarker Detection
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving Sepsis Campaign: International guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013, 39, 165–228. [Google Scholar] [CrossRef] [PubMed]
- American Thoracic Society, Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 388. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, B.I. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J. Infect. Dis. 1990, 161, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.J.; van Zanten, E.; Akkerboom, V.; Wisselink, G.; van Slochteren, K.; de Boer, R.F.; Hendrix, R.; Friedrich, A.W.; Rossen, J.W.; Kooistra-Smid, A. Targeted next-generation sequencing of the 16S-23S rRNA region for culture-independent bacterial identification-increased discrimination of closely related species. Sci. Rep. 2017, 7, 1–2. [Google Scholar] [CrossRef]
- Ko, J.; Park, S.G.; Lee, S.; Wang, X.; Mun, C.; Kim, S.; Kim, D.H.; Choo, J. Culture-free detection of bacterial pathogens on plasmonic nanopillar arrays using rapid Raman mapping. ACS Appl. Mater. Interfaces 2018, 10, 6831–6840. [Google Scholar] [CrossRef]
- Shrivastava, S.; Lee, W.I.; Lee, N.E. Culture-free, highly sensitive, quantitative detection of bacteria from minimally processed samples using fluorescence imaging by smartphone. Biosens. Bioelectron. 2018, 109, 90–97. [Google Scholar] [CrossRef]
- Wang, J.C.; Tung, Y.C.; Ichiki, K.; Sakamoto, H.; Yang, T.H.; Suye, S.I.; Chuang, H.S. Culture-free detection of methicillin-resistant Staphylococcus aureus by using self-driving diffusometric DNA nanosensors. Biosens. Bioelectron. 2020, 148, 111817. [Google Scholar] [CrossRef]
- Fenollar, F.; Raoult, D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int. J. Antimicrob. Agents 2007, 30, 7–15. [Google Scholar] [CrossRef]
- Şen Karaman, D.; Ercan, U.K.; Bakay, E.; Topaloğlu, N.; Rosenholm, J.M. Evolving technologies and strategies for combating antibacterial resistance in the advent of the postantibiotic era. Adv. Funct. Mater. 2020, 30, 1908783. [Google Scholar] [CrossRef]
- Ashton, L.; Lau, K.; Winder, C.L.; Goodacre, R. Raman spectroscopy: Lighting up the future of microbial identification. Future Microbiol. 2011, 6, 991–997. [Google Scholar] [CrossRef]
- Kotanen, C.N.; Martinez, L.; Alvarez, R.; Simecek, J.W. Surface enhanced Raman scattering spectroscopy for detection and identification of microbial pathogens isolated from human serum. Sens. Bio Sens. Res. 2016, 8, 20–26. [Google Scholar] [CrossRef]
- Singh, V.K.; Sharma, J.; Pathak, A.K.; Ghany, C.T.; Gondal, M.A. Laser-induced breakdown spectroscopy (LIBS): A novel technology for identifying microbes causing infectious diseases. Biophys. Rev. 2018, 10, 1221–1239. [Google Scholar] [CrossRef]
- Saari, S.; Järvinen, S.; Reponen, T.; Mensah-Attipoe, J.; Pasanen, P.; Toivonen, J.; Keskinen, J. Identification of single microbial particles using electro-dynamic balance assisted laser-induced breakdown and fluorescence spectroscopy. Aerosol Sci. Technol. 2016, 50, 126–132. [Google Scholar] [CrossRef]
- Cheeseman, S.; Shaw, Z.L.; Vongsvivut, J.; Crawford, R.J.; Dupont, M.F.; Boyce, K.J.; Gangadoo, S.; Bryant, S.J.; Bryant, G.; Cozzolino, D.; et al. Analysis of pathogenic bacterial and yeast biofilms using the combination of synchrotron ATR-FTIR microspectroscopy and chemometric approaches. Molecules 2021, 26, 3890. [Google Scholar] [CrossRef]
- Bosch, A.; Serra, D.; Prieto, C.; Schmitt, J.; Naumann, D.; Yantorno, O. Characterization of Bordetella pertussis growing as biofilm by chemical analysis and FT-IR spectroscopy. Appl. Microbiol. Biotechnol. 2006, 71, 736–747. [Google Scholar] [CrossRef]
- Wang, H.; Ding, S.; Wang, G.; Xu, X.; Zhou, G. In situ characterization and analysis of Salmonella biofilm formation under meat processing environments using a combined microscopic and spectroscopic approach. Int. J. Food Microbiol. 2013, 167, 293–302. [Google Scholar] [CrossRef]
- Chen, P.; Wang, J.J.; Hong, B.; Tan, L.; Yan, J.; Zhang, Z.; Liu, H.; Pan, Y.; Zhao, Y. Characterization of mixed-species biofilm formed by Vibrio parahaemolyticus and Listeria monocytogenes. Front. Microbiol. 2019, 10, 2543. [Google Scholar] [CrossRef]
- Van Duuren, J.B.; Müsken, M.; Karge, B.; Tomasch, J.; Wittmann, C.; Häussler, S.; Brönstrup, M. Use of single-frequency impedance spectroscopy to characterize the growth dynamics of biofilm formation in Pseudomonas aeruginosa. Sci. Rep. 2017, 7, 5223. [Google Scholar] [CrossRef]
- Yang, K.; Li, H.Z.; Zhu, X.; Su, J.Q.; Ren, B.; Zhu, Y.G.; Cui, L. Rapid antibiotic susceptibility testing of pathogenic bacteria using heavy-water-labeled single-cell Raman spectroscopy in clinical samples. Anal. Chem. 2019, 91, 6296–6303. [Google Scholar] [CrossRef]
- Zarnowiec, P.; Lechowicz, L.; Czerwonka, G.; Kaca, W. Fourier transform infrared spectroscopy (FTIR) as a tool for the identification and differentiation of pathogenic bacteria. Curr. Med. Chem. 2015, 22, 1710–1718. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Asai, T.; Nakaya, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; et al. Raman imaging of pathogenic Candida auris: Visualization of structural characteristics and machine-learning identification. Front. Microbiol. 2021, 12, 769597. [Google Scholar] [CrossRef] [PubMed]
