Advancing Eco-Sustainable Bioremediation for Hydrocarbon Contaminants: Challenges and Solutions
Abstract
:1. Introduction
2. Polycyclic Aromatic Hydrocarbons (PAHs) and Aliphatic Hydrocarbons
3. Hydrocarbon Toxicity and Ecosystem Impact
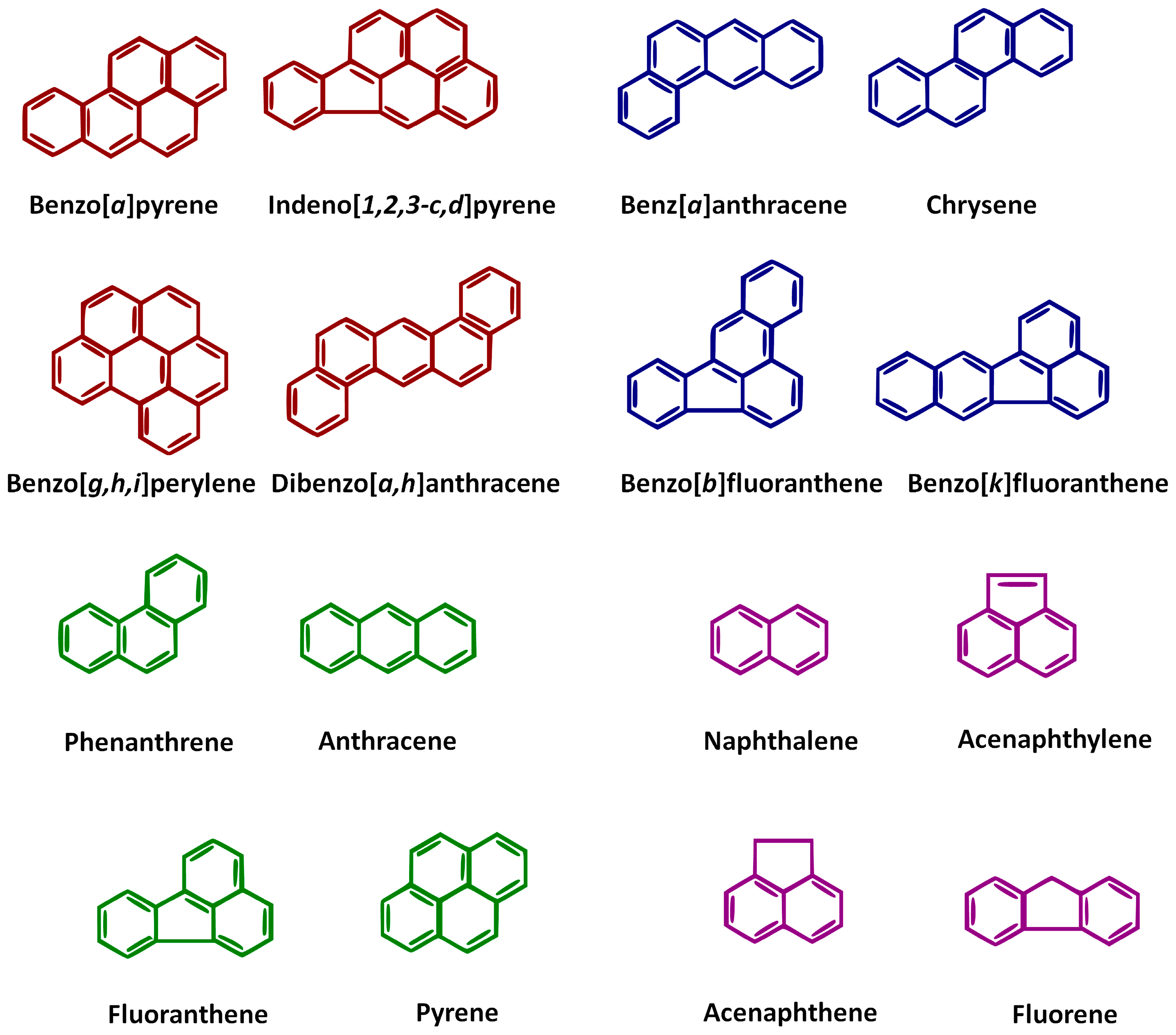
4. Variability in Hydrocarbon Properties: Implications for Bioremediation
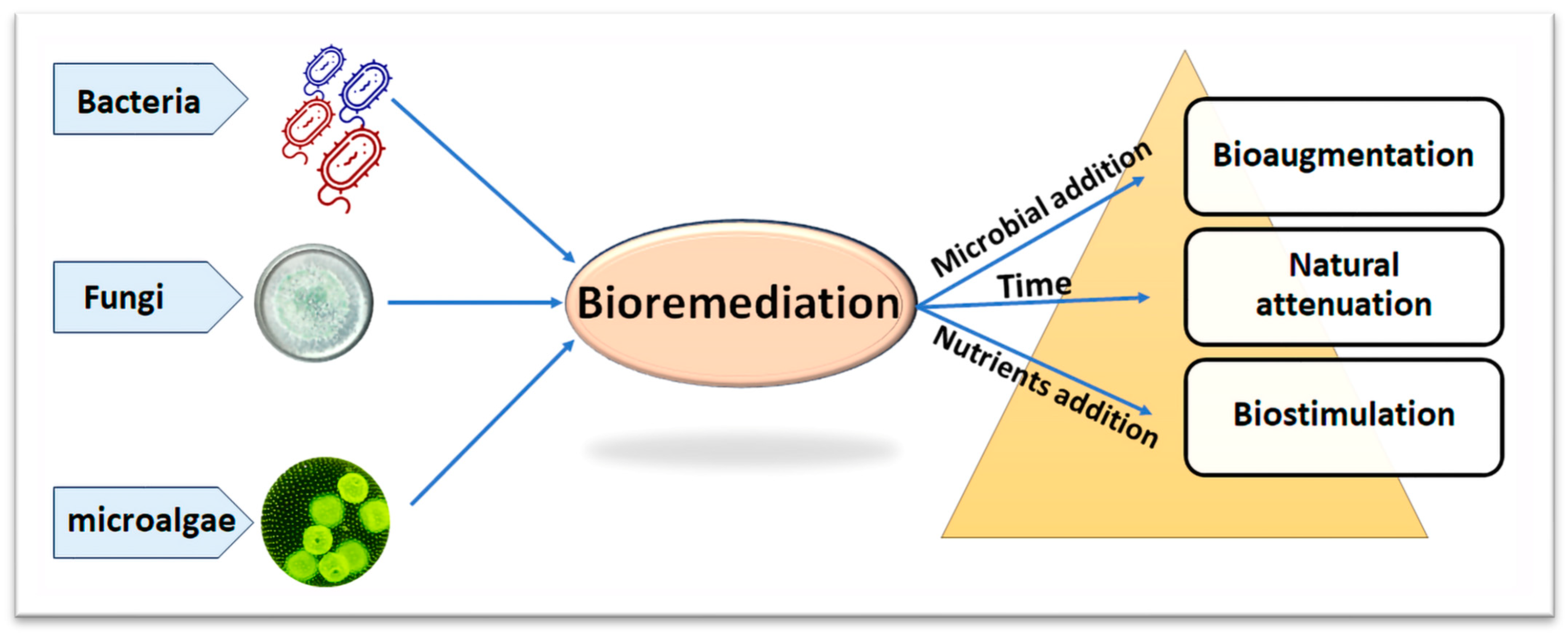
5. Bioremediation of Petroleum Hydrocarbons: A Green Solution
6. Microbial Involvement in the Biodegradation of Hydrocarbons
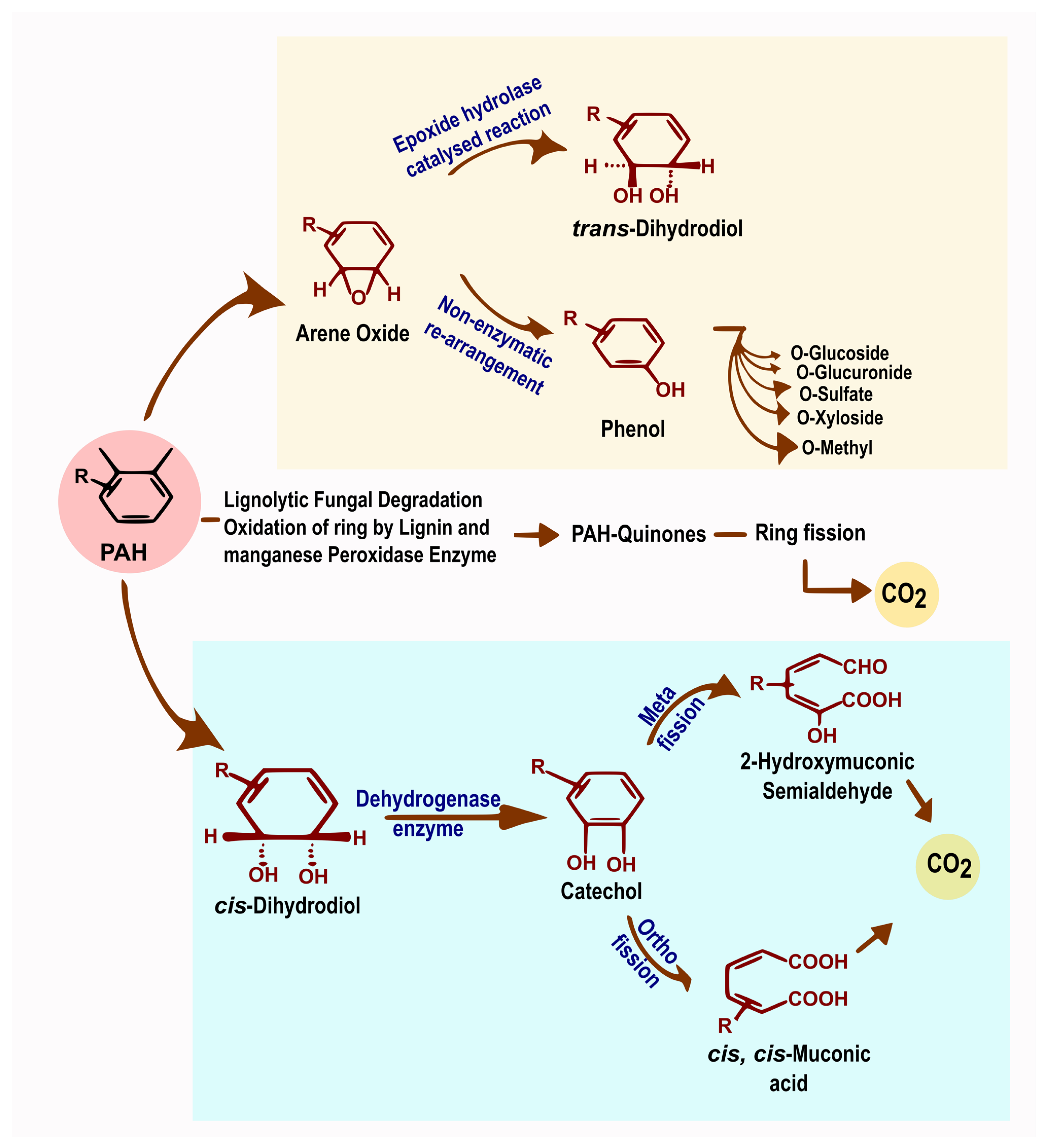
6.1. Bacterial Degradation of Hydrocarbons
6.2. Fungi with Hydrocarbon Degradation Capabilities
6.3. Microalgae Role in Petroleum Remediation
6.4. Genetically Modified Organisms (GMOs) for Bioremediation
6.5. Synergistic Approaches for Sustainable Hydrocarbon Contaminant Remediation
7. Influences on Petroleum Hydrocarbon Degradation
7.1. Factors Influencing Degradation
7.2. Enhancement of Bioremediation Conditions
7.2.1. Soil Conditions
7.2.2. Bioreactors
8. Hydrocarbon Degradation Pathways
8.1. Enzymatic Responses and Hydrocarbon Degradation Genes
8.1.1. Hydrocarbon Degradation Enzymes
8.1.2. Hydrocarbon Degradation Genes
9. Recent Advances in Biodegradation and Bioremediation Research
10. Challenges and Future Prospects in Bioremediation
11. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giri, K.; Rai, J. Bacterial Metabolism of Petroleum Hydrocarbons. Biotechnology 2014, 11, 73–93. [Google Scholar]
- Abdullah, S.R.S.; Al-Baldawi, I.A.; Almansoory, A.F.; Purwanti, I.F.; Al-Sbani, N.H.; Sharuddin, S.S.N. Plant-assisted remediation of hydrocarbons in water and soil: Application, mechanisms, challenges and opportunities. Chemosphere 2020, 247, 125932. [Google Scholar] [CrossRef]
- Logeshwaran, P.; Megharaj, M.; Chadalavada, S.; Bowman, M.; Naidu, R. Petroleum hydrocarbons (PH) in groundwater aquifers: An overview of environmental fate, toxicity, microbial degradation and risk-based remediation approaches. Environ. Technol. Innov. 2018, 10, 175–193. [Google Scholar] [CrossRef]
- Joutey, N.T.; Bahafid, W.; Sayel, H.; El Ghachtouli, N. Biodegradation: Involved microorganisms and genetically engineered microorganisms. Biodegrad.-Life Sci. 2013, 1, 289–320. [Google Scholar]
- Liu, S.-c.; Sun, S.-j.; Cui, P.; Ding, Y.-f. Molecular modification of fluoroquinolone-biodegrading enzymes based on molecular docking and homology modelling. Int. J. Environ. Res. Public Health 2019, 16, 3407. [Google Scholar] [CrossRef] [PubMed]
- Ossai, I.C.; Ahmed, A.; Hassan, A.; Hamid, F.S. Remediation of soil and water contaminated with petroleum hydrocarbon: A review. Environ. Technol. Innov. 2020, 17, 100526. [Google Scholar] [CrossRef]
- Poorsoleiman, M.S.; Hosseini, S.A.; Etminan, A.; Abtahi, H.; Koolivand, A. Bioremediation of Petroleum Hydrocarbons by using a two-step inoculation composting process scaled-up from a mineral-based medium: Effect of biostimulation of an indigenous bacterial strain. Waste Biomass Valorization 2021, 12, 2089–2096. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Balaganesh, P.; Vasudevan, M.; Natarajan, N.; Chauhan, A.; Arora, J.; Ranjan, A.; Rajput, V.D.; Sushkova, S.; Minkina, T. Bioremediation of Hydrocarbon Pollutants: Recent Promising Sustainable Approaches, Scope, and Challenges. Sustainability 2023, 15, 5847. [Google Scholar] [CrossRef]
- Lawal, A.T. Polycyclic aromatic hydrocarbons. A review. Cogent Environ. Sci. 2017, 3, 1339841. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, L.; Zhang, H.; Xing, W.; Wang, Y.; Bai, P.; Zhang, L.; Hayakawa, K.; Toriba, A.; Wei, Y. Assessing approaches of human inhalation exposure to polycyclic aromatic hydrocarbons: A review. Int. J. Environ. Res. Public Health 2021, 18, 3124. [Google Scholar] [CrossRef]
- Mallah, M.A.; Changxing, L.; Mallah, M.A.; Noreen, S.; Liu, Y.; Saeed, M.; Xi, H.; Ahmed, B.; Feng, F.; Mirjat, A.A. Polycyclic aromatic hydrocarbon and its effects on human health: An overeview. Chemosphere 2022, 296, 133948. [Google Scholar] [CrossRef]
