Comparative Analysis of the Interaction between the Antiviral Drug Umifenovir and Umifenovir Encapsulated in Phospholipids Micelles (Nanosome/Umifenovir) with dsDNA as a Model for Pharmacogenomic Analysis by Electrochemical Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Electrochemical Equipment
2.3. Preparation of Electrochemical Sensors
3. Results and Discussion
3.1. Electrochemical Behavior of Umi on SPE/SWCNT
3.2. Electrochemical Behavior of DNA on SPE/SWCNT
3.3. Investigation of the Interaction between Umi or NUmi and dsDNA
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jena, N. Drug targets, mechanisms of drug action, and therapeutics against SARS-CoV-2. Chem. Phys. Impact 2021, 2, 100011. [Google Scholar] [CrossRef]
- Wang, X.; Cao, R.; Zhang, H.; Liu, J.; Xu, M.; Hu, H.; Li, Y.; Zhao, L.; Li, W.; Sun, X.; et al. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Disc. 2020, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Pécheur, E.-I.; Borisevich, V.; Halfmann, P.; Morrey, J.; Smee, D.; Prichard, M.; Mire, C.; Kawaoka, Y.; Geisbert, T.; Polyak, S. The synthetic antiviral drug arbidol inhibits globally prevalent pathogenic viruses. J. Virol. 2016, 90, 3086–3092. [Google Scholar] [CrossRef] [PubMed]
- Proskurnina, E.; Izmailov, D.; Sozarukova, M.; Zhuravleva, T.; Leneva, I.; Poromov, A. Antioxidant potential of antiviral drug umifenovir. Molecules 2020, 25, 1577. [Google Scholar] [CrossRef]
- Deng, P.; Zhong, D.; Yu, K.; Zhang, Y.; Wang, T.; Chen, X. Pharmacokinetics, Metabolism, and Excretion of the Antiviral Drug Arbidol in Humans. Antimicrob. Agents Chemother. 2013, 57, 1743–1755. [Google Scholar] [CrossRef]
- Tarighi, P.; Eftekhari, S.; Chizari, M.; Sabernavaei, M.; Jafari, D.; Mirzabeigi, P. A review of potential suggested drugs for coronavirus disease (COVID-19) treatment. Eur. J. Pharmacol. 2021, 895, 173890. [Google Scholar] [CrossRef]
- Kumar, D.; Trived, N. Disease-drug and drug-drug interaction in COVID-19: Risk and assessment. Biomed. Pharmacother. 2021, 139, 111642. [Google Scholar] [CrossRef]