- Sinha, M.; Jupe, J.; Mack, H.; Coleman, T.P.; Lawrence, S.M.; Fraley, S.I. Emerging Technologies for Molecular Diagnosis of Sepsis. Clin. Microbiol. Rev. 2018, 31, e00089-17. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.; Dornuf, F.; Klein, O.; Mäntele, W. IR spectroscopy goes to the hospital: Progress in reagent-free blood analysis and haemodialysis monitoring. FTIR Spectrosc. Microbiol. Med. Diagn. 2011, 20, 46. [Google Scholar]
- Lopez-Reyes, G.; Pérez, F.R. A method for the automated Raman spectra acquisition. J. Raman Spectrosc. 2017, 48, 1654–1664. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Zhao, W. Emerging Microtechnologies and Automated Systems for Rapid Bacterial Identification and Antibiotic Susceptibility Testing. SLAS Technol. 2017, 22, 585. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, Y.; Tang, H.; Zong, N.; Jiang, X. Microfluidics for biomedical analysis. Small Methods 2020, 4, 1900451. [Google Scholar] [CrossRef]
- Cowcher, D.P.; Xu, Y.; Goodacre, R. Portable, quantitative detection of Bacillus bacterial spores using surface-enhanced Raman scattering. Anal. Chem. 2013, 85, 3297–3302. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Carbonnelle, E.; Raskine, L. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Bio Trib. Mag. 2011, 39, 35–42. [Google Scholar] [CrossRef]
- Madonna, A.J.; van Cuyk, S.; Voorhees, K.J. Detection of Escherichia coli using immunomagnetic separation and bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 257–263. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Černáková, L.; Kadam, S.; Kaushik, K.S.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stępień, K.; Leszczewicz, M.; et al. Advances in chemical and biological methods to identify microorganisms—From past to present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef]
- Braga, P.A.; Tata, A.; dos Santos, V.G.; Barreiro, J.R.; Schwab, N.V.; dos Santos, M.V.; Eberlin, M.N.; Ferreira, C.R. Bacterial identification: From the agar plate to the mass spectrometer. RSC Adv. 2013, 3, 994–1008. [Google Scholar] [CrossRef]
- Trivedi, N.; Dubey, A. Degradation studies of pendimethalin by indigenous soil bacterium Pseudomonas strain PD1 using spectrophotometric scanning and FTIR. Arch. Microbiol. 2021, 203, 4499–4507. [Google Scholar] [CrossRef]
- Myers, J.A.; Curtis, B.S.; Curtis, W.R. Improving accuracy of cell and chromophore concentration measurements using optical density. BMC Biophys. 2013, 6, 4. [Google Scholar] [CrossRef]
- McBirney, S.E.; Trinh, K.; Wong-Beringer, A.; Armani, A.M. Wavelength-normalized spectroscopic analysis of Staphylococcus aureus and Pseudomonas aeruginosa growth rates. Biomed. Opt. Express 2016, 7, 4034–4042. [Google Scholar] [CrossRef]
- Zhang, Y.; Hong, H.; Myklejord, D.V.; Cai, W. Molecular imaging with SERS-active nanoparticles. Small. 2011, 7, 3261–3269. [Google Scholar] [CrossRef]
- Cui, S.; Zhang, S.; Yue, S. Raman Spectroscopy and Imaging for Cancer Diagnosis. J. Healthc. Eng. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227. [Google Scholar] [CrossRef]
- Neng, J.; Harpster, M.H.; Wilson, W.C.; Johnson, P.A. Surface-enhanced Raman scattering (SERS) detection of multiple viral antigens using magnetic capture of SERS-active nanoparticles. Biosens. Bioelectron. 2013, 41, 316–321. [Google Scholar] [CrossRef]
- Ho, C.S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef]
- Efrima, S.; Zeiri, L. Understanding SERS of bacteria. J. Raman Spectr. 2009, 40, 277–288. [Google Scholar] [CrossRef]
- Kloß, S.; Rösch, P.; Pfister, W.; Kiehntopf, M.; Popp, J. Toward culture-free Raman spectroscopic identification of pathogens in ascitic fluid. Anal. Chem. 2015, 87, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.P.; Ngo-Thi, N.A.; van Vreeswijk, T.; Stämmler, M.; Endtz, H.P.; Bruining, H.A.; Naumann, D.; Puppels, G.J. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 2003, 41, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hu, Q.; Li, C.; Zhang, F.; Gu, H.; Wang, X.; Li, S.; Xue, L.; Madl, T.; Zhang, Y.; et al. Wide-Range, Rapid, and Specific Identification of Pathogenic Bacteria by Surface-Enhanced Raman Spectroscopy. ACS Sens. 2021, 6, 2911–2919. [Google Scholar] [CrossRef]
- Singh, K.S.; Majik, M.S.; Tilvi, S. Vibrational spectroscopy for structural characterization of bioactive compounds. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2014; Volume 65, pp. 115–148. [Google Scholar]
- Gieroba, B.; Krysa, M.; Wojtowicz, K.; Wiater, A.; Pleszczyńska, M.; Tomczyk, M.; Sroka-Bartnicka, A. The FT-IR and Raman Spectroscopies as Tools for Biofilm Characterization Created by Cariogenic Streptococci. Int. J. Mol. Sci. 2020, 21, 3811. [Google Scholar] [CrossRef]
- Ozaki, Y. Infrared spectroscopy—Mid-infrared, near-infrared, and far-infrared/terahertz spectroscopy. Anal. Sci. 2021, 37, 1193–1212. [Google Scholar] [CrossRef]
- How an FTIR Spectrometer Operates—Chemistry LibreTexts. 2022. Available online: https://chem.libretexts.org/Book-shelves/Physical_and_Theoretical_Chemistry_Textbook_Maps/Supplemental_Modules_(Physical_and_Theoretical_Chemis-try)/Spectroscopy/Vibrational_Spectroscopy/Infrared_Spectroscopy/How_an_FTIR_Spectrometer_Operates (accessed on 23 February 2022).