- Singh, S.; Kang, S.H.; Mulchandani, A.; Chen, W. Bioremediation: Environmental clean-up through pathway engineering. Curr. Opin. Biotechnol. 2008, 19, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Lange, I.; Kotiukov, P.; Lebedeva, Y. Analyzing Physical-Mechanical and Hydrophysical Properties of Sandy Soils Exposed to Long-Term Hydrocarbon Contamination. Sustainability 2023, 15, 3599. [Google Scholar] [CrossRef]
- Mahjoubi, M.; Cappello, S.; Souissi, Y.; Jaouani, A.; Cherif, A. Microbial bioremediation of petroleum hydrocarbon–contaminated marine environments. Recent Insights Pet. Sci. Eng. 2018, 325, 325–350. [Google Scholar]
- Sharma, I. Bioremediation techniques for polluted environment: Concept, advantages, limitations, and prospects. In Trace Metals in the Environment-New Approaches and Recent Advances; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar]
- Blenis, N.; Hue, N.; Maaz, T.M.; Kantar, M. Biochar production, modification, and its uses in soil remediation: A review. Sustainability 2023, 15, 3442. [Google Scholar] [CrossRef]
- Adams, G.O.; Fufeyin, P.T.; Okoro, S.E.; Ehinomen, I. Bioremediation, biostimulation and bioaugmention: A review. Int. J. Environ. Bioremediat. Biodegrad. 2015, 3, 28–39. [Google Scholar]
- Sayara, T.; Sánchez, A. Bioremediation of PAH-contaminated soils: Process enhancement through composting/compost. Appl. Sci. 2020, 10, 3684. [Google Scholar] [CrossRef]
- Saravanan, A.; Kumar, P.S.; Vo, D.-V.N.; Jeevanantham, S.; Karishma, S.; Yaashikaa, P. A review on catalytic-enzyme degradation of toxic environmental pollutants: Microbial enzymes. J. Hazard. Mater. 2021, 419, 126451. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Gholami, A.; Omidifar, N.; Zarei, M.; Bahrani, S.; Yousefi, K.; Chiang, W.-H.; Babapoor, A. Bioinorganic synthesis of polyrhodanine stabilized Fe3O4/Graphene oxide in microbial supernatant media for anticancer and antibacterial applications. Bioinorg. Chem. Appl. 2021, 2021, 9972664. [Google Scholar] [CrossRef]
- Azhdari, R.; Mousavi, S.M.; Hashemi, S.A.; Bahrani, S.; Ramakrishna, S. Decorated graphene with aluminum fumarate metal organic framework as a superior non-toxic agent for efficient removal of Congo Red dye from wastewater. J. Environ. Chem. Eng. 2019, 7, 103437. [Google Scholar] [CrossRef]
- Eze, M.O.; Hose, G.C.; George, S.C.; Daniel, R. Diversity and metagenome analysis of a hydrocarbon-degrading bacterial consortium from asphalt lakes located in Wietze, Germany. AMB Express 2021, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Gayathiri, E.; Prakash, P.; Karmegam, N.; Varjani, S.; Awasthi, M.K.; Ravindran, B. Biosurfactants: Potential and eco-friendly material for sustainable agriculture and environmental safety—A review. Agronomy 2022, 12, 662. [Google Scholar] [CrossRef]
- Fathepure, B.Z. Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front. Microbiol. 2014, 5, 173. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Zarei, M.; Hashemi, S.A.; Ramakrishna, S.; Chiang, W.-H.; Lai, C.W.; Gholami, A. Gold nanostars-diagnosis, bioimaging and biomedical applications. Drug Metab. Rev. 2020, 52, 299–318. [Google Scholar] [CrossRef]
- Mohkam, M.; Rasoul-Amini, S.; Shokri, D.; Berenjian, A.; Rahimi, F.; Sadraeian, M.; Khalvati, B.; Gholami, A.; Ghasemi, Y. Characterization and in vitro probiotic assessment of potential indigenous Bacillus strains isolated from soil rhizosphere. Minerva Biotecnol. 2016, 28, 19–28. [Google Scholar]
- Lin, C.; Gan, L.; Chen, Z.-L. Biodegradation of naphthalene by strain Bacillus fusiformis (BFN). J. Hazard. Mater. 2010, 182, 771–777. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. A new look on factors affecting microbial degradation of petroleum hydrocarbon pollutants. Int. Biodeterior. Biodegrad. 2017, 120, 71–83. [Google Scholar] [CrossRef]
- Mahiudddin, M.; Fakhruddin, A. Degradation of phenol via meta cleavage pathway by Pseudomonas fluorescens PU1. Int. Sch. Res. Netw. 2012, 2012, 741820. [Google Scholar]
- Gholami, A.; Mohammadi, F.; Ghasemi, Y.; Omidifar, N.; Ebrahiminezhad, A. Antibacterial activity of SPIONs versus ferrous and ferric ions under aerobic and anaerobic conditions: A preliminary mechanism study. IET Nanobiotechnol. 2020, 14, 155–160. [Google Scholar] [CrossRef]