- Yang, C.; Ke, C.; Yue, D.; Li, W.; Hu, Z.; Liu, W.; Hu, S.; Wang, S.; Liu, J. Effectiveness of arbidol for COVID-19 prevention in health professionals. Front. Public Health 2020, 8, 249. [Google Scholar] [CrossRef]
- Hanai, T. Quantitative in silico analysis of SARS-CoV-2 S-RBD omicron mutant transmissibility. Talanta 2022, 240, 123206. [Google Scholar] [CrossRef]
- Crommelin, D.; van Hoogevest, P.; Storm, G. The role of liposomes in clinical nanomedicine development. What now? Now what? J. Control. Release 2020, 318, 256–263. [Google Scholar] [CrossRef]
- Alavi, M.; Karimi, N.; Safaei, M. Application of Various Types of Liposomes in Drug Delivery Systems. Adv. Pharm. Bull. 2017, 7, 3–9. [Google Scholar] [CrossRef]
- Sharma, D.; Ali, A.; Trivedi, L. An Updated Review on: Liposomes as drug delivery system. PharmaTutor 2018, 6, 50–62. [Google Scholar] [CrossRef]
- Ipatova, O.; Torkhovskaya, T.; Medvedeva, N.; Prozorovsky, V.; Ivanova, N.; Shironin, A. Bioavailability of oral drugs and the methods for its improvement. Biochem. Moscow Suppl. Ser. B 2010, 4, 82–94. [Google Scholar] [CrossRef]
- Tereshkina, Y.; Torkhovskaya, T.; Tikhonova, E.; Kostryukova, L.; Sanzhakov, M.; Korotkevich, E.; Khudoklinova, Y.; Orlova, N.; Kolesanova, E. Nanoliposomes as drug delivery systems: Safety concerns. J. Drug Target. 2022, 30, 313–325. [Google Scholar] [CrossRef]
- Shumyantseva, V.; Kuzikov, A.; Masamrekh, R.; Bulko, T.; Archakov, A. From electrochemistry to enzyme kinetics of cytochrome P450. Biosens. Bioelectron. 2018, 121, 192–204. [Google Scholar] [CrossRef]
- Kurbanoglu, S.; Dogan-Topal, B.; Rodriguez, E.; Bozal-Palabiyik, B.; Ozkan, S.; Uslu, B. Advances in electrochemical DNA biosensors and their interaction mechanism with pharmaceuticals. J. Electroanal. Chem. 2016, 775, 8–26. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Shadjou, N. Pharmacogenomic study using bio- and nanobioelectrochemistry: Drug–DNA interaction. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 61, 1002–1017. [Google Scholar] [CrossRef]
- Nikitin, M. Non-complementary strand commutation as a fundamental alternative for information processing by DNA and gene regulation. Nat. Chem. 2023, 15, 70–82. [Google Scholar] [CrossRef]
