Phytochemicals from Piper betle (L.) as Putative Modulators of a Novel Network-Derived Drug Target for Coronary Artery Disease: An In Silico Study
Abstract
:1. Introduction
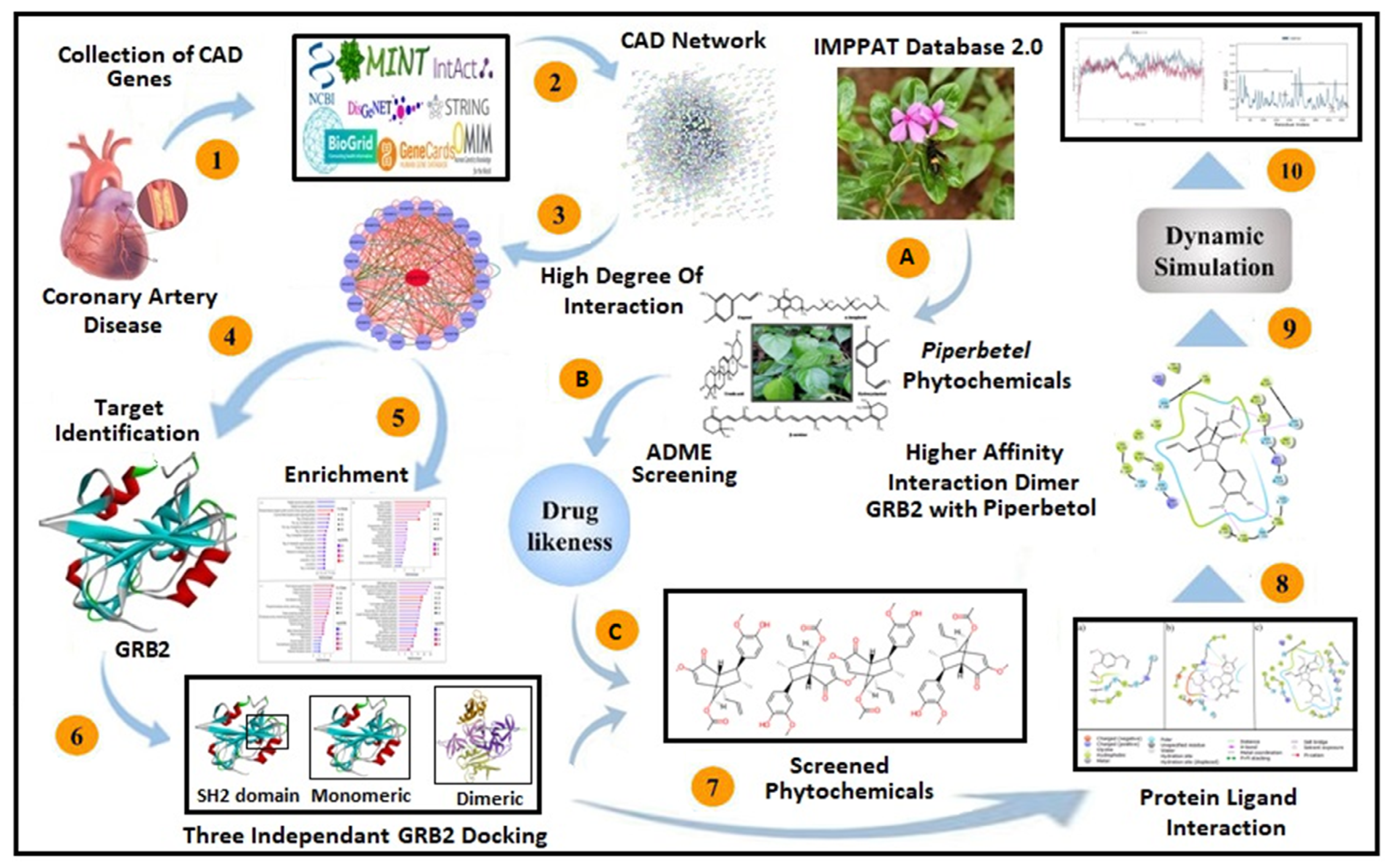
2. Materials and Methods
2.1. Collection and Construction of the CAD Network
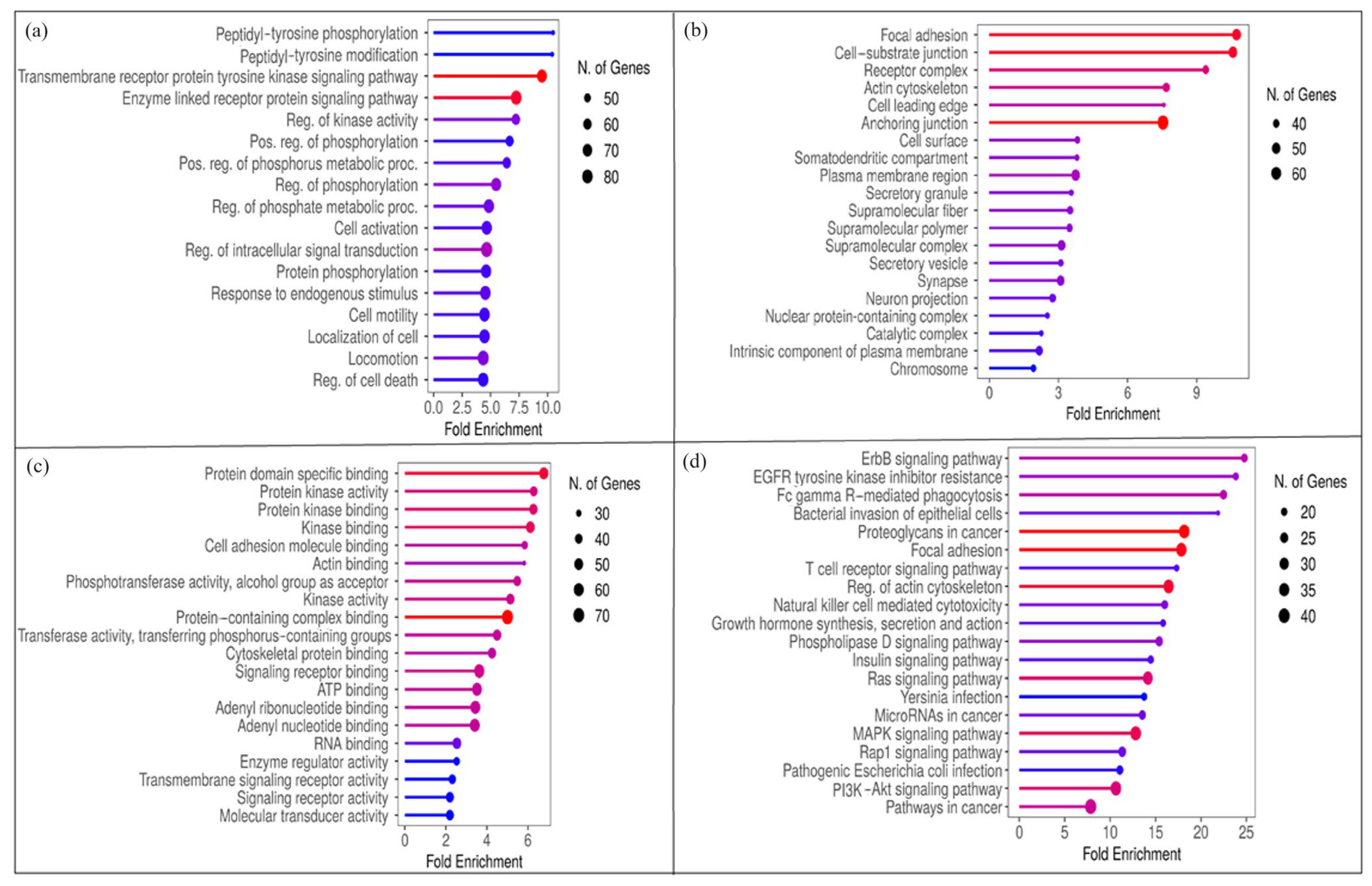
2.2. Network Assessment and Enrichment Analysis
2.3. Ligand Collection and ADMET Profiling
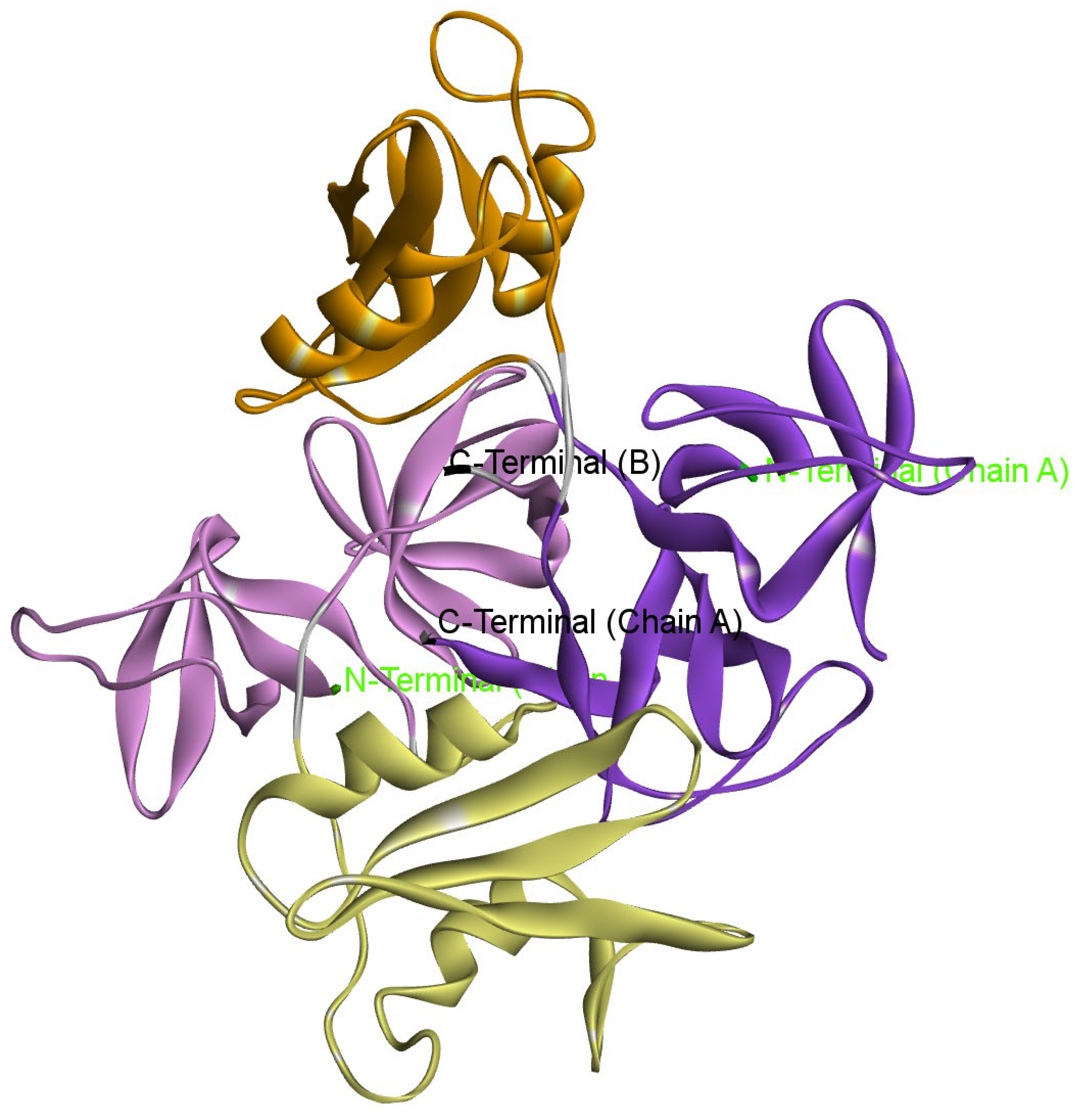
2.4. Protein Preparation
2.5. Molecular Docking
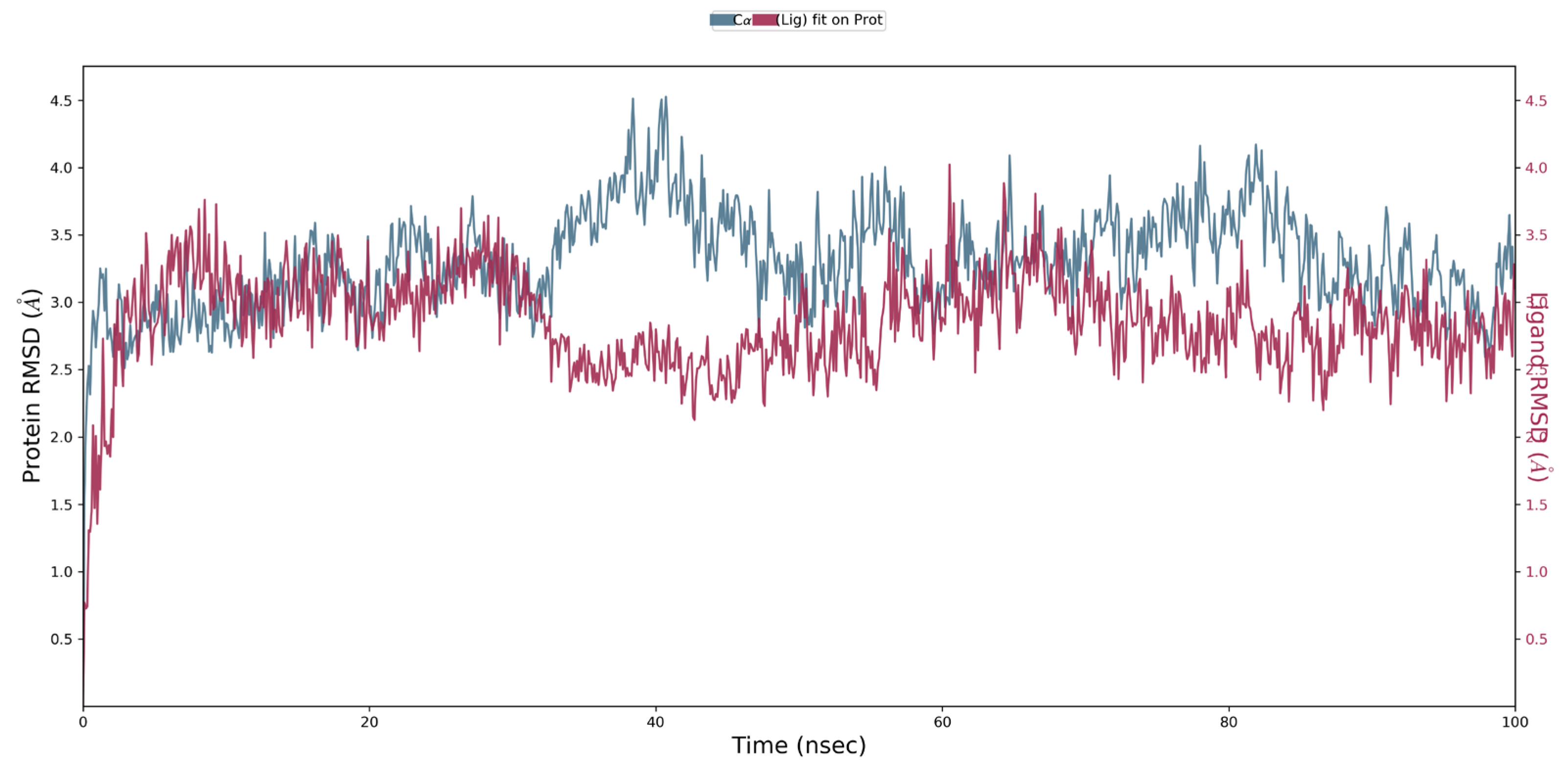
2.6. Molecular Dynamics Simulation
3. Results
3.1. Target Selection and Enrichment Analysis
3.2. ADMET and Docking Analysis