- Pandolfo, E.; Barra Caracciolo, A.; Rolando, L. Recent advances in bacterial degradation of hydrocarbons. Water 2023, 15, 375. [Google Scholar] [CrossRef]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, P.; Bestetti, G.; Galli, E.; Zannoni, D. Microbiologia Ambientale ed Elementi di Ecologia Microbica; CEA: Singapore, 2008. [Google Scholar]
- Huang, L.; Batterman, S.A. Multimedia model for polycyclic aromatic hydrocarbons (PAHs) and nitro-PAHs in Lake Michigan. Environ. Sci. Technol. 2014, 48, 13817–13825. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review. Front. Microbiol. 2016, 1, 1369. [Google Scholar] [CrossRef] [PubMed]
- Lebov, J.; Grieger, K.; Womack, D.; Zaccaro, D.; Whitehead, N.; Kowalcyk, B.; MacDonald, P.D. A framework for One Health research. One Health 2017, 3, 44–50. [Google Scholar] [CrossRef]
- Nzila, A. Current status of the degradation of aliphatic and aromatic petroleum hydrocarbons by thermophilic microbes and future perspectives. Int. J. Environ. Res. Public Health 2018, 15, 2782. [Google Scholar] [CrossRef]
- Salari, M.; Rahmanian, V.; Hashemi, S.A.; Chiang, W.-H.; Lai, C.W.; Mousavi, S.M.; Gholami, A. Bioremediation treatment of polyaromatic hydrocarbons for environmental sustainability. Water 2022, 14, 3980. [Google Scholar] [CrossRef]
- Premnath, N.; Mohanrasu, K.; Rao, R.G.R.; Dinesh, G.; Prakash, G.S.; Ananthi, V.; Ponnuchamy, K.; Muthusamy, G.; Arun, A. A crucial review on polycyclic aromatic Hydrocarbons-Environmental occurrence and strategies for microbial degradation. Chemosphere 2021, 280, 130608. [Google Scholar] [CrossRef]
- Eldos, H.I.; Zouari, N.; Saeed, S.; Al-Ghouti, M.A. Recent advances in the treatment of PAHs in the environment: Application of nanomaterial-based technologies. Arab. J. Chem. 2022, 15, 103918. [Google Scholar] [CrossRef]
- Khan, A.; Ahsan, A.; Farooq, M.A.; Naveed, M.; Li, H. Role of polycyclic aromatic hydrocarbons as EDCs in metabolic disorders. In Endocrine Disrupting Chemicals-Induced Metabolic Disorders and Treatment Strategies; Springer: Berlin/Heidelberg, Germany, 2021; pp. 323–341. [Google Scholar]
- Garay-Narváez, L.; Arim, M.; Flores, J.D.; Ramos-Jiliberto, R. The more polluted the environment, the more important biodiversity is for food web stability. Oikos 2013, 122, 1247–1253. [Google Scholar] [CrossRef]
- Ma, H.; Pu, S.; Liu, S.; Bai, Y.; Mandal, S.; Xing, B. Microplastics in aquatic environments: Toxicity to trigger ecological consequences. Environ. Pollut. 2020, 261, 114089. [Google Scholar] [CrossRef]
- Nagkirti, P.; Shaikh, A.; Vasudevan, G.; Paliwal, V.; Dhakephalkar, P. Bioremediation of terrestrial oil spills: Feasibility Assessment. In Optimization and applicability of bioprocesses; Springer: Berlin/Heidelberg, Germany, 2017; pp. 141–173. [Google Scholar]
- Samanta, S.K.; Singh, O.V.; Jain, R.K. Polycyclic aromatic hydrocarbons: Environmental pollution and bioremediation. TRENDS Biotechnol. 2002, 20, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Kuppusamy, S.; Maddela, N.R.; Megharaj, M.; Venkateswarlu, K. Ecological Impacts of Total Petroleum Hydrocarbons. In Total Petroleum Hydrocarbons: Environmental Fate, Toxicity, and Remediation; Kuppusamy, S., Maddela, N.R., Megharaj, M., Venkateswarlu, K., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 95–138. [Google Scholar] [CrossRef]
- Devatha, C.; Vishnu Vishal, A.; Purna Chandra Rao, J. Investigation of physical and chemical characteristics on soil due to crude oil contamination and its remediation. Appl. Water Sci. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Maddela, N.R.; Megharaj, M.; Venkateswarlu, K. Total petroleum hydrocarbons. In Environmental Fate, Toxicity, and Remediation; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Truskewycz, A.; Gundry, T.D.; Khudur, L.S.; Kolobaric, A.; Taha, M.; Aburto-Medina, A.; Ball, A.S.; Shahsavari, E. Petroleum hydrocarbon contamination in terrestrial ecosystems—Fate and microbial responses. Molecules 2019, 24, 3400. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Zheng, B.; Li, X.; Zhao, X.; Dionysiou, D.D.; Liu, Y. Influencing factors and health risk assessment of polycyclic aromatic hydrocarbons in groundwater in China. J. Hazard. Mater. 2021, 402, 123419. [Google Scholar] [CrossRef]