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771. [Google Scholar] [CrossRef]
- Singh, R.; Hong, S.; Jang, J. Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 2017, 7, 42771. [Google Scholar] [CrossRef]
- Al-Qadiri, H.M.; Lin, M.; Cavinato, A.G.; Rasco, B.A. Fourier transform infrared spectroscopy, detection and identification of Escherichia coli O157:H7 and Alicyclobacillus strains in apple juice. Int. J. Food Microbiol. 2006, 111, 73–80. [Google Scholar] [CrossRef]
- Donlan, R.M.; Piede, J.A.; Heyes, C.D.; Sanii, L.; Murga, R.; Edmonds, P.; El-Sayed, I.; El-Sayed, M.A. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl. Environ. Microbiol. 2004, 70, 4980–4988. [Google Scholar] [CrossRef]
- Stewart, G.N. The charges produced by the growth of bacteria in the molecular concentration and electrical conductivity of culture media. J. Exp. Med. 1899, 4, 235. [Google Scholar] [CrossRef]
- Brosel-Oliu, S.; Uria, N.; Abramova, N.; Bratov, A. Impedimetric sensors for bacteria detection. Biosens. Micro Nanoscale Appl. 2015, 24, 257–288. [Google Scholar]
- Lagier, J.C.; Edouard, S.; Pagnier, I.; Mediannikov, O.; Drancourt, M.; Raoult, D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin. Microbiol. Rev. 2015, 28, 208. [Google Scholar] [CrossRef]
- Dean, D.A.; Ramanathan, T.; Machado, D.; Sundararajan, R. Electrical impedance spectroscopy study of biological tissues. J. Electrost. 2008, 66, 165–177. [Google Scholar] [CrossRef]
- Yang, L.; Bashir, R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol. Adv. 2008, 26, 135–150. [Google Scholar] [CrossRef]
- Jahnke, H.G.; Heimann, A.; Azendorf, R.; Mpoukouvalas, K.; Kempski, O.; Robitzki, A.A.; Charalampaki, P. Impedance spectroscopy—An outstanding method for label-free and real-time discrimination between brain and tumor tissue in vivo. Biosens. Bioelectron. 2013, 46, 8–14. [Google Scholar] [CrossRef]
- Permeh, S.; Lau, K.; Duncan, M. Characterization of biofilm formation and coating degradation by electrochemical impedance spectroscopy. Coatings 2019, 9, 518. [Google Scholar] [CrossRef]
- Gogichaeva, N.V.; Williams, T.; Alterman, M.A. MALDI TOF/TOF tandem mass spectrometry as a new tool for amino acid analysis. J. Am. Soc. Mass Spectrom. 2007, 18, 279–284. [Google Scholar] [CrossRef]
- Goloborodko, A.A.; Gorshkov, M.V.; Good, D.M.; Zubarev, R.A. Sequence scrambling in shotgun proteomics is negligible. J. Am. Soc. Mass Spectrom. 2011, 22, 1121–1124. [Google Scholar] [CrossRef]
- Juiz, P.M.; Almela, M.; Melción, C.; Campo, I.; Esteban, C.; Pitart, C.; Marco, F.; Vila, J. A comparative study of two different methods of sample preparation for positive blood cultures for the rapid identification of bacteria using MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1353–1358. [Google Scholar] [CrossRef]
- Hou, T.Y.; Chiang-Ni, C.; Teng, S.H. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Haiko, J.; Savolainen, L.E.; Hilla, R.; Pätäri-Sampo, A. Identification of urinary tract pathogens after 3-hours urine culture by MALDI-TOF mass spectrometry. J. Microbiol. Methods 2016, 129, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Rychert, J. Benefits and limitations of MALDI-TOF mass spectrometry for the identification of microorganisms. J. Infect. Epidemiol. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Lasch, P.; Nattermann, H.; Erhard, M.; Stämmler, M.; Grunow, R.; Bannert, N.; Appel, B.; Naumann, D. MALDI-TOF mass spectrometry compatible inactivation method for highly pathogenic microbial cells and spores. Anal. Chem. 2008, 80, 2026–2034. [Google Scholar] [CrossRef]
- Jeong, Y.S.; Choi, S.; Chong, E.; Kim, J.H.; Kim, S.J. Rapid detection of B acillus spore aerosol particles by direct in situ analysis using MALDI-TOF mass spectrometry. Lett. Appl. Microbiol. 2014, 59, 177–183. [Google Scholar] [CrossRef]
- Johansson, Å.; Nagy, E.; Sóki, J. Detection of carbapenemase activities of Bacteroides fragilis strains with matrix-assisted laser desorption ionization—time of flight mass spectrometry (MALDI-TOF MS). Anaerobe 2014, 26, 49–52. [Google Scholar] [CrossRef]
- Hoyos-Mallecot, Y.; Cabrera-Alvargonzalez, J.; Miranda-Casas, C.; Rojo-Martín, M.D.; Liebana-Martos, C.; Navarro-Marí, J. MALDI-TOF MS, a useful instrument for differentiating metallo-β-lactamases in Enterobacteriaceae and Pseudomonas spp. Lett. Appl. Microbiol. 2014, 58, 325–329. [Google Scholar] [CrossRef]