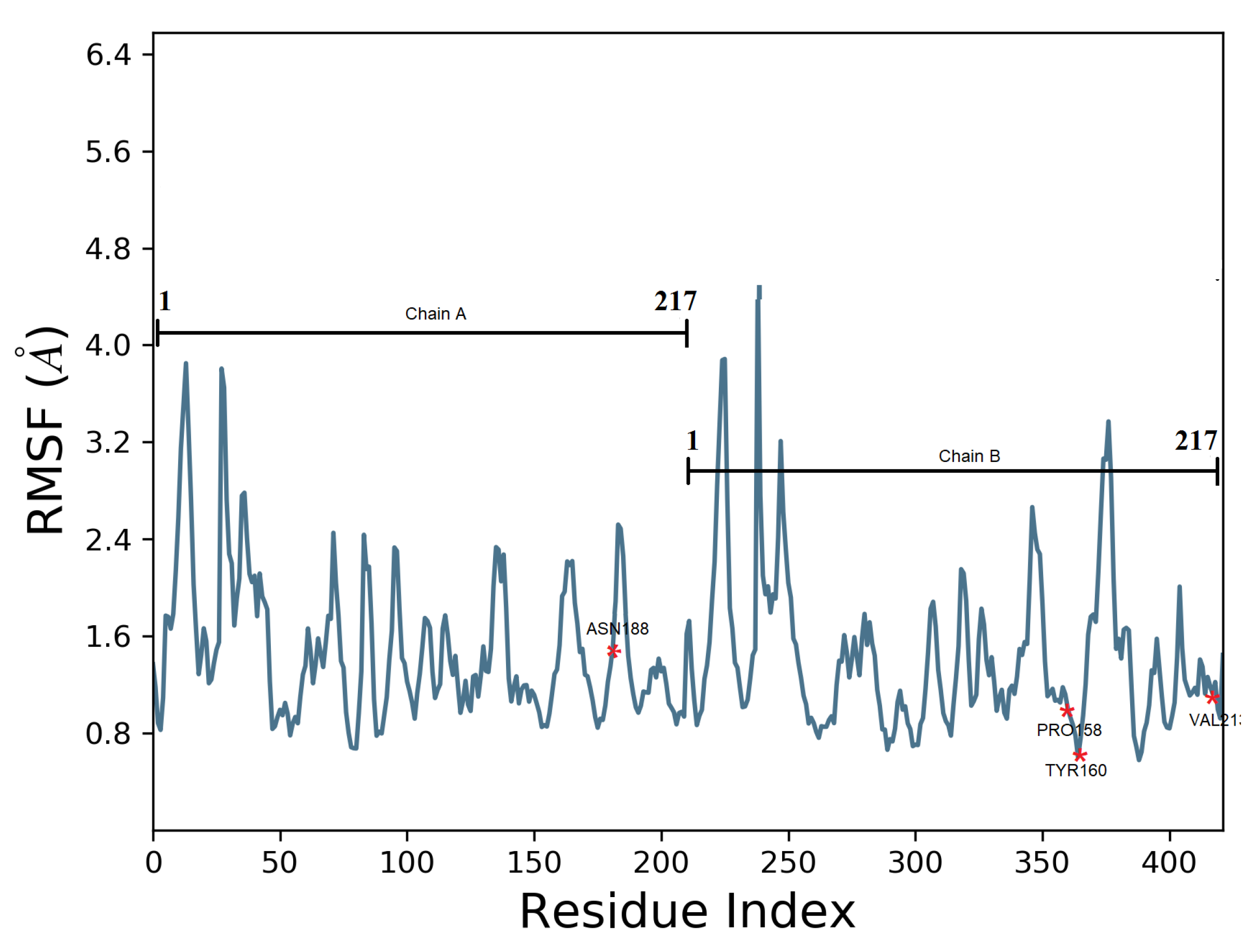
3.3. MD Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monisha, K.G.; Prabu, P.; Chokkalingam, M.; Murugesan, R.; Milenkovic, D.; Ahmed, S.S.S.J. Clinical utility of brain-derived neurotrophic factor as a biomarker with left ventricular echocardiographic indices for potential diagnosis of coronary artery disease. Sci. Rep. 2020, 10, 16359. [Google Scholar] [CrossRef]
- Ralapanawa, U.; Sivakanesan, R. Epidemiology and the magnitude of coronary artery disease and acute coronary syndrome: A narrative review. J. Epidemiol. Glob. Health 2021, 11, 169. [Google Scholar] [CrossRef]
- Hajar, R. Risk factors for coronary artery disease: Historical perspectives. Heart Views 2017, 18, 109. [Google Scholar] [CrossRef]