- Abdel-Shafy, H.I.; Mansour, M.S. A review on polycyclic aromatic hydrocarbons: Source, environmental impact, effect on human health and remediation. Egypt. J. Pet. 2016, 25, 107–123. [Google Scholar] [CrossRef]
- Kebede, G.; Tafese, T.; Abda, E.M.; Kamaraj, M.; Assefa, F. Factors influencing the bacterial bioremediation of hydrocarbon contaminants in the soil: Mechanisms and impacts. J. Chem. 2021, 2021, 23362. [Google Scholar] [CrossRef]
- Gargouri, B.; Karray, F.; Mhiri, N.; Aloui, F.; Sayadi, S. Bioremediation of petroleum hydrocarbons-contaminated soil by bacterial consortium isolated from an industrial wastewater treatment plant. J. Chem. Technol. Biotechnol. 2014, 89, 978–987. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Z.-W.; Liu, R.-X.; Ju, H.-Y.; Bian, X.-K.; Zhang, W.-Z.; Zhang, C.-B.; Yang, T.; Guo, B.; Xiao, C.-L. Research progress in bioremediation of petroleum pollution. Environ. Sci. Pollut. Res. 2021, 28, 46877–46893. [Google Scholar] [CrossRef]
- Ortega-Calvo, J.J. Strategies to Increase Bioavailability and Uptake of Hydrocarbons. In Cellular Ecophysiology of Microbe; Krell, T., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–12. [Google Scholar] [CrossRef]
- Haritash, A.; Kaushik, C. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef]
- Phale, P.S.; Basu, A.; Majhi, P.D.; Deveryshetty, J.; Vamsee-Krishna, C.; Shrivastava, R. Metabolic diversity in bacterial degradation of aromatic compounds. Omics A J. Integr. Biol. 2007, 11, 252–279. [Google Scholar] [CrossRef]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef] [PubMed]
- Borgne, S.L. Introduction to Microorganisms Utilizing Nitrogen and Sulfur Containing Heterocyclic Hydrocarbons. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 2123–2127. [Google Scholar] [CrossRef]
- Gunasekera, T.S.; Bowen, L.L.; Radwan, O.; Striebich, R.C.; Ruiz, O.N. Genomic and transcriptomic characterization revealed key adaptive mechanisms of Marinobacter hydrocarbonoclasticus NI9 for proliferation and degradation of jet fuel. Int. Biodeterior. Biodegrad. 2022, 175, 105502. [Google Scholar] [CrossRef]
- Conlon, R.; Wang, M.; Germaine, X.L.; Mali, R.; Dowling, D.; Germaine, K.J. Ecopiling: Beneficial Soil Bacteria, Plants, and Optimized Soil Conditions for Enhanced Remediation of Hydrocarbon Polluted Soil. In Good Microbes in Medicine, Food Production, Biotechnology, Bioremediation, and Agriculture; Wiley Online Library: Hoboken, NJ, USA, 2022; pp. 337–347. [Google Scholar]
- Abdel-Shafy, H.I.; Mansour, M.S.M. Microbial Degradation of Hydrocarbons in the Environment: An Overview. In Microbial Action on Hydrocarbons; Kumar, V., Kumar, M., Prasad, R., Eds.; Springer: Singapore, 2018; pp. 353–386. [Google Scholar] [CrossRef]
- Foght, J. Anaerobic biodegradation of aromatic hydrocarbons: Pathways and prospects. Microb. Physiol. 2008, 15, 93–120. [Google Scholar] [CrossRef] [PubMed]
- Ibrar, M.; Khan, S.; Hasan, F.; Yang, X. Biosurfactants and chemotaxis interplay in microbial consortium-based hydrocarbons degradation. Environ. Sci. Pollut. Res. 2022, 29, 24391–24410. [Google Scholar] [CrossRef] [PubMed]
- Hamouda, R.A.E.F.; Sorour, N.M.; Yeheia, D.S. Biodegradation of crude oil by Anabaena oryzae, Chlorella kessleri and its consortium under mixotrophic conditions. Int. Biodeterior. Biodegrad. 2016, 112, 128–134. [Google Scholar] [CrossRef]
- Safdel, M.; Anbaz, M.A.; Daryasafar, A.; Jamialahmadi, M. Microbial enhanced oil recovery, a critical review on worldwide implemented field trials in different countries. Renew. Sustain. Energy Rev. 2017, 74, 159–172. [Google Scholar] [CrossRef]
- Tiwari, S.; Tripathi, A.; Gaur, R. Bioremediation of plant refuges and xenobiotics. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Springer: Berlin/Heidelberg, Germany, 2017; pp. 85–142. [Google Scholar]
