Analysis of the Unintended Effects of the Bacillus thuringiensis Insecticidal Protein in Genetically Modified Rice Using Untargeted Transcriptomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction, PCR Analysis, and Test Strips
2.3. RNA Extraction, Library Preparation, and Sequencing
2.4. RNA-Seq Data Analysis
3. Results
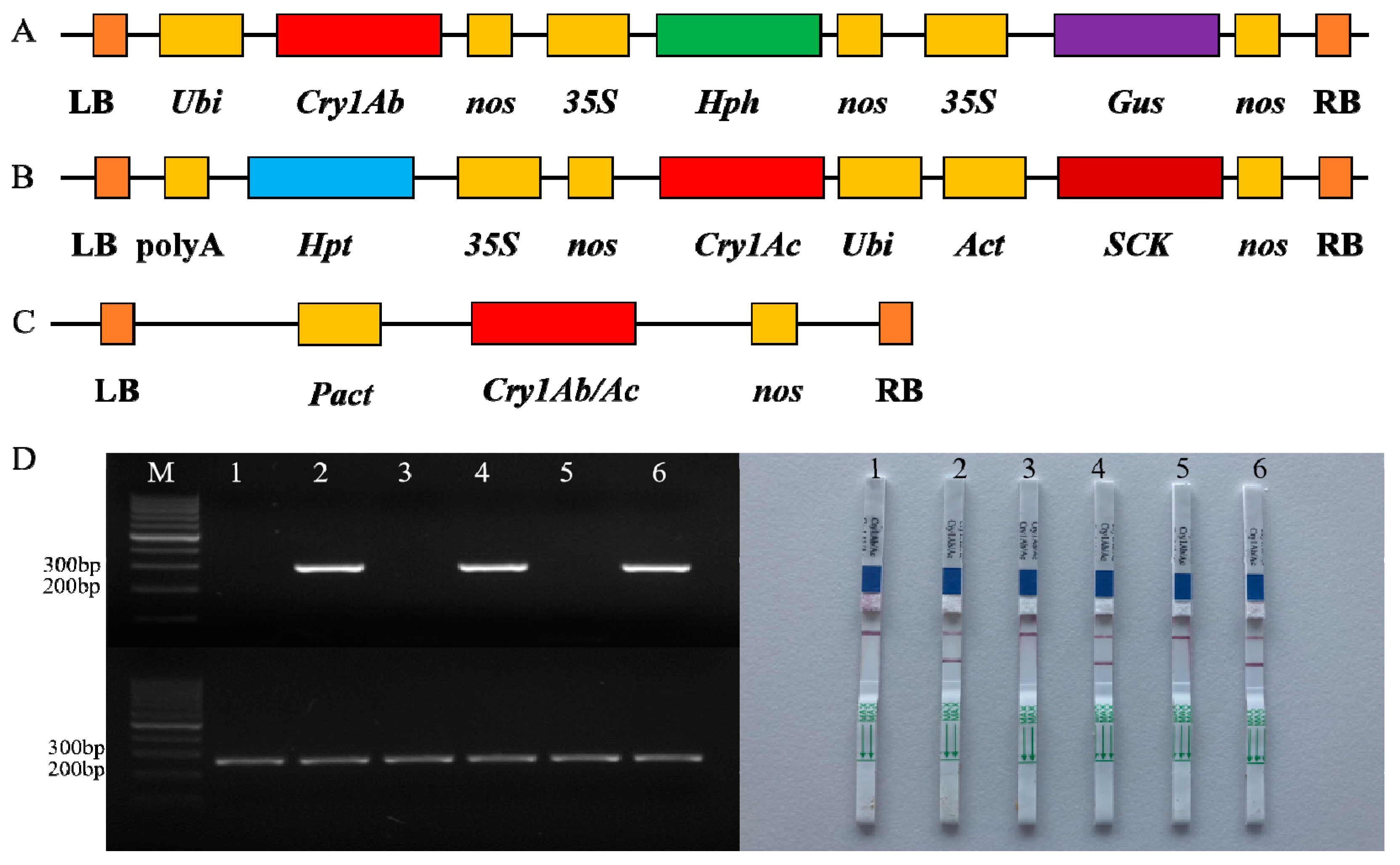
3.1. Bt Protein Expression in GM Rice
3.2. Evaluation of Rice Transcriptome Sequencing Data
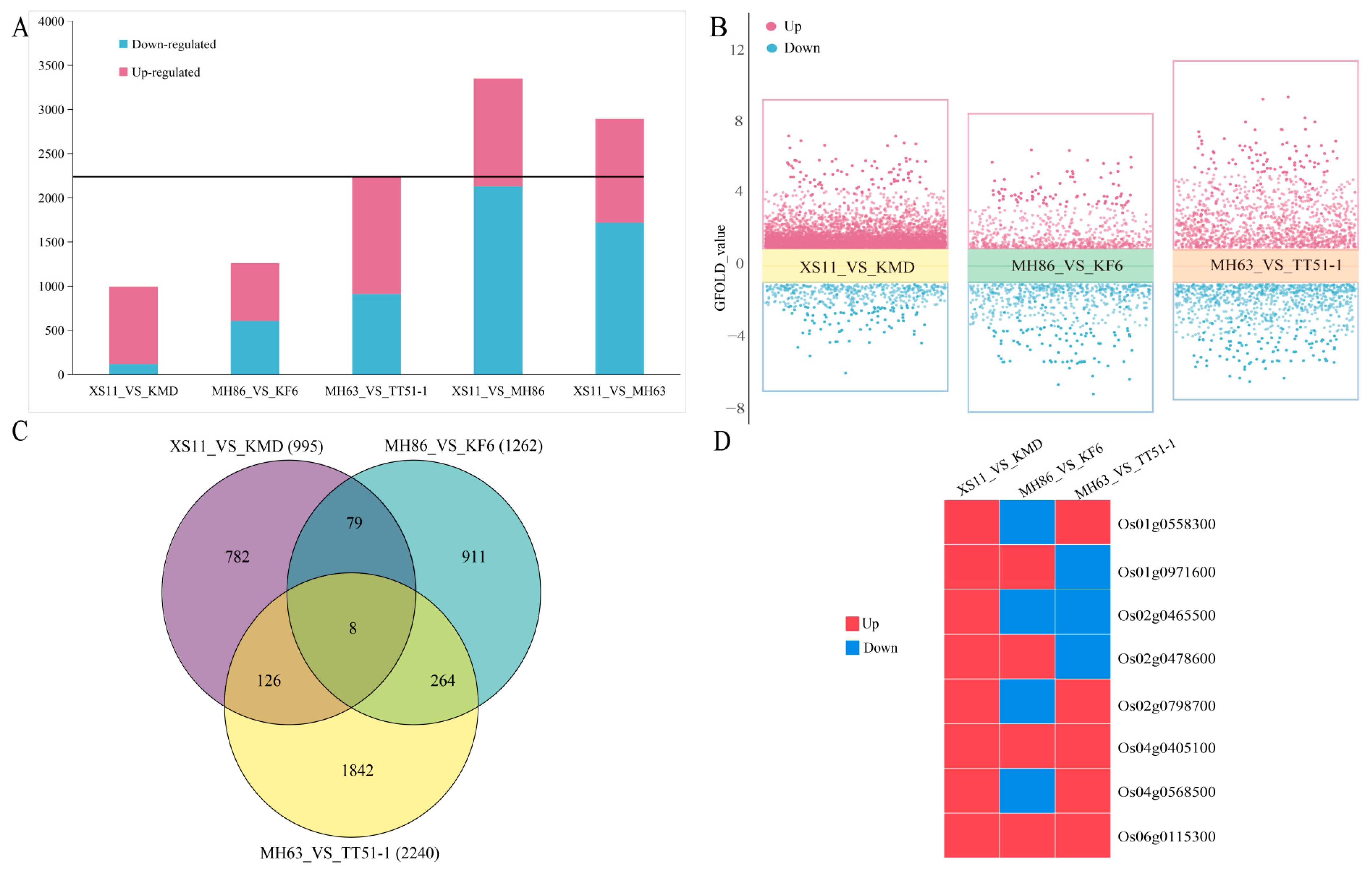
3.3. DEGs between the Transgenic and Non-Transgenic Rice Varieties
3.4. Identification of DEGs
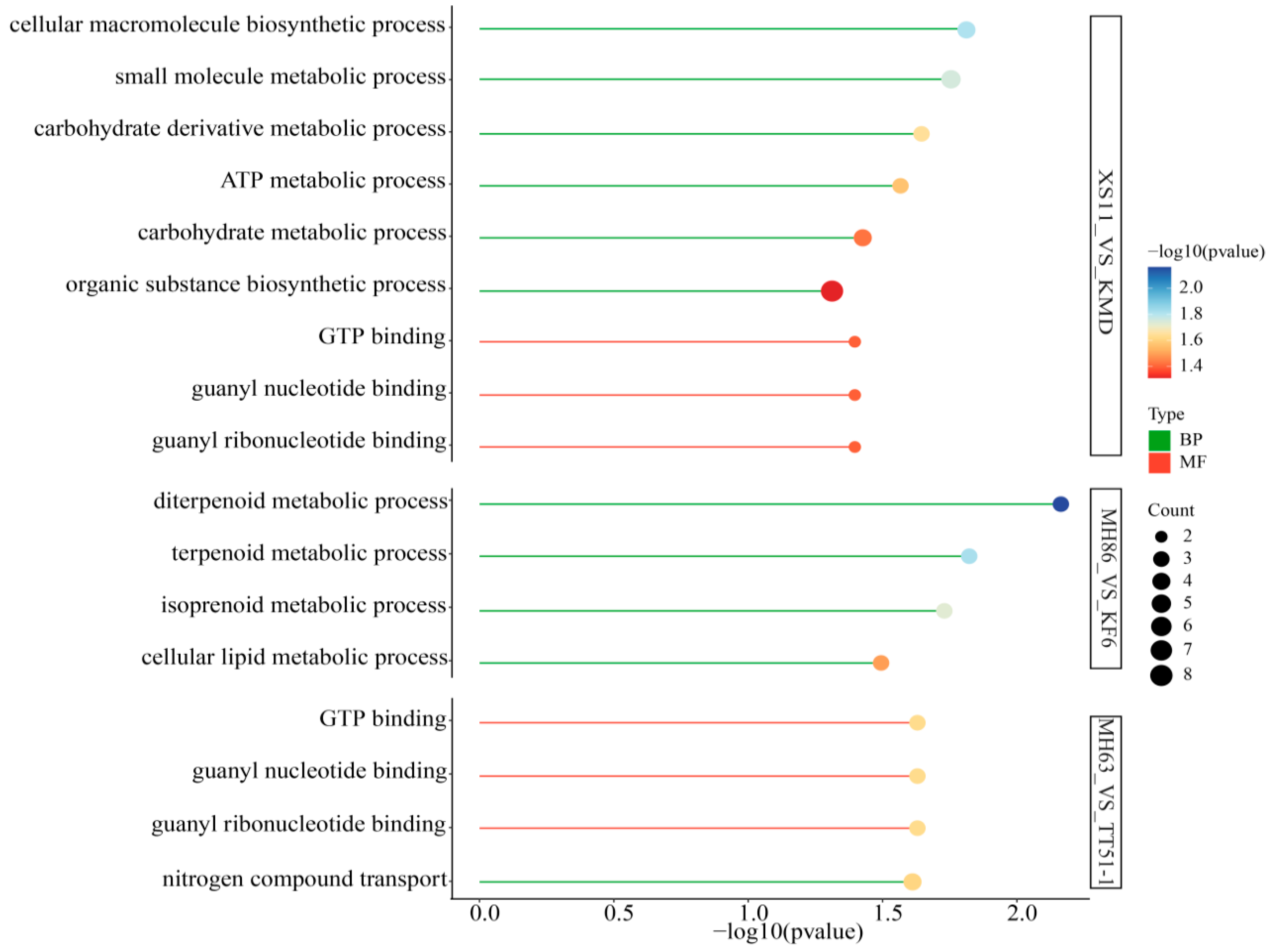
3.5. GO and KEGG Analysis of DEGs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Hallerman, E.M.; Liu, Q.; Wu, K.; Peng, Y. The development and status of Bt rice in China. Plant Biotechnol. J. 2016, 14, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, K.; He, X.; Zhu, B.; Liang, Z.; Li, H.; Luo, Y. Comparison of nutritional quality between Chinese indica rice with sck and cry1Ac genes and its nontransgenic counterpart. J. Food Sci. 2007, 72, S420–S424. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Hallerman, E.; Peng, Y.; Li, Y. Development of Bt Rice and Bt Maize in China and Their Efficacy in Target Pest Control. Int. J. Mol. Sci. 2016, 17, 1561. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Tan, Y.; Han, X.; Sun, Z.; Duan, M. Research Progress and Developing Tendency in Insect-resistant Transgenic Rice. Hybrid Rice 2011, 26, 1–6. [Google Scholar] [CrossRef]
- Chen, M.; Shelton, A.; Ye, G.Y. Insect-resistant genetically modified rice in China: From research to commercialization. Annu. Rev. Entomol. 2011, 56, 81–101. [Google Scholar] [CrossRef]
- Barros, E.; Lezar, S.; Anttonen, M.J.; van Dijk, J.P.; Rohlig, R.M.; Kok, E.J.; Engel, K.H. Comparison of two GM maize varieties with a near-isogenic non-GM variety using transcriptomics, proteomics and metabolomics. Plant Biotechnol. J. 2010, 8, 436–451. [Google Scholar] [CrossRef]