- Hart, P.J.; Wey, E.; McHugh, T.D.; Balakrishnan, I.; Belgacem, O. A method for the detection of antibiotic resistance markers in clinical strains of Escherichia coli using MALDI mass spectrometry. J. Microbiol. Methods 2015, 111, 1–8. [Google Scholar] [CrossRef]
- Ferone, M.; Gowen, A.; Fanning, S.; Scannell AG, M. Microbial detection and identification methods: Bench top assays to omics approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1–24. [Google Scholar] [CrossRef]
- Harz, M.; Rösch, P.; Popp, J. Vibrational spectroscopy—A powerful tool for the rapid identification of microbial cells at the single-cell level. Cytom. Part A 2009, 75, 104–113. [Google Scholar] [CrossRef]
- Roggo, Y.; Chalus, P.; Maurer, L.; Lema-Martinez, C.; Edmond, A.; Jent, N. A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J. Pharm. Biomed. Anal. 2007, 44, 683–700. [Google Scholar] [CrossRef]
- Hamprecht, J.; Corsten, D.; Noll, M.; Meier, E. Controlling the sustainability of food supply chains. Supply Chain. Manag. Int. J. 2005, 10, 7–10. [Google Scholar] [CrossRef]
- Curto, B.; Moreno, V.; García-Esteban, J.A.; Blanco, F.J.; González, I.; Vivar, A.; Revilla, I. Accurate prediction of sensory attributes of cheese using near-infrared spectroscopy based on artificial neural network. Sensors 2020, 20, 3566. [Google Scholar] [CrossRef]
- Li, B.; Lin, Y.; Yu, W.; Wilson, D.I.; Young, B.R. Application of mechanistic modelling and machine learning for cream cheese fermentation pH prediction. J. Chem. Technol. Biotechnol. 2020, 96, 125–133. [Google Scholar] [CrossRef]
- Sipos, A. A knowledge-based system as a sustainable software application for the supervision and intelligent control of an alcoholic fermentation process. Sustainability 2020, 12, 10205. [Google Scholar] [CrossRef]
- Viejo, C.G.; Torrico, D.D.; Dunshea, F.R.; Fuentes, S. Development of artificial neural network models to assess beer acceptability based on sensory properties using a robotic pourer: A comparative model approach to achieve an artificial intelligence system. Beverages 2019, 5, 33. [Google Scholar] [CrossRef]
- Deng, X.; Cao, S.; Horn, A.L. Emerging Applications of Machine Learning in Food Safety. Annu. Rev. Food Sci. Technol. 2021, 12, 513–538. [Google Scholar] [CrossRef]
- Vajdi, M.; Varidi, M.J.; Varidi, M.; Mohebbi, M. Using electronic nose to recognize fish spoilage with an optimum classifier. J. Food Meas. Charact. 2019, 13, 1205–1217. [Google Scholar] [CrossRef]
- Gutiérrez, P.; Godoy, S.E.; Torres, S.; Oyarzún, P.; Sanhueza, I.; Díaz-García, V.; Contreras-Trigo, B.; Coelho, P. Improved anti-biotic detection in raw milk using machine learning tools over the absorption spectra of a problem-specific nanobiosensor. Sensors 2020, 20, 4552. [Google Scholar] [CrossRef]
- Karami, H.; Rasekh, M.; Mirzaee-Ghaleh, E. Application of the E-nose machine system to detect adulterations in mixed edible oils using chemometrics methods. J. Food Process Preserv. 2020, 44, 1–12. [Google Scholar] [CrossRef]
- Ayari, F.; Mirzaee- Ghaleh, E.; Rabbani, H.; Heidarbeigi, K. Using an E-nose machine for detecting the adulteration of margarine in cow ghee. J. Food Process. Eng. 2018, 41, e12806. [Google Scholar] [CrossRef]
- Amigo, J.M.; Martí, I.; Gowen, A. Hyperspectral imaging and chemometrics: A perfect combination for the analysis of food structure, composition and quality. Data Handl. Sci. Technol. 2013, 28, 343–370. [Google Scholar]
- Spyrelli, E.D.; Papachristou, C.K.; Nychas, G.J.E.; Panagou, E.Z. Microbiological Quality Assessment of Chicken Thigh Fillets Using Spectroscopic Sensors and Multivariate Data Analysis. Foods 2021, 10, 2723. [Google Scholar] [CrossRef] [PubMed]
- Cebrián, E.; Núñez, F.; Rodríguez, M.; Grassi, S.; González-Mohino, A. Potential of Near Infrared Spectroscopy as a Rapid Method to Discriminate OTA and Non-OTA-Producing Mould Species in a Dry-Cured Ham Model System. Toxins 2021, 13, 620. [Google Scholar] [CrossRef] [PubMed]
- Alexandrakis, D.; Downey, G.; Scannell, A.G.M. Rapid non-destructive detection of spoilage of intact chicken breast muscle using near-infrared and Fourier transform mid-infrared spectroscopy and multivariate statistics. Food Bioprocess Technol. 2012, 5, 338–347. [Google Scholar] [CrossRef]
- Azadshahraki, F.; Sharifi, K.; Jamshidi, B.; Karimzadeh, R.; Naderi, H. Diagnosis of Early Blight Disease in Tomato Plant based on Visible/Near-Infrared Spectroscopy and Principal Components Analysis-Artificial Neural Network Prior to Visual Disease Symptoms. J. Agric. Mach. 2022, 12, 81–94. [Google Scholar]