- Touliabah, H.E.-S.; El-Sheekh, M.M.; Ismail, M.M.; El-Kassas, H. A review of microalgae-and cyanobacteria-based biodegradation of organic pollutants. Molecules 2022, 27, 1141. [Google Scholar] [CrossRef]
- Karigar, C.S.; Rao, S.S. Role of microbial enzymes in the bioremediation of pollutants: A review. Enzym. Res 2011, 2011, 805187. [Google Scholar] [CrossRef]
- Sarsaiya, S.; Awasthi, S.K.; Jain, A.; Mishra, S.; Jia, Q.; Shu, F.; Li, J.; Duan, Y.; Singh, R.; Awasthi, M.K.; et al. Recent Developments in the Treatment of Petroleum Hydrocarbon and Oily Sludge from the Petroleum Industry. In Biological Processing of Solid Waste; CRC Press: Boca Raton, FL, USA, 2019; pp. 277–310. [Google Scholar] [CrossRef]
- Varner, P.M.; Gunsch, C.K. Properties affecting transfer and expression of degradative plasmids for the purpose of bioremediation. Biodegradation 2021, 32, 361–375. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef]
- Liu, X.; Li, Z.; Zhang, C.; Tan, X.; Yang, X.; Wan, C.; Lee, D.-J. Enhancement of anaerobic degradation of petroleum hydrocarbons by electron intermediate: Performance and mechanism. Bioresour. Technol. 2020, 295, 122305. [Google Scholar] [CrossRef] [PubMed]
- Bahrani, S.; Hashemi, S.A.; Mousavi, S.M.; Azhdari, R. Zinc-based metal–organic frameworks as nontoxic and biodegradable platforms for biomedical applications: Review study. Drug Metab. Rev. 2019, 51, 356–377. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Kalashgrani, M.Y.; Gholami, A.; Omidifar, N.; Babapoor, A.; Vijayakameswara Rao, N.; Chiang, W.-H. Recent Advances in Plasma-Engineered Polymers for Biomarker-Based Viral Detection and Highly Multiplexed Analysis. Biosensors 2022, 12, 286. [Google Scholar] [CrossRef] [PubMed]
- Sakshi; Haritash, A.K. A comprehensive review of metabolic and genomic aspects of PAH-degradation. Arch. Microbiol. 2020, 202, 2033–2058. [Google Scholar] [CrossRef] [PubMed]
- Parhamfar, M.; Abtahi, H.; Godini, K.; Saeedi, R.; Sartaj, M.; Villaseñor, J.; Coulon, F.; Kumar, V.; Soltanighias, T.; Ghaznavi-Rad, E. Biodegradation of heavy oily sludge by a two-step inoculation composting process using synergistic effect of indigenous isolated bacteria. Process Biochem. 2020, 91, 223–230. [Google Scholar] [CrossRef]
- Weelink, S.A.; Van Eekert, M.H.; Stams, A.J. Degradation of BTEX by anaerobic bacteria: Physiology and application. Rev. Environ. Sci. Bio/Technol. 2010, 9, 359–385. [Google Scholar] [CrossRef]
- Noh, S.-L.; Choi, J.-M.; An, Y.-J.; Park, S.-S.; Cho, K.-S. Anaerobic biodegradation of toluene coupled to sulfate reduction in oil-contaminated soils: Optimum environmental conditions for field applications. J. Environ. Sci. Health Part A 2003, 38, 1087–1097. [Google Scholar] [CrossRef]
- Purnomo, A.; Mauliddawati, V.; Khoirudin, M.; Yonda, A.; Nawfa, R.; Putra, S. Bio-decolorization and novel bio-transformation of methyl orange by brown-rot fungi. Int. J. Environ. Sci. Technol. 2019, 16, 7555–7564. [Google Scholar] [CrossRef]
- Haripriyan, U.; Gopinath, K.; Arun, J.; Govarthanan, M. Bioremediation of organic pollutants: A mini review on current and critical strategies for wastewater treatment. Arch. Microbiol. 2022, 204, 286. [Google Scholar] [CrossRef]
- Sharma, S.; Pandey, L.M. Biodegradation kinetics of binary mixture of hexadecane and phenanthrene by the bacterial microconsortium. Bioresour. Technol. 2022, 358, 127408. [Google Scholar] [CrossRef]
- Poorsoleiman, M.S.; Hosseini, S.A.; Etminan, A.; Abtahi, H.; Koolivand, A. Effect of two-step bioaugmentation of an indigenous bacterial strain isolated from oily waste sludge on petroleum hydrocarbons biodegradation: Scaling-up from a liquid mineral medium to a two-stage composting process. Environ. Technol. Innov. 2020, 17, 100558. [Google Scholar] [CrossRef]
- Yong, J.J.J.Y.; Chew, K.W.; Khoo, K.S.; Show, P.L.; Chang, J.-S. Prospects and development of algal-bacterial biotechnology in environmental management and protection. Biotechnol. Adv. 2021, 47, 107684. [Google Scholar] [CrossRef] [PubMed]