- Sigolaeva, L.; Bulko, T.; Kozin, M.; Zhang, W.; Köhler, M.; Romanenko, I.; Yuan, J.; Schacher, F.; Pergushov, D.; Shumyantseva, V. Long-term stable poly(ionic liquid)/MWCNTs inks enable enhanced surface modification for electrooxidative detection and quantification of dsDNA. Polymer 2019, 168, 95–103. [Google Scholar] [CrossRef]
- Krejcova- Sirlova, Z.; Barek, J.; Vyskocil, V. Voltammetric studies of the interaction of genotoxic 2-nitrofluorene with DNA. Bioelectrochemistry 2023, 149, 108326. [Google Scholar] [CrossRef]
- Shumyantseva, V.; Bulko, T.; Tikhonova, E.; Sanzhakov, M.; Kuzikov, A.; Masamrekh, R.; Pergushov, D.; Schacher, F.; Sigolaeva, L. Electrochemical studies of the interaction of rifampicin and nanosome/rifampicin with dsDNA. Bioelectrochemistry 2021, 140, 107736. [Google Scholar] [CrossRef] [PubMed]
- Morawska, K.; Popławski, T.; Ciesielski, W.; Smarzewska, S. Electrochemical and spectroscopic studies of the interaction of antiviral drug Tenofovir with single and double stranded DNA. Bioelectrochemistry 2018, 123, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Bolat, G. Investigation of poly(CTAB-MWCNTs) composite based electrochemical DNA biosensor and interaction study with anticancer drug Irinotecan. Microchem. J. 2020, 159, 105426. [Google Scholar] [CrossRef]
- Graves, D.; Velea, L. Intercalative Binding of Small Molecules to Nucleic Acids. Curr. Org. Chem. 2005, 4, 915–929. [Google Scholar] [CrossRef]
- Hatamluyi, B.; Es’haghi, Z. Quantitative Biodetection of Anticancer Drug Rituxan with DNA Biosensor Modified PAMAM Dendrimer/Reduced Graphene Oxide Nanocomposite. Electroanalysis 2018, 30, 1659–1668. [Google Scholar] [CrossRef]
- Rupar, J.; Aleksić, M.; Dobričić, V.; Brborić, J.; Čudina, O. An electrochemical study of 9-chloroacridine redox behavior and its interaction with double-stranded DNA. Bioelectrochemistry 2020, 135, 107579. [Google Scholar] [CrossRef]