- Rahi, S.; Mobli, H.; Jamshidi, B.; Azizi, A.; Sharifi, M. Achieving a robust Vis/NIR model for microbial contamination detection of Persian leek by spectral analysis based on genetic, iPLS algorithms and VIP scores. Postharvest Biol. Technol. 2021, 175, 111413. [Google Scholar] [CrossRef]
- Sirisomboon, C.D.; Wongthip, P.; Sirisomboon, P. Potential of near infrared spectroscopy as a rapid method to detect aflatoxins in brown rice. J. Near Infrared Spectrosc. 2019, 27, 232–240. [Google Scholar] [CrossRef]
- Tao, F.; Yao, H.; Hruska, Z.; Liu, Y.; Rajasekaran, K.; Bhatnagar, D. Detection of aflatoxin B1 on corn kernel surfaces using visible-near infrared spectroscopy. J. Near Infrared Spectrosc. 2020, 28, 59–69. [Google Scholar] [CrossRef]
- Pan, W.; Zhao, J.; Chen, Q. Classification of foodborne pathogens using near infrared (NIR) laser scatter imaging system with multivariate calibration. Sci. Rep. 2015, 5, 9524. [Google Scholar] [CrossRef]
- Duan, C.; Chen, C.; Khan, M.N.; Liu, Y.; Zhang, R.; Lin, H.; Cao, L. Non-destructive determination of the total bacteria in flounder fillet by portable near infrared spectrometer. Food Control 2014, 42, 18–22. [Google Scholar] [CrossRef]
- Tito, N.B.; Rodemann, T.; Powell, S.M. Use of near infrared spectroscopy to predict microbial numbers on Atlantic salmon. Food Microbiol. 2012, 32, 431–436. [Google Scholar] [CrossRef]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef]
- El-Bouri, K.; Johnston, S.; Rees, E.; Thomas, I.; Bome-Mannathoko, N.; Jones, C.; Reid, M.; Ben-Ismaeil, B.; Davies, A.P.; Harris, L.G.; et al. Comparison of bacterial identification by MALDI-TOF mass spectrometry and conventional diagnostic microbiology methods: Agreement, speed and cost implications. Br. J. Biomed. Sci. 2012, 69, 47–55. [Google Scholar] [CrossRef]
- Yeh, Y.T.; Gulino, K.; Zhang, Y.; Sabestien, A.; Chou, T.W.; Zhou, B.; Lin, Z.; Albert, I.; Lu, H.; Swaminathan, V.; et al. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. USA 2020, 117, 895–901. [Google Scholar] [CrossRef]
- Ye, J.; Yeh, Y.T.; Xue, Y.; Wang, Z.; Zhang, N.; Liu, H.; Zhang, K.; Yu, Z.; Roder, A.; Lopez, N.P.; et al. Accurate Virus Identification with Interpretable Raman Signatures by Machine Learning. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rebrošová, K.; Bernatová, S.; Šiler, M.; Uhlirova, M.; Samek, O.; Ježek, J.; Holá, V.; Růžička, F.; Zemanek, P. Raman spectroscopy-a tool for rapid differentiation among microbes causing urinary tract infections. Anal. Chim. Acta 2022, 1191, 339292. [Google Scholar] [CrossRef]
- Zhao, Y.; Tian, S.; Yu, L.; Zhang, Z.; Zhang, W. Analysis and Classification of Hepatitis Infections Using Raman Spectroscopy and Multiscale Convolutional Neural Networks. J. Appl. Spectrosc. 2021, 88, 441–451. [Google Scholar] [CrossRef]
- Tiwari, D.; Jakhmola, S.; Pathak, D.K.; Kumar, R.; Jha, H.C. Temporal In Vitro Raman Spectroscopy for Monitoring Replication Kinetics of Epstein-Barr Virus Infection in Glial Cells. ACS Omega 2020, 5, 29547–29560. [Google Scholar] [CrossRef]
- Zyubin, A.; Lavrova, A.; Manicheva, O.; Dogonadze, M.; Belik, V.; Samusev, I. Raman spectroscopy for glutathione measurements in Mycobacterium tuberculosis strains with different antibiotic resistance. J. Raman Spectrosc. 2021, 52, 1661–1666. [Google Scholar] [CrossRef]
- Barker, K.R.; Santino, M.; LiPuma, J.J.; Tullis, E.; Muller, M.P.; Matukas, L.M.; Tadros, M. Fourier Transform Infrared Spectroscopy for Typing Burkholderia cenocepacia ET12 Isolates. Microbiol. Spectr. 2021, 9, e0183121. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Liu, Z.; Peng, Y.; Hou, G.; Chen, W.; Xiao, R. Automatic and sensitive detection of West Nile virus non-structural protein 1 with a portable SERS-LFIA detector. Mikrochim. Acta 2021, 188, 206. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Purrà, M.; Carré-Camps, M.; de Puig, H.; Bosch, I.; Gehrke, L.; Hamad-Schifferli, K. Surface-Enhanced Raman Spectroscopy-Based Sandwich Immunoassays for Multiplexed Detection of Zika and Dengue Viral Biomarkers. ACS Infect. Dis. 2017, 3, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; Cooper, A.; Mabbott, S.; Bradley, B.; Asiala, S.; Jamieson, L.; Clucas, C.; Capewell, P.; Marchesi, F.; Gibbins, M.P.; et al. Raman spectroscopic analysis of skin as a diagnostic tool for Human African Trypanosomiasis. PLoS Pathog. 2021, 17, e1010060. [Google Scholar] [CrossRef]