- Gupte, A.; Tripathi, A.; Patel, H.; Rudakiya, D.; Gupte, S. Bioremediation of polycyclic aromatic hydrocarbon (PAHs): A perspective. Open Biotechnol. J. 2016, 10, 363–378. [Google Scholar] [CrossRef]
- Jafari, A.; Zamankhan, P.; Mousavi, S.M.; Henttinen, K. Multiscale modeling of fluid turbulence and flocculation in fiber suspensions. J. Appl. Phys. 2006, 100, 034901. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, K.; Li, B.-G.; Mbadinga, S.M.; Zhou, L.; Liu, J.-F.; Yang, S.-Z.; Gu, J.-D.; Mu, B.-Z. Simulation of in situ oil reservoir conditions in a laboratory bioreactor testing for methanogenic conversion of crude oil and analysis of the microbial community. Int. Biodeterior. Biodegrad. 2019, 136, 24–33. [Google Scholar] [CrossRef]
- Cruz, A.; Cavaleiro, A.J.; Paulo, A.M.; Louvado, A.; Alves, M.M.; Almeida, A.; Cunha, Â. Microbial remediation of organometals and oil hydrocarbons in the marine environment. In Marine Pollution and Microbial Remediation; Springer: Berlin/Heidelberg, Germany, 2017; pp. 41–66. [Google Scholar]
- Kuttiyathil, M.S.; Mohamed, M.M.; Al-Zuhair, S. Using microalgae for remediation of crude petroleum oil–water emulsions. Biotechnol. Prog. 2021, 37, e3098. [Google Scholar] [CrossRef]
- Deborah Gnana Selvam, A.; Thatheyus, A.J. Microbial Degradation of Petroleum Hydrocarbons: An Overview. In Microbial Action on Hydrocarbons; Kumar, V., Kumar, M., Prasad, R., Eds.; Springer: Singapore, 2018; pp. 485–503. [Google Scholar] [CrossRef]
- Gwak, J.-H.; Awala, S.I.; Nguyen, N.-L.; Yu, W.-J.; Yang, H.-Y.; von Bergen, M.; Jehmlich, N.; Kits, K.D.; Loy, A.; Dunfield, P.F. Sulfur and methane oxidation by a single microorganism. Proc. Natl. Acad. Sci. USA 2022, 119, e2114799119. [Google Scholar] [CrossRef]
- Gupta, G.; Kumar, V.; Pal, A. Microbial degradation of high molecular weight polycyclic aromatic hydrocarbons with emphasis on pyrene. Polycycl. Aromat. Compd. 2019, 39, 124–138. [Google Scholar] [CrossRef]
- Sho, M.; Hamel, C.; Greer, C.W. Two distinct gene clusters encode pyrene degradation in Mycobacterium sp. strain S65. FEMS Microbiol. Ecol. 2004, 48, 209–220. [Google Scholar] [CrossRef]
- Peng, J.; Zhang, Y.; Su, J.; Qiu, Q.; Jia, Z.; Zhu, Y.-G. Bacterial communities predominant in the degradation of 13C4-4, 5, 9, 10-pyrene during composting. Bioresour. Technol. 2013, 143, 608–614. [Google Scholar] [CrossRef]
- Ruiz, O.N.; Radwan, O.; Striebich, R.C. GC–MS hydrocarbon degradation profile data of Pseudomonas frederiksbergensis SI8, a bacterium capable of degrading aromatics at low temperatures. Data Brief 2021, 35, 106864. [Google Scholar] [CrossRef] [PubMed]
- Posada-Baquero, R.; Jiménez-Volkerink, S.N.; García, J.L.; Vila, J.; Cantos, M.; Grifoll, M.; Ortega-Calvo, J.J. Rhizosphere-enhanced biosurfactant action on slowly desorbing PAHs in contaminated soil. Sci. Total Environ. 2020, 720, 137608. [Google Scholar] [CrossRef] [PubMed]
- Medić, A.; Lješević, M.; Inui, H.; Beškoski, V.; Kojić, I.; Stojanović, K.; Karadžić, I. Efficient biodegradation of petroleum n-alkanes and polycyclic aromatic hydrocarbons by polyextremophilic Pseudomonas aeruginosa san ai with multidegradative capacity. RSC Adv. 2020, 10, 14060–14070. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, W.; Zhou, W.; Wang, Y.; Deng, M.; Zhou, S. Synergistic degradation of pyrene by Pseudomonas aeruginosa PA06 and Achromobacter sp. AC15 with sodium citrate as the co-metabolic carbon source. Ecotoxicology 2021, 30, 1487–1498. [Google Scholar] [CrossRef]
- Rolando, L.; Vila, J.; Baquero, R.P.; Castilla-Alcantara, J.C.; Caracciolo, A.B.; Ortega-Calvo, J.-J. Impact of bacterial motility on biosorption and cometabolism of pyrene in a porous medium. Sci. Total Environ. 2020, 717, 137210. [Google Scholar] [CrossRef]
- Imam, A.; Suman, S.K.; Kanaujia, P.K.; Ray, A. Biological machinery for polycyclic aromatic hydrocarbons degradation: A review. Bioresour. Technol. 2022, 343, 126121. [Google Scholar] [CrossRef]
- Song, X.; Xu, Y.; Li, G.; Zhang, Y.; Huang, T.; Hu, Z. Isolation, characterization of Rhodococcus sp. P14 capable of degrading high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons. Mar. Pollut. Bull. 2011, 62, 2122–2128. [Google Scholar] [CrossRef]