- Vyskočil, V.; Blašková, M.; Hájková, A.; Horáková, E.; Krejčová, Z.; Stávková, K.; Wang, J. Electrochemical DNA biosensors—Useful diagnostic tools for the detection of damage to DNA caused by organic xenobiotics (A Review). Sens. Electroanal. 2012, 7, 141–162. [Google Scholar]
- Ramotowska, S.; Ciesielska, A.; Makowski, M. What can electrochemical methods offer in determining DNA–drug interactions? Molecules 2021, 26, 3478. [Google Scholar] [CrossRef]
- Held, P. Nucleic Acid Purity Assessment Using A260/A280 Ratios. In Application Note; BioTek Instruments Incorporation: Winooski, VT, USA, 2001; pp. 1–5. [Google Scholar]
- Smith, P.; Jin, P.; Rahman, K. Strategies for drug repurposing against coronavirus targets. Curr. Res. Pharmacol. Drug Discov. 2022, 3, 100072. [Google Scholar] [CrossRef]
- Carrara, S.; Baj-Rossi, C.; Boero, C.; De Micheli, G. Do Carbon Nanotubes contribute to Electrochemical Biosensing? Electrochim. Acta 2014, 128, 102–112. [Google Scholar] [CrossRef]
- Nouri-Nigje, E.; Bischoff, R.; Bruins, A.; Permentier, H. Electrochemistry in the mimicry of oxidative drug metabolism by cytochrome P450s. Curr. Drug Metab. 2011, 12, 359–371. [Google Scholar] [CrossRef]
- Jurva, U.; Wikström, H.; Weidolf, L.; Bruins, A. Comparison between electrochemistry/mass spectrometry and cytochrome P450 catalyzed oxidation reactions. Rapid Commun. Mass Spectrom. 2003, 17, 800–810. [Google Scholar] [CrossRef]
- Gosser, D. Cyclic Voltammtry; VCH: New York, NY, USA, 1994. [Google Scholar]
- Compton, B.; Banks, C. Understanding Voltammetry, 2nd ed.; Imperial College Press: London, UK, 2011. [Google Scholar]
- Brett, C.M.A.; Oliveira Brett, A.M. Electrochemistry: Principles, Methods, and Applications, 1st ed.; Oxford Science University Publications: Oxford, UK, 1993; p. 219. [Google Scholar] [CrossRef]
- Miller, J.; Miller, J.C. Statistics and Chemometrics for Analytical Chemistry, 7th ed.; Pearson: London, UK, 2018; Available online: https://www.pearson.com/uk/educators/higher-education-educators/program/Miller-Statistics-and-Chemometrics-forAnalytical-Chemistry-7th-Edition/PGM2061238.html (accessed on 10 May 2020).
- Ferapontova, E. DNA electrochemistry and electrochemical sensor for nucleic acids. Ann. Rev. Anal. Chem. 2018, 11, 197–218. [Google Scholar] [CrossRef]