- Goh, B.; Ching, K.; Magalhães, R.J.S.; Ciocchetta, S.; Edstein, M.D.; Maciel-de-Freitas, R.; Sikulu-Lord, M.T. The application of spectroscopy techniques for diagnosis of malaria parasites and arboviruses and surveillance of mosquito vectors: A systematic review and critical appraisal of evidence. PLoS Negl. Trop. Dis. 2021, 15, e0009218. [Google Scholar] [CrossRef]
- Sanchez, L.; Ermolenkov, A.; Tang, X.T.; Tamborindeguy, C.; Kurouski, D. Non-invasive diagnostics of Liberibacter disease on tomatoes using a hand-held Raman spectrometer. Planta 2020, 251, 64. [Google Scholar] [CrossRef]
- Vallejo-Pérez, M.R.; Sosa-Herrera, J.A.; Navarro-Contreras, H.R.; Álvarez-Preciado, L.G.; Rodríguez-Vázquez, Á.G.; Lara-Ávila, J.P. Raman Spectroscopy and Machine-Learning for Early Detection of Bacterial Canker of Tomato: The Asymptomatic Disease Condition. Plants 2021, 10, 1542. [Google Scholar] [CrossRef]
- Strycker, B.D.; Han, Z.; Duan, Z.; Commer, B.; Wang, K.; Shaw, B.D.; Sokolov, A.V.; Scully, M.O. Identification of toxic mold species through Raman spectroscopy of fungal conidia. PLoS ONE 2020, 15, e0242361. [Google Scholar] [CrossRef]
- Saif, F.A.; Yaseen, S.A.; Alameen, A.S.; Mane, S.B.; Undre, P.B. Identification and characterization of Aspergillus species of fruit rot fungi using microscopy, FT-IR, Raman and UV-Vis spectroscopy. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2021, 246, 119010. [Google Scholar] [CrossRef]
- Mandrile, L.; Rotunno, S.; Miozzi, L.; Vaira, A.M.; Giovannozzi, A.M.; Rossi, A.M.; Noris, E. Nondestructive Raman Spectroscopy as a Tool for Early Detection and Discrimination of the Infection of Tomato Plants by Two Economically Important Viruses. Anal. Chem. 2019, 91, 9025–9031. [Google Scholar] [CrossRef]
- Huang, C.C.; Hsu, Z.H.; Lai, Y.S. Raman spectroscopy for virus detection and the implementation of unorthodox food safety. Trends Food Sci. Technol. 2021, 116, 525–532. [Google Scholar] [CrossRef]
- Petersen, M.; Yu, Z.; Lu, X. Application of Raman Spectroscopic Methods in Food Safety: A Review. Biosensors 2021, 11, 187. [Google Scholar] [CrossRef]
- Deidda, F.; Cionci, N.B.; Cordovana, M.; Campedelli, I.; Fracchetti, F.; Di Gioia, D.; Ambretti, S.; Pane, M. Bifidobacteria Strain Typing by Fourier Transform Infrared Spectroscopy. Front. Microbiol. 2021, 12, 692975. [Google Scholar] [CrossRef]
- Yakes, B.J.; Ellsworth, Z.; Karunathilaka, S.R.; Crump, E. Evaluation of Portable Sensor and Spectroscopic Devices for Seafood Decomposition Determination. Food Anal. Methods 2021, 14, 2346–2356. [Google Scholar] [CrossRef]
- Wang, K.; Chen, L.; Ma, X.; Ma, L.; Chou, K.C.; Cao, Y.; Khan, I.; Gölz, G.; Lu, X. Arcobacter Identification and Species Determination Using Raman Spectroscopy Combined with Neural Networks. Appl. Environ. Microbiol. 2020, 86, e00924-20. [Google Scholar] [CrossRef]
- Du, Y.; Han, D.; Liu, S.; Sun, X.; Ning, B.; Han, T.; Wang, J.; Gao, Z. Raman spectroscopy-based adversarial network combined with SVM for detection of foodborne pathogenic bacteria. Talanta 2022, 237, 122901. [Google Scholar] [CrossRef]
- Yan, S.; Wang, S.; Qiu, J.; Li, M.; Li, D.; Xu, D.; Li, D.; Liu, Q. Raman spectroscopy combined with machine learning for rapid detection of food-borne pathogens at the single-cell level. Talanta 2021, 226, 122195. [Google Scholar] [CrossRef]
- Xu, J.L.; Herrero-Langreo, A.; Lamba, S.; Ferone, M.; Scannell, A.; Caponigro, V.; Gowen, A.A. Characterisation and Classification of Foodborne Bacteria Using Reflectance FTIR Microscopic Imaging. Molecules 2021, 26, 6318. [Google Scholar] [CrossRef]
- Lu, J.; Chen, J.; Liu, C.; Zeng, Y.; Sun, Q.; Li, J.; Shen, Z.; Chen, S.; Zhang, R. Identification of antibiotic resistance and virulence-encoding factors in Klebsiella pneumoniae by Raman spectroscopy and deep learning. Microb. Biotechnol. 2022, 15, 1270–1280. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Caliskan, A.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Drug-resistant Staphylococcus aureus bacteria detection by combining surface-enhanced Raman spectroscopy (SERS) and deep learning techniques. Sci. Rep. 2021, 11, 18444. [Google Scholar] [CrossRef]
- Chen, X.; Tang, M.; Liu, Y.; Huang, J.; Liu, Z.; Tian, H.; Zheng, Y.; de la Chapelle, M.L.; Zhang, Y.; Fu, W. Surface-enhanced Raman scattering method for the identification of methicillin-resistant Staphylococcus aureus using positively charged silver nanoparticles. Mikrochim. Acta 2019, 186, 102. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chen, L.; Chou, K.C.; Lu, X. Campylobacter jejuni Antimicrobial Resistance Profiles and Mechanisms Determined Using a Raman Spectroscopy-Based Metabolomic Approach. Appl. Environ. Microbiol. 2021, 87, e0038821. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Song, Y.; Xu, X.; Peng, D.; Wang, J.; Qie, X.; Lin, K.; Yu, M.; Ge, M.; Wang, Y.; et al. Development of a Fast Raman-Assisted Antibiotic Susceptibility Test (FRAST) for the Antibiotic Resistance Analysis of Clinical Urine and Blood Samples. Anal. Chem. 2021, 93, 5098–5106. [Google Scholar] [CrossRef] [PubMed]