- McGaughey, W.H. Insect Resistance to the Biological Insecticide Bacillus thuringiensis. Science 1985, 229, 193–195. [Google Scholar] [CrossRef]
- Snow, A.A.; Palma, P.M. Commercialization of transgenic plants: Potential ecological risks. Bioscience 1997, 47, 86–96. [Google Scholar] [CrossRef]
- ISAAA. Global Status of Commercialized Biotech/GM Crops in 2019: Biotech Crops Drive SocioEconomic Development and Sustainable Environment in the New Frontier. In ISAAA Brief; No. 55; ISAAA: Ithaca, NY, USA, 2019. [Google Scholar]
- Li, P.; Xue, Y.; Shi, J.; Pan, A.; Tang, X.; Ming, F. The response of dominant and rare taxa for fungal diversity within different root environments to the cultivation of Bt and conventional cotton varieties. Microbiome 2018, 6, 184. [Google Scholar] [CrossRef]
- Deng, P.J.; Zhou, X.Y.; Yang, D.Y.; Hou, H.L.; Yang, X.K.; Li, Y.H.; Yang, Y.C.; Wang, X.L.; Fang, S.S.; Wu, S.Q.; et al. The definition, source, manifestation and assessment of unintended effects in genetically modified plants. J. Sci. Food Agric. 2008, 88, 2401–2413. [Google Scholar] [CrossRef]
- Kuiper, H.A.; Kleter, G.A.; Noteborn, H.P.; Kok, E.J. Assessment of the food safety issues related to genetically modified foods. Plant J. 2001, 27, 503–528. [Google Scholar] [CrossRef] [PubMed]
- Cellini, F.; Chesson, A.; Colquhoun, I.; Constable, A.; Davies, H.V.; Engel, K.H.; Gatehouse, A.M.; Karenlampi, S.; Kok, E.J.; Leguay, J.J.; et al. Unintended effects and their detection in genetically modified crops. Food Chem. Toxicol. 2004, 42, 1089–1125. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ye, S.; Chen, J.; Wang, L.; Li, Y.; Ge, L.; Wu, G.; Song, L.; Wang, C.; Sun, Y.; et al. Combined metagenomic and metabolomic analyses reveal that Bt rice planting alters soil C-N metabolism. ISME Commun. 2023, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Krause, S.M.B.; Nather, A.; Ortiz Cortes, V.; Mullins, E.; Kessel, G.J.T.; Lotz, L.A.P.; Tebbe, C.C. No Tangible Effects of Field-Grown Cisgenic Potatoes on Soil Microbial Communities. Front. Bioeng. Biotechnol. 2020, 8, 603145. [Google Scholar] [CrossRef] [PubMed]