- Findik, M.; Bingol, H.; Erdem, A. Hybrid nanoflowers modified pencil graphite electrodes developed for electrochemical monitoring of interaction between Mitomycin C and DNA. Talanta 2021, 222, 121647. [Google Scholar] [CrossRef]
- Bagni, G.; Osella, D.; Sturchio, E.; Macsini, M. Deoxyribonucleic acid (DNA) biosensors for environmental risk assessment and drug studies. Anal. Chim. Acta 2006, 573, 81–89. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.-M.; Oliveira-Brett, A.-M. DNA Electrochemical Biosensors for In Situ Probing of Pharmaceutical Drug Oxidative DNA Damage. Sensors 2021, 21, 1125. [Google Scholar] [CrossRef]
- Erdem, A.; Muti, M.; Papakonstantinou, P.; Canavar, E.; Karadeniz, H.; Congur, G.; Sharma, S. Graphene oxide integrated sensor for electrochemical monitoring of mitomycin C–DNA interaction. Analyst 2012, 137, 2129–2135. [Google Scholar] [CrossRef]
- Eksin, E.; Zor, E.; Erdem, A.; Bingöl, H. Electrochemical monitoring of biointeraction by graphene-based material modified pencil graphite electrode. Biosens. Bioelectron. 2017, 92, 207–214. [Google Scholar] [CrossRef]
- Fotouhi, L.; Bahmani, F. MWCNT Modified Glassy Carbon Electrode: Probing Furazolidone-DNA Interactions and DNA Determination. Electroanalysis 2013, 25, 757–764. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–vis, fluorescence spectroscopies and cyclic voltammetry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Dogan-Topal, B.; Bozal-Palabiyik, B.; Ozkan, S.; Uslu, B. Investigation of anticancer drug lapatinib and its interaction with dsDNA by electrochemical and spectroscopic techniques. Sens. Actuators B Chem. 2014, 194, 185–194. [Google Scholar] [CrossRef]
- Temerk, Y.; Ibrahim, M.; Ibrahim, H.; Kotb, M. Interactions of an anticancer drug lomustine with single and double stranded DNA at physiological conditions analysed by electrochemical and spectroscopic methods. J. Electroanal. Chem. 2016, 769, 62–71. [Google Scholar] [CrossRef]