- Fernández-López, C.; Posada-Baquero, R.; García, J.L.; Castilla-Alcantara, J.C.; Cantos, M.; Ortega-Calvo, J.J. Root-mediated bacterial accessibility and cometabolism of pyrene in soil. Sci. Total Environ. 2021, 760, 143408. [Google Scholar] [CrossRef]
- Chebbi, A.; Hentati, D.; Zaghden, H.; Baccar, N.; Rezgui, F.; Chalbi, M.; Sayadi, S.; Chamkha, M. Polycyclic aromatic hydrocarbon degradation and biosurfactant production by a newly isolated Pseudomonas sp. strain from used motor oil-contaminated soil. Int. Biodeterior. Biodegrad. 2017, 122, 128–140. [Google Scholar] [CrossRef]
- Elufisan, T.O.; Rodríguez-Luna, I.C.; Oyedara, O.O.; Sánchez-Varela, A.; Hernández-Mendoza, A.; Gonzalez, E.D.; Paz-González, A.D.; Muhammad, K.; Rivera, G.; Villalobos-Lopez, M.A. The Polycyclic Aromatic Hydrocarbon (PAH) degradation activities and genome analysis of a novel strain Stenotrophomonas sp. Pemsol isolated from Mexico. PeerJ 2020, 8, e8102. [Google Scholar] [CrossRef]
- Ahmed, A.W.; Alzubaidi, F.S.; Hamza, S.J. Biodegradation of crude oil in contaminated water by local isolates of Enterobacter cloacae. Iraqi J. Sci. 2014, 55, 1025–1033. [Google Scholar]
- Van Beilen, J.B.; Marín, M.M.; Smits, T.H.; Röthlisberger, M.; Franchini, A.G.; Witholt, B.; Rojo, F. Characterization of two alkane hydroxylase genes from the marine hydrocarbonoclastic bacterium Alcanivorax borkumensis. Environ. Microbiol. 2004, 6, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Laczi, K.; Kis, Á.; Horváth, B.; Maróti, G.; Hegedüs, B.; Perei, K.; Rákhely, G. Metabolic responses of Rhodococcus erythropolis PR4 grown on diesel oil and various hydrocarbons. Appl. Microbiol. Biotechnol. 2015, 99, 9745–9759. [Google Scholar] [CrossRef] [PubMed]
- Orro, A.; Cappelletti, M.; D’Ursi, P.; Milanesi, L.; Di Canito, A.; Zampolli, J.; Collina, E.; Decorosi, F.; Viti, C.; Fedi, S. Genome and phenotype microarray analyses of Rhodococcus sp. BCP1 and Rhodococcus opacus R7: Genetic determinants and metabolic abilities with environmental relevance. PLoS ONE 2015, 10, e0139467. [Google Scholar] [CrossRef] [PubMed]
- Schuler, L.; Jouanneau, Y.; Ní Chadhain, S.M.; Meyer, C.; Pouli, M.; Zylstra, G.J.; Hols, P.; Agathos, S.N. Characterization of a ring-hydroxylating dioxygenase from phenanthrene-degrading Sphingomonas sp. strain LH128 able to oxidize benz [a] anthracene. Appl. Microbiol. Biotechnol. 2009, 83, 465–475. [Google Scholar] [CrossRef]
- Ní Chadhain, S.M.; Moritz, E.M.; Kim, E.; Zylstra, G.J. Identification, cloning, and characterization of a multicomponent biphenyl dioxygenase from Sphingobium yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 2007, 34, 605–613. [Google Scholar] [CrossRef]
- Dudhagara, D.R.; Dave, B.P. Mycobacterium as polycyclic aromatic hydrocarbons (PAHs) degrader. Mycobacterium-Res. Dev. 2018, 1, 1–9. [Google Scholar]
- Lo Piccolo, L.; De Pasquale, C.; Fodale, R.; Puglia, A.M.; Quatrini, P. Involvement of an alkane hydroxylase system of Gordonia sp. strain SoCg in degradation of solid n-alkanes. Appl. Environ. Microbiol. 2011, 77, 1204–1213. [Google Scholar] [CrossRef]
- Throne-Holst, M.; Markussen, S.; Winnberg, A.; Ellingsen, T.E.; Kotlar, H.-K.; Zotchev, S.B. Utilization of n-alkanes by a newly isolated strain of Acinetobacter venetianus: The role of two AlkB-type alkane hydroxylases. Appl. Microbiol. Biotechnol. 2006, 72, 353–360. [Google Scholar] [CrossRef]
- Banat, I.; Rahman, K.; Thahira-Rahman, J. Bioremediation of hydrocarbon pollution using biosurfactant producing oil degrading bacteria. WIT Trans. Ecol. Environ. 2002, 59, 221–230. [Google Scholar]
- Chandra, S.; Sharma, R.; Singh, K.; Sharma, A. Application of bioremediation technology in the environment contaminated with petroleum hydrocarbon. Ann. Microbiol. 2013, 63, 417–431. [Google Scholar] [CrossRef]
- Desai, A.; Vyas, P. Petroleum and Hydrocarbon Microbiology; Department of Microbiology, M. S. University of Baroda: Vadodara, India, 2008. [Google Scholar]