- Batista, R.; Saibo, N.; Lourenco, T.; Oliveiro, M.M. Microarray analyses reveal that plant mutagenesis may induce more transcriptomic changes than transgene insertion. Proc. Natl. Acad. Sci. USA 2008, 105, 3640–3645. [Google Scholar] [CrossRef]
- Coll, A.; Nadal, A.; Collado, R.; Capellades, G.; Kubista, M.; Messeguer, J.; Pla, M. Natural variation explains most transcriptomic changes among maize plants of MON810 and comparable non-GM varieties subjected to two N-fertilization farming practices. Plant Mol. Biol. 2010, 73, 349–362. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Kawahara, Y.; Itoh, T.; Takaiwa, F. A Whole-Genome Analysis of a Transgenic Rice Seed-Based Edible Vaccine Against Cedar Pollen Allergy. DNA Res. 2013, 20, 623–631. [Google Scholar] [CrossRef][Green Version]
- Coll, A.; Nadal, A.; Rossignol, M.; Puigdomenech, P.; Pla, M. Proteomic analysis of MON810 and comparable non-GM maize varieties grown in agricultural fields. Transgenic Res. 2011, 20, 939–949. [Google Scholar] [CrossRef]
- Koh, J.; Chen, G.; Yoo, M.J.; Zhu, N.; Dufresne, D.; Erickson, J.E.; Shao, H.; Chen, S. Comparative Proteomic Analysis of Brassica napus in Response to Drought Stress. J. Proteome Res. 2015, 14, 3068–3081. [Google Scholar] [CrossRef]
- Okazaki, Y.; Saito, K. Recent advances of metabolomics in plant biotechnology. Plant Biotechnol. Rep. 2012, 6, 1–15. [Google Scholar] [CrossRef]
- Catchpole, G.S.; Beckmann, M.; Enot, D.P.; Mondhe, M.; Zywicki, B.; Taylor, J.; Hardy, N.; Smith, A.; King, R.D.; Kell, D.B.; et al. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modifiedand conventional potato crops. Proc. Natl. Acad. Sci. USA 2005, 102, 14458–14462. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, C.; Xu, H.; Yuan, K.; Lu, X.; Zhu, Z.; Wu, Y.; Xu, G. Metabolic profiling of transgenic rice with cryIAc and sck genes: An evaluation of unintended effects at metabolic level by using GC-FID and GC-MS. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2009, 877, 725–732. [Google Scholar] [CrossRef]