- Muti, M.; Muti, M. Electrochemical monitoring of the interaction between anticancer drug and DNA in the presence of antioxidant. Talanta 2018, 178, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Agafonova, L.; Tikhonova, E.; Sanzhakov, M.; Kostryukova, L.; Shumyantseva, V. Electrochemical Studies of the Interaction of Phospholipid Nanoparticles with dsDNA. Processes 2022, 10, 2324. [Google Scholar] [CrossRef]
- Carrara, S.; Cavallini, A.; Erokhin, V.; De Micheli, G. Multi-panel drugs detection in human serum for personalized therapy. Biosens. Bioelectron. 2011, 26, 3914–3919. [Google Scholar] [CrossRef]
- Mohammad, S.; Choi, Y.; Chung, J.; Cedrone, E.; Neun, B.; Dobrovolskaia, M.; Yang, X.; Guo, W.; Chew, Y.; Kim, J.; et al. Nanocomplexes of doxorubicin and DNA fragments for efficient and safe cancer chemotherapy. J. Control. Release 2023, 354, 91–108. [Google Scholar] [CrossRef]
- Evtugyn, G.; Porfireva, A.; Belyakova, S. Electrochemical DNA sensors for drug determination. J. Pharmac. Biomed. Anal. 2022, 221, 115058. [Google Scholar] [CrossRef]
- Hashemi, M.; Ghadyani, F.; Hasani, S.; Olyaee, Y.; Raei, B.; Khodadadi, M.; Ziyarani, M.; Basti, F.; Tavakolpournegari, A.; Matinahmadi, A.; et al. Nanoliposomes for doxorubicin delivery: Reversing drug resistance, stimuli-responsive carriers and clinical translation. J. Drug Deliv. Sci. Technol. 2023, 80, 104112. [Google Scholar] [CrossRef]



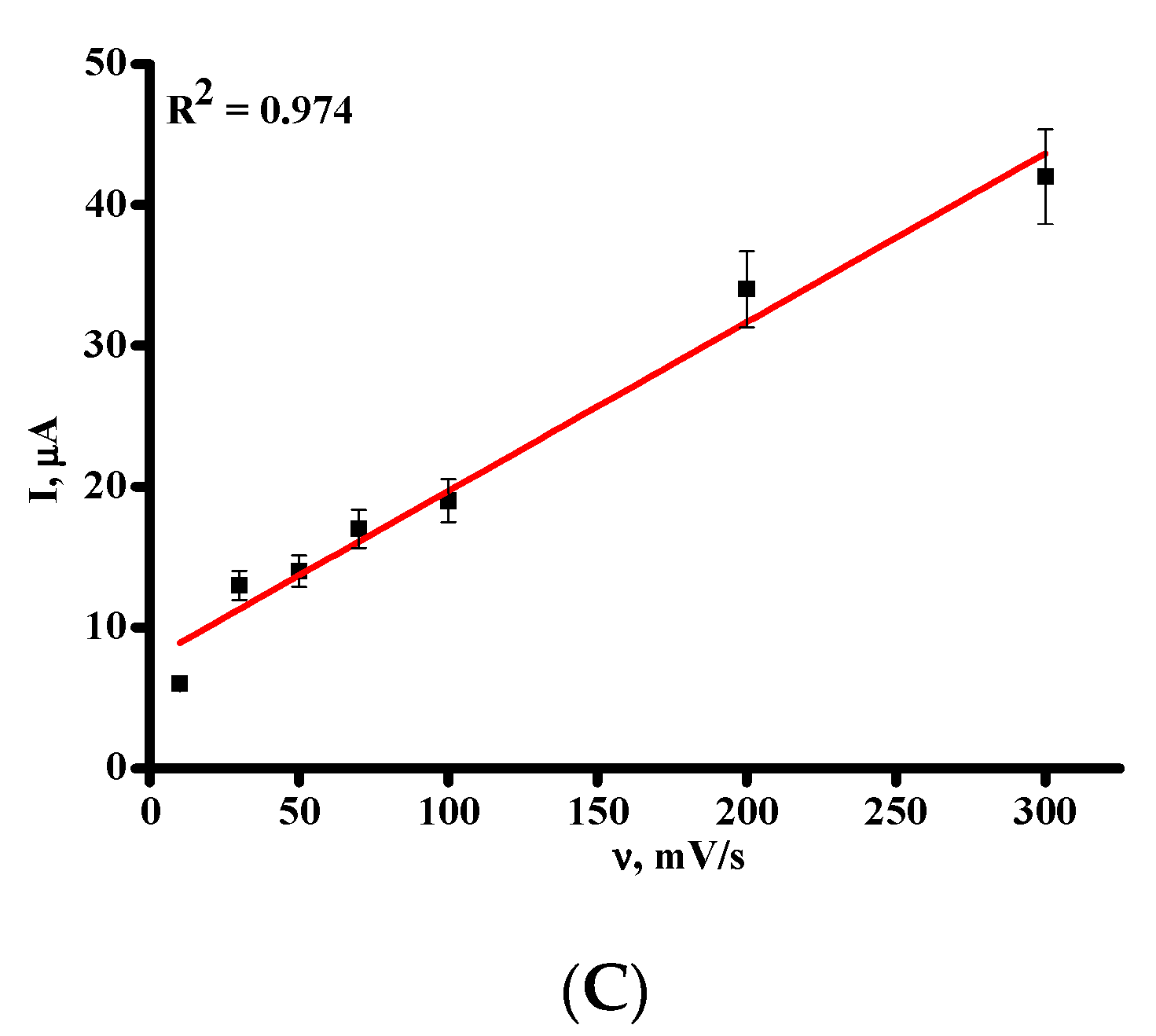
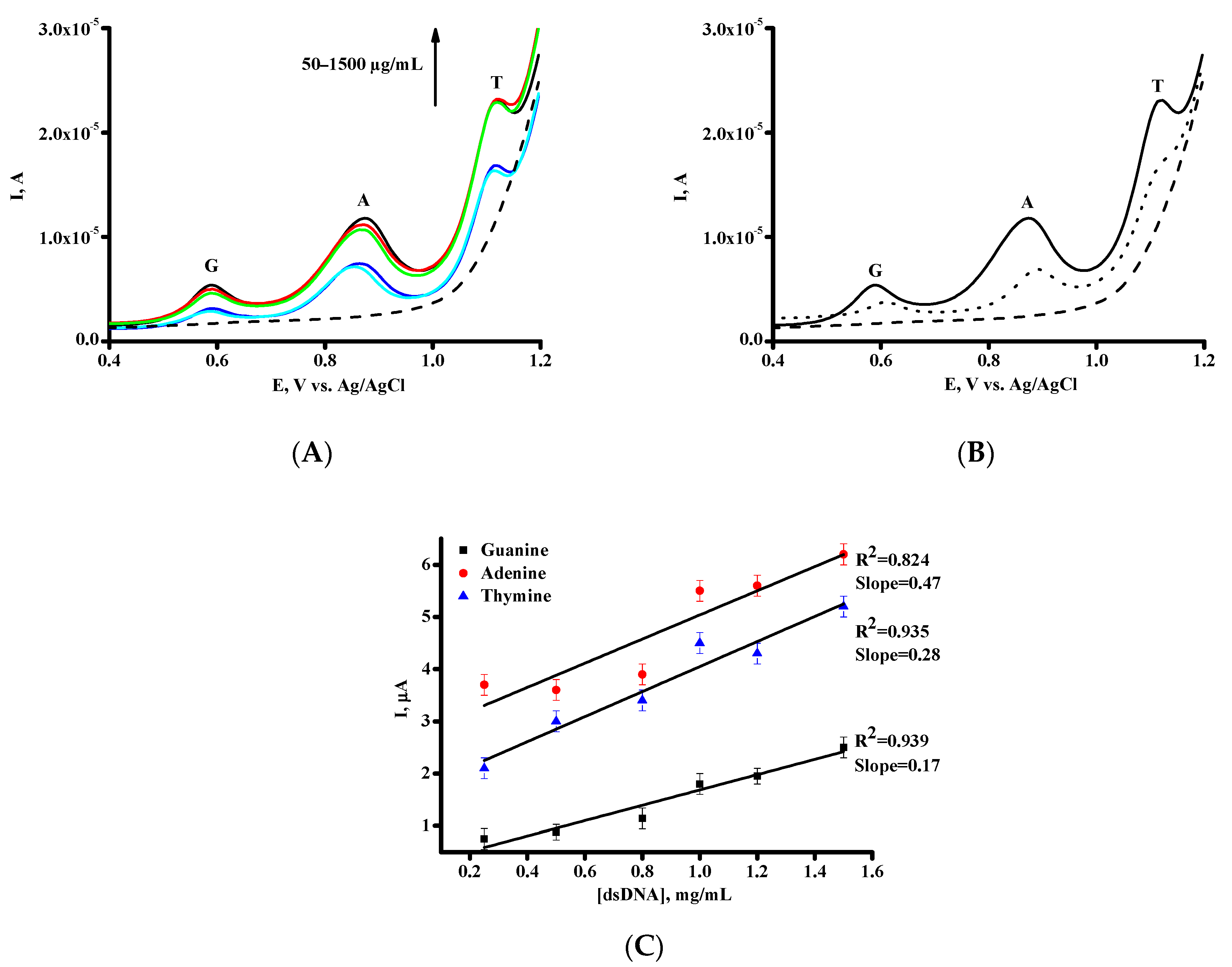
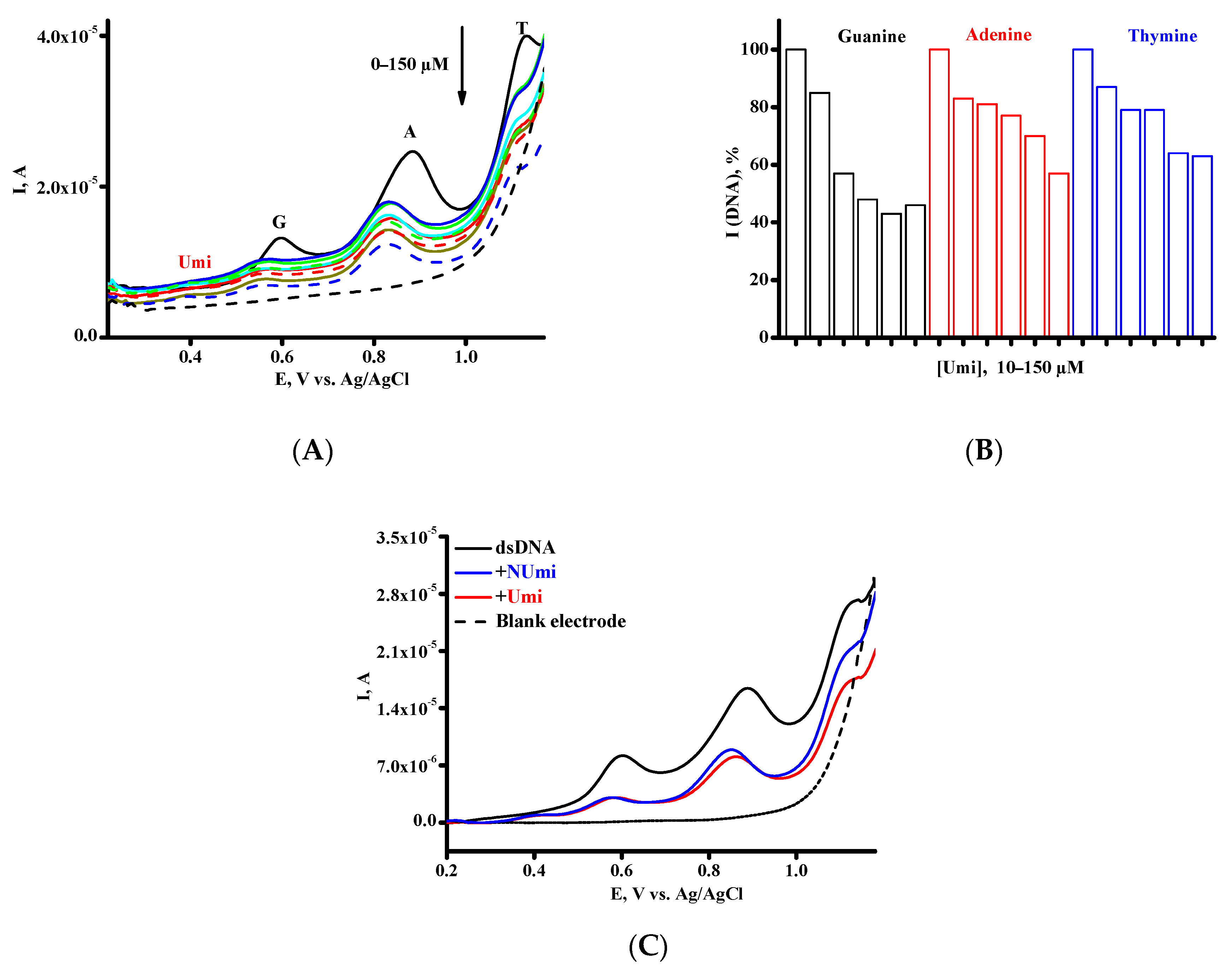

| Parameters | DPV |
|---|---|
| Eox, V | 0.388 ± 0.002 |
| Sensitivity, µA/µM (Slope) | 0.53 |
| Linear range, µM | 50–500 |
| Limit of quantification LOQ, µM | 50 |
| Limit of detection LOD, µM | 17 |
| Equation for linear regression * | Iox = 0.0062 ± 0.0002 [Umi] − 0.076 ± 0.059 |
| Correlation coefficient, R2 | 0.992 |
| Parameters | Guanine (G) | Adenine (A) | Thymine (T) |
|---|---|---|---|
| Eox, V | 0.59 ± 0.01 | 0.88 ± 0.01 | 1.12 ± 0.05 |
| Sensitivity, µA/(µg × mL−1) | 0.17 | 0.47 | 0.28 |
| Linear range, µg/mL | 50–1500 | 50–1500 | 50–1500 |
| Equation for linear regression * | IG = 1.47 ± 0.17 [dsDNA] + 0.22 ± 0.16 | IA = 2.31 ± 0.47 [dsDNA] + 2.72 ± 0.45 | IT = 2.40 ± 0.28 [dsDNA] + 1.65 ± 0.27 |