- Gannesen, A.V.; Zdorovenko, E.L.; Botchkova, E.A.; Hardouin, J.; Massier, S.; Kopitsyn, D.S.; Gorbachevskii, M.V.; Kadykova, A.A.; Shashkov, A.S.; Zhurina, M.V.; et al. Composition of the Biofilm Matrix of Cutibacterium acnes Acneic Strain RT5. Front. Microbiol. 2019, 10, 1284. [Google Scholar] [CrossRef]
- Do, H.; Kwon, S.R.; Fu, K.; Morales-Soto, N.; Shrout, J.D.; Bohn, P.W. Electrochemical Surface-Enhanced Raman Spectroscopy of Pyocyanin Secreted by Pseudomonas aeruginosa Communities. Langmuir ACS J. Surf. Colloids 2019, 35, 7043–7049. [Google Scholar] [CrossRef]
- Horiue, H.; Sasaki, M.; Yoshikawa, Y.; Toyofuku, M.; Shigeto, S. Raman spectroscopic signatures of carotenoids and polyenes enable label-free visualization of microbial distributions within pink biofilms. Sci. Rep. 2020, 10, 7704. [Google Scholar] [CrossRef]
- Kriem, L.S.; Wright, K.; Ccahuana-Vasquez, R.A.; Rupp, S. Mapping of a Subgingival Dual-Species Biofilm Model Using Confocal Raman Microscopy. Front. Microbiol. 2021, 12, 729720. [Google Scholar] [CrossRef]
- World Health Organization. Clinical Management of COVID-19: Living Guideline, 13 January 2023; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- Wood, B.R.; Kochan, K.; Bedolla, D.E.; Salazar-Quiroz, N.; Grimley, S.L.; Perez-Guaita, D.; Baker, M.J.; Vongsvivut, J.; Tobin, M.J.; Bambery, K.R.; et al. Infrared Based Saliva Screening Test for COVID-19. Angew. Chem. 2021, 60, 17102–17107. [Google Scholar] [CrossRef]
- Huang, J.; Wen, J.; Zhou, M.; Ni, S.; Le, W.; Chen, G.; Wei, L.; Zeng, Y.; Qi, D.; Pan, M.; et al. On-Site Detection of SARS-CoV-2 Antigen by Deep Learning-Based Surface-Enhanced Raman Spectroscopy and Its Biochemical Foundations. Anal. Chem. 2021, 93, 9174–9182. [Google Scholar] [CrossRef]
- Yadav, S.; Sadique, M.A.; Ranjan, P.; Kumar, N.; Singhal, A.; Srivastava, A.K.; Khan, R. SERS Based Lateral Flow Immunoassay for Point-of-Care Detection of SARS-CoV-2 in Clinical Samples. ACS Appl. Bio Mater. 2021, 4, 2974–2995. [Google Scholar] [CrossRef]
- Zavyalova, E.; Ambartsumyan, O.; Zhdanov, G.; Gribanyov, D.; Gushchin, V.; Tkachuk, A.; Rudakova, E.; Nikiforova, M.; Kuznetsova, N.; Popova, L.; et al. SERS-Based Aptasensor for Rapid Quantitative Detection of SARS-CoV-2. Nanomaterials 2021, 11, 1394. [Google Scholar] [CrossRef]
- Desai, S.; Mishra, S.V.; Joshi, A.; Sarkar, D.; Hole, A.; Mishra, R.; Dutt, S.; Chilakapati, M.K.; Gupta, S.; Dutt, A. Raman spectroscopy-based detection of RNA viruses in saliva: A preliminary report. J. Biophotonics 2020, 13, e202000189. [Google Scholar] [CrossRef]
- Guleken, Z.; Jakubczyk, P.; Wiesław, P.; Krzysztof, P.; Bulut, H.; Öten, E.; Depciuch, J.; Tarhan, N. Characterization of Covid-19 infected pregnant women sera using laboratory indexes, vibrational spectroscopy, and machine learning classifications. Talanta 2022, 237, 122916. [Google Scholar] [CrossRef]
- Guleken, Z.; Tok, Y.T.; Jakubczyk, P.; Paja, W.; Pancerz, K.; Shpotyuk, Y.; Cebulski, J.; Depciuch, J. Development of novel spectroscopic and machine learning methods for the measurement of periodic changes in COVID-19 antibody level. Measurement 2022, 196, 111258. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, Y.; Zughaier, S.M. Highly Sensitive Detection and Differentiation of Endotoxins Derived from Bacterial Pathogens by Surface-Enhanced Raman Scattering. Biosensors 2021, 11, 234. [Google Scholar] [CrossRef]
- Ge, X.; Pereira, F.C.; Mitteregger, M.; Berry, D.; Zhang, M.; Wagner, M.; Cheng, J.X. SRS-FISH: High-Throughput Platform Linking Microbiome Function to Identity at the Single Cell Level. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Cui, D.; Kong, L.; Chen, S.; Xie, W.; Zhang, C. Classification and Identification of Archaea Using Single-Cell Raman Ejection and Artificial Intelligence: Implications for Investigating Uncultivated Microorganisms. Anal. Chem. 2021, 93, 17012–17019. [Google Scholar] [CrossRef]
- Ren, Y.; Ji, Y.; Teng, L.; Zhang, H. Using Raman spectroscopy and chemometrics to identify the growth phase of Lactobacillus casei Zhang during batch culture at the single-cell level. Microb. Cell Factories 2017, 16, 233. [Google Scholar] [CrossRef]