- Findley, A.S.; Richards, A.L.; Petrini, C.; Alazizi, A.; Doman, E.; Shanku, A.G.; Davis, G.O.; Hauff, N.; Sorokin, Y.; Wen, X.; et al. Interpreting coronary artery disease risk through gene–environment interactions in gene regulation. Genetics 2019, 213, 651–663. [Google Scholar] [CrossRef]
- Tantry, U.S.; Navarese, E.P.; Bliden, K.P.; Gurbel, P.A. Acetylsalicylic acid and clopidogrel hyporesponsiveness following acute coronary syndromes. Kardiol. Pol. 2018, 76, 1312–1319. [Google Scholar] [CrossRef]
- Shaito, A.; Thuan, D.T.; Phu, H.T.; Nguyen, T.H.; Hasan, H.; Halabi, S.; Abdelhady, S.; Nasrallah, G.K.; Eid, A.H.; Pintus, G. Herbal medicine for cardiovascular diseases: Efficacy, mechanisms, and safety. Front. Pharmacol. 2020, 7, 422. [Google Scholar] [CrossRef] [PubMed]
- Sofowora, A.; Ogunbodede, E.; Onayade, A. The role and place of medicinal plants in the strategies for disease prevention. Afr. J. Tradit. Complement. Altern. Med. 2013, 10, 210–229. [Google Scholar] [CrossRef]
- Nayaka, N.M.; Sasadara, M.M.; Sanjaya, D.A.; Yuda, P.E.; Dewi, N.L.; Cahyaningsih, E.; Hartati, R. Piper betle (L): Recent review of antibacterial and antifungal properties, safety profiles, and commercial applications. Molecules 2021, 26, 2321. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, S.; Ravichandran, D.; Lakshmanan, D.K.; Ravichandran, G.; Arumugam, V.; Raju, K.; Geetha, K.; Thilagar, S. Evaluation of anti-rheumatic activity of Piper betle L. (Betelvine) extract using in silico, in vitro and in vivo approaches. Bio. Org. Chem. 2020, 103, 104227. [Google Scholar] [CrossRef]
- Ng, P.L.; Rajab, N.F.; Then, S.M.; Yusof, Y.A.; Ngah, W.Z.; Pin, K.Y.; Looi, M.L. Piper betle leaf extract enhances the cytotoxicity effect of 5-fluorouracil in inhibiting the growth of HT29 and HCT116 colon cancer cells. J. Zhejiang Univ. Sci. B 2014, 15, 692. [Google Scholar] [CrossRef]
- Arya, D.S.; Arora, S.; Malik, S.; Nepal, S.; Kumari, S.; Ojha, S. Effect of Piper betle on cardiac function, marker enzymes, and oxidative stress in isoproterenol-induced cardiotoxicity in rats. Toxicol. Mech. Methods 2010, 20, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Mohanraj, K.; Karthikeyan, B.S.; Vivek-Ananth, R.P.; Chand, R.P.B.; Aparna, S.R.; Mangalapandi, P.; Samal, A. IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry and Therapeutics. Sci. Rep. 2018, 8, 4329. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comp. Chem. 2005, 26, 1752. [Google Scholar] [CrossRef]
- Schrödinger. QikProp User Manual; Schrödinger Press: Cambridge, MA, USA, 2015; Available online: http://gohom.win/ManualHom/Schrodinger/Schrodinger_2015-2_docs/qikprop/qikprop_user_manual.pdf (accessed on 2 August 2023).