- Wang, X.J.; Zhang, X.; Yang, J.T.; Wang, Z.X. Effect on transcriptome and metabolome of stacked transgenic maize containing insecticidal cry and glyphosate tolerance epsps genes. Plant J. 2018, 93, 1007–1016. [Google Scholar] [CrossRef]
- Ladics, G.S.; Bartholomaeus, A.; Bregitzer, P.; Doerrer, N.G.; Gray, A.; Holzhauser, T.; Jordan, M.; Keese, P.; Kok, E.; Macdonald, P.; et al. Genetic basis and detection of unintended effects in genetically modified crop plants. Transgenic Res. 2015, 24, 587–603. [Google Scholar] [CrossRef] [PubMed]
- Schnell, J.; Steele, M.; Bean, J.; Neuspiel, M.; Girard, C.; Dormann, N.; Pearson, C.; Savoie, A.; Bourbonniere, L.; Macdonald, P. A comparative analysis of insertional effects in genetically engineered plants: Considerations for pre-market assessments. Transgenic Res. 2015, 24, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bednář, M. DNA microarray technology and application. Diagn. Med. Technol. 2000, 6, 796–800. [Google Scholar]
- Velculescu, V.E.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Serial Analysis of Gene Expression. Science 1995, 270, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Zhu, T.; Li, B.; Li, T. Application research of combined transcriptome with metabolome in plants. J. Shanxi Agric. Univ. 2022, 42, 1–13. [Google Scholar] [CrossRef]
- Peng, C.; Chen, X.; Wang, X.; Xu, X.; Wei, W.; Wang, C.; Xu, J. Comparative analysis of miRNA expression profiles in transgenic and non-transgenic rice using miRNA-Seq. Sci. Rep. 2018, 8, 338. [Google Scholar] [CrossRef]
- Guo, B.F.; Hong, H.L.; Sun, L.P.; Guo, Y.; Qiu, L.J. Transcriptome analysis reveals differing response and tolerance mechanism of EPSPS and GAT genes among transgenic soybeans. Mol. Biol. Rep. 2021, 48, 7351–7360. [Google Scholar] [CrossRef]