- Yan, S.; Qiu, J.; Guo, L.; Li, D.; Xu, D.; Liu, Q. Development overview of Raman-activated cell sorting devoted to bacterial detection at single-cell level. Appl. Microbiol. Biotechnol. 2021, 105, 1315–1331. [Google Scholar] [CrossRef]
- Song, Y.; Kaster, A.K.; Vollmers, J.; Song, Y.; Davison, P.A.; Frentrup, M.; Preston, G.M.; Thompson, I.P.; Murrell, J.C.; Yin, H.; et al. Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea. Microb. Biotechnol. 2017, 10, 125–137. [Google Scholar] [CrossRef]
- García-Timermans, C.; Props, R.; Zacchetti, B.; Sakarika, M.; Delvigne, F.; Boon, N. Raman Spectroscopy-Based Measurements of Single-Cell Phenotypic Diversity in Microbial Populations. mSphere 2020, 5, e00806-20. [Google Scholar] [CrossRef] [PubMed]
- Skolrood, L.; Wang, Y.; Zhang, S.; Wei, Q. Single-molecule and particle detection on true portable microscopy platforms. Sens. Actuators Rep. 2022, 4, 100063. [Google Scholar] [CrossRef]


| Application | Chemometric Method | Main Finding | Reference |
|---|---|---|---|
| Early detection of blight disease in tomato with Vis-NIR spectroscopy | PCA-ANN | Early detection of blight disease and the associated pathogen type was achieved with about 93–100% accuracy | [88] |
| Detection of E. coli contamination in Persian leek with Vis/NIR spectroscopy | PLSDA with Genetic Algorithm (GA), interval PLS, variable influence on projection scores | GA exhibited high sensitivity (100%) and specificity (98%) and low classification error (0.8) in E. coli detection | [89] |
| Estimation of total viable counts and Pseudomonas spp. in chicken thigh fillets with FTIR and MSI | PLSR, LDA, QDA, SVM, and QSVM | SVM coupled with multispectral imaging showed the highest performance with about 94.4% overall accuracy | [85] |
| Detection of ochratoxin A-producing fungi from non-ochratoxin-producing fungi in dried meat with NIRS | PCA with SVM-DA | The SVM-DA model could differentiate between ochratoxin and non-ochratoxin-producing fungi with 86% specificity and 85% accuracy | [86] |
| Detection of aflatoxin B1 in corn kernel using Vis-NIRS | PCA-LDA and PLS-DA | Both discriminant and classification models exhibited over 90% accurate performance | [91] |
| Detection of aflatoxin contamination in brown rice with NIRS | PLSR | The model showed good predictive performance with a prediction coefficient of 0.95% | [90] |
| Classification of foodborne pathogens (E. coli, S. aureus, S. typhimurium, and mixed bacteria) using NIR-LSIS | Linear (PLSDA, KNN, and LDA) and nonlinear (BPANN, OSELM, and SVM) | Nonlinear methods performed better than linear methods, with OSELM exhibiting a performance accuracy of 95% | [92] |
| Quantification of total bacteria in fish fillets with a portable NIR spectrometer | PLS, GA combined with BPANN | GA combined with BPANN exhibited a better efficiency of prediction (about 96% accuracy) than PLS | [93] |
| Non-invasive and non-destructive detection of spoilage in chicken breast muscles via NIRS and FTIR | PCA, PLS-DA, and outer product analysis (OPA) | OPA performed better compared to PCA and PLS-DA in discriminating between bacterial loads | [87] |
| Detection and prediction of microbial spoilage in salmon with NIRS | PCA and PLS | The validation curve exhibited a large error of R2 = 0.64, although the calibration equation presented a good R2 of 0.95 | [94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandian, S.; Lakshmi, S.A.; Priya, A.; Balasubramaniam, B.; Zaukuu, J.-L.Z.; Durgadevi, R.; Abe-Inge, V.; Sohn, S.-I. Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview. Processes 2023, 11, 1191. https://doi.org/10.3390/pr11041191
Pandian S, Lakshmi SA, Priya A, Balasubramaniam B, Zaukuu J-LZ, Durgadevi R, Abe-Inge V, Sohn S-I. Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview. Processes. 2023; 11(4):1191. https://doi.org/10.3390/pr11041191
Chicago/Turabian StylePandian, Subramani, Selvaraj Alagu Lakshmi, Arumugam Priya, Boopathi Balasubramaniam, John-Lewis Zinia Zaukuu, Ravindran Durgadevi, Vincent Abe-Inge, and Soo-In Sohn. 2023. "Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview" Processes 11, no. 4: 1191. https://doi.org/10.3390/pr11041191
APA StylePandian, S., Lakshmi, S. A., Priya, A., Balasubramaniam, B., Zaukuu, J.-L. Z., Durgadevi, R., Abe-Inge, V., & Sohn, S.-I. (2023). Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview. Processes, 11(4), 1191. https://doi.org/10.3390/pr11041191