- Maignan, S.; Guilloteau, J.P.; Fromage, N.; Arnoux, B.; Becquart, J.; Ducruix, A. Crystal structure of the mammalian Grb2 adaptor. Science 1995, 268, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Nioche, P.; Liu, W.Q.; Broutin, I.; Charbonnier, F.; Latreille, M.T.; Vidal, M.; Roques, B.; Garbay, C.; Ducruix, A. Crystal structures of the SH2 domain of Grb2: Highlight on the binding of a new high-affinity inhibitor. J. Mol. Biol. 2002, 315, 1167–1177. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Shaw, D.E. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 84-es. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Evans, D.J.; Holian, B.L. The Nose-Hoover Thermostat. J. Chem. Phys. 1985, 83, 4069–4074. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Pello Lazaro, A.M.; Blanco-Colio, L.M.; Franco Pelaez, J.A.; Tunon, J. Anti-Inflammatory drugs in patients with ischemic heart disease. J. Clin. Med. 2021, 10, 2835. [Google Scholar] [CrossRef]
- Serafini, M.; Peluso, I. Functional foods for health: The interrelated antioxidant and anti-inflammatory role of fruits, vegetables, herbs, spices and cocoa in humans. Curr. Pharm. Des. 2016, 22, 6701–6715. [Google Scholar] [CrossRef]
- Seo, J.; Lee, U.; Seo, S.; Wibowo, A.E.; Pongtuluran, O.B.; Lee, K.; Han, S.B.; Cho, S. Anti-inflammatory and antioxidant activities of methanol extract of Piper betle Linn. (Piper betle L.) leaves and stems by inhibiting NF-κB/MAPK/Nrf2 signaling pathways in RAW 264.7 macrophages. Biomed. Pharmacother. 2022, 155, 113734. [Google Scholar] [CrossRef]
- Biswas, P.; Anand, U.; Saha, S.C.; Kant, N.; Mishra, T.; Masih, H.; Bar, A.; Pandey, D.K.; Jha, N.K.; Majumder, M.; et al. Betelvine (Piper betle L.): A comprehensive insight into its ethnopharmacology, phytochemistry, and pharmacological, biomedical and therapeutic attributes. J. Cell. Mol. Med. 2022, 26, 3083–3119. [Google Scholar] [CrossRef]
- Alam, B.; Akter, F.; Parvin, N.; Pia, R.S.; Akter, S.; Chowdhury, J.; Sifath-E-Jahan, K.; Haque, E. Antioxidant, analgesic and anti-inflammatory activities of the methanolic extract of Piper betle leaves. Avicenna J. Phytomed. 2013, 3, 112. [Google Scholar]
- Dasgupta, N.; De, B. Antioxidant activity of Piper betle L. leaf extract in vitro. Food Chem. 2004, 88, 219–224. [Google Scholar] [CrossRef]
- Mitra, P.; Kalailingam, P.; Tan, H.B.; Thanabalu, T. Overexpression of GRB2 enhances epithelial to mesenchymal transition of A549 cells by upregulating SNAIL expression. Cells 2018, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Proctor, B.M.; Ren, J.; Chen, Z.; Schneider, J.G.; Coleman, T.; Lupu, T.S.; Semenkovich, C.F.; Muslin, A.J. Grb2 is required for atherosclerotic lesion formation. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 1361–1367. [Google Scholar] [CrossRef]
- Dong, Y.; Liu, J.; Ma, J.; Quan, J.; Bao, Y.; Cui, Y. The possible correlation between serum GRB2 levels and carotid atherosclerosis in patients with type 2 diabetes mellitus. Front. Endocrinol. 2022, 13, 963191. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, X.; Wang, X.; Cui, S.; Liu, R.; Liu, J.; Fu, B.; Gong, M.; Wang, C.; Shi, Y.; et al. Grb2 Induces Cardiorenal Syndrome Type 3: Roles of IL-6, Cardiomyocyte Bioenergetics, and Akt/mTOR Pathway. Front. Cell Dev. Biol. 2021, 9, 630412. [Google Scholar] [CrossRef]