- Shu, Q.; Ye, G.; Cui, H.; Xiang, Y.; Gao, M. Development of Transgenic Bacillus thuriengiensis Rice Resistant to Rice Stem Borers and Leaf Folders. J. Zhejiang Agric. Univ. 1998, 24, 579–580. [Google Scholar]
- Shu, Q.; Ye, G.; Cui, H.; Cheng, X.; Xiang, Y.; Wu, D.; Gao, M.; Xia, Y.; Hu, C.; Sardana, R.; et al. Transgenic rice plants with a synthetic cry1Ab gene from Bacillus thuringiensis were highly resistant to eight lepidopteran rice pest species. Mol. Breed. 2000, 6, 433–439. [Google Scholar] [CrossRef]
- Xiang, Y.; Liang, Z.; Gao, M.; Shu, Q.; Ye, G.; Cheng, X.; Altosaar, I. Agrobacterium-mediated Transformation of Insecticidal Bacillus thuringiensis cryIA(b) and cryIA(c) Genes and Their Expression in Rice. Chin. J. Biotechnol. 1999, 15, 495–500. [Google Scholar] [CrossRef]
- Rong, J.; Song, Z.; Su, J.; Xia, H.; Lu, B.R.; Wang, F. Low frequency of transgene flow from Bt/CpTI rice to its nontransgenic counterparts planted at close spacing. New Phytol. 2005, 168, 559–566. [Google Scholar] [CrossRef]
- Tu, J.; Zhang, G.; Datta, K.; Xu, C.; He, Y.; Zhang, Q.; Khush, G.S.; Datta, S.K. Field performance of transgenic elite commercial hybrid rice expressing Bacillus thuringiensis δ-endotoxin. Nat. Biotechnol. 2000, 18, 1101–1104. [Google Scholar] [CrossRef]
- Feng, J.; Meyer, C.A.; Wang, Q.; Liu, J.S.; Shirley Liu, X.; Zhang, Y. GFOLD: A generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 2012, 28, 2782–2788. [Google Scholar] [CrossRef]
- Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; Harris, M.A.; Hill, D.P.; Issel-Tarver, L.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Miki, B.; Abdeen, A.; Manabe, Y.; MacDonald, P. Selectable marker genes and unintended changes to the plant transcriptome. Plant Biotechnol. J. 2009, 7, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yang, X.; Tzin, V.; Peng, Y.; Romeis, J.; Li, Y. Plant breeding involving genetic engineering does not result in unacceptable unintended effects in rice relative to conventional cross-breeding. Plant J. 2020, 103, 2236–2249. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Kato, K.; Asuka, H.; Sugioka, Y.; Mochizuki, T.; Nishiuchi, T.; Miyahara, T.; Kodama, H.; Ohta, D. Multi-omics Analyses of Non-GM Tomato Scion Engrafted on GM Rootstocks. Food Safety 2023, 11, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Miyahara, T.; Nishiuchi, T.; Fujikawa, N.; Oguchi, T.; Kikuchi, A.; Taoka, K.-I.; Ogawa, T.; Honda, K.; Yamaguchi, Y.; Mochizuki, T.; et al. Omics Profiles of Non-GM Tubers from Transgrafted Potato with a GM Scion. Food Saf. 2023, 11, D-22-00010. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, W.; Li, Y.; Liu, F.; Han, W.; Li, J. Transcriptomic and proteomic responses to brown plant hopper (Nilaparvata lugens) in cultivated and Bt-transgenic rice (Oryza sativa) and wild rice (O. rufipogon). J. Proteom. 2021, 232, 104051. [Google Scholar] [CrossRef]
- Wang, C.; Lu, W.; Lin, Y.; Luo, J. Development and Application of Transcriptome Sequencing. Eucalypt Sci. Technol. 2018, 35, 20–26. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Li, L.; Yang, Y.; Ding, Y.; Guan, H.; Wang, X.; Zhang, A.; Wen, H. Negligible transcriptome and metabolome alterations in RNAi insecticidal maize against Monolepta hieroglyphica. Plant Cell Rep. 2020, 39, 1539–1547. [Google Scholar] [CrossRef]
- Kogel, K.H.; Voll, L.M.; Schafer, P.; Jansen, C.; Wu, Y.; Langen, G.; Imani, J.; Hofmann, J.; Schmiedl, A.; Sonnewald, S.; et al. Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific variances. Proc. Natl. Acad. Sci. USA 2010, 107, 6198–6203. [Google Scholar] [CrossRef]
- Long, Y.; Xu, W.; Liu, C.; Dong, M.; Pei, X.; Chen, R.; Jin, W.; Liu, W.; Li, L. Genetically modified soybean lines exhibit less transcriptomic variation compared to natural varieties. GM Crops Food 2023, 14, 1–11. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, J.; Li, Y.; Chen, X.; Jian, G.; Peng, Y.; Qi, F. Transcriptome Analysis Revealing the Absence of Unintended Effects Both in Aerial and Underground Portions of Transgenic Rice (Oryza sativa L.) Huahui 1. J. Agric. Biotechnol. 2015, 23, 876–887. [Google Scholar]