| Correlation coefficient, R2 | 0.939 | 0.824 | 0.935 |
| Drug-DNA Complex | Kb, M−1 | ||
|---|---|---|---|
| G Peak | A Peak | T Peak | |
| [Umi × DNA] | 2.8 × 104 | 5.7 × 104 | 4.7 × 104 |
| [NUmi × DNA] | 1.1 × 105 | 4.4 × 104 | 8.2 × 103 |
| Drug-DNA Complex | ΔG, kJ/mol | ||
|---|---|---|---|
| G | A | T | |
| [Umi × DNA] | −25.30 | −27.05 | −26.58 |
| [NUmi × DNA] | −28.68 | −26.41 | −22.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shumyantseva, V.V.; Bulko, T.V.; Agafonova, L.E.; Pronina, V.V.; Kostryukova, L.V. Comparative Analysis of the Interaction between the Antiviral Drug Umifenovir and Umifenovir Encapsulated in Phospholipids Micelles (Nanosome/Umifenovir) with dsDNA as a Model for Pharmacogenomic Analysis by Electrochemical Methods. Processes 2023, 11, 922. https://doi.org/10.3390/pr11030922
Shumyantseva VV, Bulko TV, Agafonova LE, Pronina VV, Kostryukova LV. Comparative Analysis of the Interaction between the Antiviral Drug Umifenovir and Umifenovir Encapsulated in Phospholipids Micelles (Nanosome/Umifenovir) with dsDNA as a Model for Pharmacogenomic Analysis by Electrochemical Methods. Processes. 2023; 11(3):922. https://doi.org/10.3390/pr11030922
Chicago/Turabian StyleShumyantseva, Victoria V., Tatiana V. Bulko, Lyubov E. Agafonova, Veronika V. Pronina, and Lyubov V. Kostryukova. 2023. "Comparative Analysis of the Interaction between the Antiviral Drug Umifenovir and Umifenovir Encapsulated in Phospholipids Micelles (Nanosome/Umifenovir) with dsDNA as a Model for Pharmacogenomic Analysis by Electrochemical Methods" Processes 11, no. 3: 922. https://doi.org/10.3390/pr11030922
APA StyleShumyantseva, V. V., Bulko, T. V., Agafonova, L. E., Pronina, V. V., & Kostryukova, L. V. (2023). Comparative Analysis of the Interaction between the Antiviral Drug Umifenovir and Umifenovir Encapsulated in Phospholipids Micelles (Nanosome/Umifenovir) with dsDNA as a Model for Pharmacogenomic Analysis by Electrochemical Methods. Processes, 11(3), 922. https://doi.org/10.3390/pr11030922