- Jorgensen, W.L. Efficient Drug Lead Discovery and Optimization. Acc. Chem. Res. 2009, 42, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Daison, F.A.; Kumar, N.; Balakrishnan, S.; Venugopal, K.; Elango, S.; Sokkar, P. Molecular dynamics studies on the bacterial membrane pore formation by small molecule antimicrobial agents. J. Chem. Inf. Model. 2021, 62, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Timsah, Z.; Suen, K.M.; Cook, N.P.; Lee, G.R.; Lin, C.C.; Gagea, M.; Marti, A.A.; Ladbury, J.E. Grb2 monomer-dimer equilibrium determines normal versus oncogenic function. Nat. Commun. 2015, 6, 7354. [Google Scholar] [CrossRef]
- Zeng, H.W.; Jiang, Y.Y.; Cai, D.G.; Bian, J.; Long, K.; Chen, Z.L. Piperbetol, methylpiperbetol, piperol A and piperol B: A new series of highly specific PAF receptor antagonists from Piper betle. Planta Med. 1997, 63, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Haslan, H.; Suhaimi, F.H.; Thent, Z.C.; Das, S. The underlying mechanism of action for various medicinal properties of Piper betle (betel). Clin. Ter. 2015, 166, 208–214. [Google Scholar]




| SH2 Domain Binding | ||
|---|---|---|
| Compound ID | Compound Name | Glide Score (Kcal/mol) |
| IMPHY006709 | Acetyleugenol | −5.12 |
| IMPHY001144 | Dillapiol | −4.95 |
| IMPHY000846 | Riboflavin | −4.58 |
| Binding score with monomeric GRB2 | ||
| IMPHY000846 | Riboflavin | −6.37 |
| IMPHY017327 | p-Menthane-1,3-diol | −5.78 |
| IMPHY001246 | Carvacrol | −5.28 |
| Binding score with dimeric GRB2 | ||
| IMPHY001073 | Piperbetol | −8.10 |
| IMPHY002191 | Piperol B | −7.68 |
| IMPHY008892 | Methyl piperbetol | −7.37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sudhan; Janakiraman; Ahmad, S.F.; Wani, A.; Ahmed, S.S.S.J. Phytochemicals from Piper betle (L.) as Putative Modulators of a Novel Network-Derived Drug Target for Coronary Artery Disease: An In Silico Study. Processes 2023, 11, 3064. https://doi.org/10.3390/pr11113064
Sudhan, Janakiraman, Ahmad SF, Wani A, Ahmed SSSJ. Phytochemicals from Piper betle (L.) as Putative Modulators of a Novel Network-Derived Drug Target for Coronary Artery Disease: An In Silico Study. Processes. 2023; 11(11):3064. https://doi.org/10.3390/pr11113064
Chicago/Turabian StyleSudhan, Janakiraman, Sheikh F. Ahmad, Abubakar Wani, and Shiek S. S. J. Ahmed. 2023. "Phytochemicals from Piper betle (L.) as Putative Modulators of a Novel Network-Derived Drug Target for Coronary Artery Disease: An In Silico Study" Processes 11, no. 11: 3064. https://doi.org/10.3390/pr11113064
APA StyleSudhan, Janakiraman, Ahmad, S. F., Wani, A., & Ahmed, S. S. S. J. (2023). Phytochemicals from Piper betle (L.) as Putative Modulators of a Novel Network-Derived Drug Target for Coronary Artery Disease: An In Silico Study. Processes, 11(11), 3064. https://doi.org/10.3390/pr11113064