- Faergeman, N.J.; Wadum, M.; Feddersen, S.; Burton, M.; Kragelund, B.B.; Knudsen, J. Acyl-CoA binding proteins; structural and functional conservation over 2000 MYA. Mol. Cell Biochem. 2007, 299, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Xiao, S.; Li, H.Y.; Tsao, S.W.; Chye, M.L. Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with heavy-metal-binding farnesylated protein AtFP6. New Phytol. 2009, 181, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Gao, W.; Chen, Q.F.; Ramalingam, S.; Chye, M.L. Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. Plant J. 2008, 54, 141–151. [Google Scholar] [CrossRef]
- Chye, M.L.; Huang, B.Q.; Zee, S.Y. Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J. 1999, 18, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.F.; Xiao, S.; Qi, W.; Mishra, G.; Ma, J.; Wang, M.; Chye, M.L. The Arabidopsis acbp1 acbp2 double mutant lacking acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytol. 2010, 186, 843–855. [Google Scholar] [CrossRef]
- Meng, W.; Su, Y.C.F.; Saunders, R.M.K.; Chye, M.L. The rice acyl-CoA-binding protein gene family: Phylogeny, expression and functional analysis. New Phytol. 2011, 189, 1170–1184. [Google Scholar] [CrossRef] [PubMed]
- Coll, A.; Nadal, A.; Collado, R.; Capellades, G.; Messeguer, J.; Mele, E.; Palaudelmas, M.; Pla, M. Gene expression profiles of MON810 and comparable non-GM maize varieties cultured in the field are more similar than are those of conventional lines. Transgenic Res. 2009, 18, 801–808. [Google Scholar] [CrossRef]
- Metzdorff, S.B.; Kok, E.J.; Knuthsen, P.; Pedersen, J. Evaluation of a non-targeted “omic” approach in the safety assessment of genetically modified plants. Plant Biol. 2006, 8, 662–672. [Google Scholar] [CrossRef]
- Ricroch, A.E.; Berge, J.B.; Kuntz, M. Evaluation of genetically engineered crops using transcriptomic, proteomic, and metabolomic profiling techniques. Plant Physiol. 2011, 155, 1752–1761. [Google Scholar] [CrossRef]



| Samples | XS11 | KMD | MH86 | KF6 | MH63 | TT51-1 |
|---|---|---|---|---|---|---|
| Raw reads | 12,112,186 | 12,632,876 | 12,336,914 | 12,370,722 | 12,390,376 | 12,301,398 |
| Clean reads | 10,463,208 | 11,262,974 | 11,164,718 | 12,213,470 | 11,011,248 | 10,927,996 |
| Clean reads rate (%) | 86.39 | 89.16 | 90.50 | 98.73 | 88.87 | 88.84 |
| Mapped reads | 8,936,099 | 9,805,073 | 9,361,273 | 10,411,687 | 9,166,135 | 9,184,202 |
| Mapped rate (%) | 85.40 | 87.06 | 83.85 | 85.25 | 83.24 | 84.04 |
| Unmapped reads | 1,527,109 | 1,457,901 | 1,803,445 | 1,801,783 | 1,845,113 | 1,743,794 |
| Unmapped rate (%) | 14.60 | 12.94 | 16.15 | 14.75 | 16.76 | 15.96 |
| GC (%) | 52 | 47 | 52 | 51 | 52 | 52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, L.; Chen, G.; Chen, X.; Wang, X.; Lu, Y.; Liang, Z.; Xu, J.; Peng, C. Analysis of the Unintended Effects of the Bacillus thuringiensis Insecticidal Protein in Genetically Modified Rice Using Untargeted Transcriptomics. Processes 2023, 11, 3202. https://doi.org/10.3390/pr11113202
Ding L, Chen G, Chen X, Wang X, Lu Y, Liang Z, Xu J, Peng C. Analysis of the Unintended Effects of the Bacillus thuringiensis Insecticidal Protein in Genetically Modified Rice Using Untargeted Transcriptomics. Processes. 2023; 11(11):3202. https://doi.org/10.3390/pr11113202
Chicago/Turabian StyleDing, Lin, Guanwei Chen, Xiaoyun Chen, Xiaofu Wang, Yuwen Lu, Zehui Liang, Junfeng Xu, and Cheng Peng. 2023. "Analysis of the Unintended Effects of the Bacillus thuringiensis Insecticidal Protein in Genetically Modified Rice Using Untargeted Transcriptomics" Processes 11, no. 11: 3202. https://doi.org/10.3390/pr11113202
APA StyleDing, L., Chen, G., Chen, X., Wang, X., Lu, Y., Liang, Z., Xu, J., & Peng, C. (2023). Analysis of the Unintended Effects of the Bacillus thuringiensis Insecticidal Protein in Genetically Modified Rice Using Untargeted Transcriptomics. Processes, 11(11), 3202. https://doi.org/10.3390/pr11113202