- Wanapaisan, P.; Laothamteep, N.; Vejarano, F.; Chakraborty, J.; Shintani, M.; Muangchinda, C.; Morita, T.; Suzuki-Minakuchi, C.; Inoue, K.; Nojiri, H. Synergistic degradation of pyrene by five culturable bacteria in a mangrove sediment-derived bacterial consortium. J. Hazard. Mater. 2018, 342, 561–570. [Google Scholar] [CrossRef]
- Ferraro, A.; Massini, G.; Miritana, V.M.; Panico, A.; Pontoni, L.; Race, M.; Rosa, S.; Signorini, A.; Fabbricino, M.; Pirozzi, F. Bioaugmentation strategy to enhance polycyclic aromatic hydrocarbons anaerobic biodegradation in contaminated soils. Chemosphere 2021, 275, 130091. [Google Scholar] [CrossRef]
- Phulpoto, I.A.; Hu, B.; Wang, Y.; Ndayisenga, F.; Li, J.; Yu, Z. Effect of natural microbiome and culturable biosurfactants-producing bacterial consortia of freshwater lake on petroleum-hydrocarbon degradation. Sci. Total Environ. 2021, 751, 141720. [Google Scholar] [CrossRef] [PubMed]
- Adebusoye, S.A.; Ilori, M.O.; Amund, O.O.; Teniola, O.D.; Olatope, S. Microbial degradation of petroleum hydrocarbons in a polluted tropical stream. World J. Microbiol. Biotechnol. 2007, 23, 1149–1159. [Google Scholar] [CrossRef]
- Bacosa, H.; Suto, K.; Inoue, C. Preferential degradation of aromatic hydrocarbons in kerosene by a microbial consortium. Int. Biodeterior. Biodegrad. 2010, 64, 702–710. [Google Scholar] [CrossRef]
- Jamal, M.T. Enrichment of potential halophilic Marinobacter consortium for mineralization of petroleum hydrocarbons and also as oil reservoir indicator in Red Sea, Saudi Arabia. Polycycl. Aromat. Compd. 2022, 42, 400–411. [Google Scholar] [CrossRef]
- Feng, L.; Jiang, X.; Huang, Y.; Wen, D.; Fu, T.; Fu, R. Petroleum hydrocarbon-contaminated soil bioremediation assisted by isolated bacterial consortium and sophorolipid. Environ. Pollut. 2021, 273, 116476. [Google Scholar] [CrossRef]
- Teng, Y.; Wang, X.; Li, L.; Li, Z.; Luo, Y. Rhizobia and their bio-partners as novel drivers for functional remediation in contaminated soils. Front. Plant Sci. 2015, 6, 32. [Google Scholar] [CrossRef]
- Vasilyeva, G.; Kondrashina, V.; Strijakova, E.; Ortega-Calvo, J.-J. Adsorptive bioremediation of soil highly contaminated with crude oil. Sci. Total Environ. 2020, 706, 135739. [Google Scholar] [CrossRef]
- Fahid, M.; Arslan, M.; Shabir, G.; Younus, S.; Yasmeen, T.; Rizwan, M.; Siddique, K.; Ahmad, S.R.; Tahseen, R.; Iqbal, S. Phragmites australis in combination with hydrocarbons degrading bacteria is a suitable option for remediation of diesel-contaminated water in floating wetlands. Chemosphere 2020, 240, 124890. [Google Scholar] [CrossRef] [PubMed]
- Arantza, S.-J.; Hiram, M.-R.; Erika, K.; Chávez-Avilés, M.N.; Valiente-Banuet, J.I.; Fierros-Romero, G. Bio- and phytoremediation: Plants and microbes to the rescue of heavy metal polluted soils. SN Appl. Sci. 2022, 4, 59. [Google Scholar] [CrossRef]
- Fernández, N.V.; Marchelli, P.; Tenreiro, R.; Chaves, S.; Fontenla, S.B. Are the rhizosphere fungal communities of Nothofagus alpina established in two different environments influenced by plant genetic diversity? For. Ecol. Manag. 2020, 473, 118269. [Google Scholar] [CrossRef]
- Al Farraj, D.A.; Hadibarata, T.; Elshikh, M.S.; Al Khulaifi, M.M.; Kristanti, R.A. Biotransformation and degradation pathway of pyrene by filamentous soil fungus Trichoderma sp. F03. Water Air Soil Pollut. 2020, 231, 1–9. [Google Scholar] [CrossRef]
- Lee, H.; Jang, Y.; Choi, Y.-S.; Kim, M.-J.; Lee, J.; Lee, H.; Hong, J.-H.; Lee, Y.M.; Kim, G.-H.; Kim, J.-J. Biotechnological procedures to select white rot fungi for the degradation of PAHs. J. Microbiol. Methods 2014, 97, 56–62. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, G.; Wu, Z.; Wen, X.; Zhong, H.; Zhong, Z.; Bian, F.; Gai, X. Agroforestry alters the rhizosphere soil bacterial and fungal communities of moso bamboo plantations in subtropical China. Appl. Soil Ecol. 2019, 143, 192–200. [Google Scholar] [CrossRef]
- Ravelet, C.; Krivobok, S.; Sage, L.; Steiman, R. Biodegradation of pyrene by sediment fungi. Chemosphere 2000, 40, 557–563. [Google Scholar] [CrossRef]
- Aranda, E.; Godoy, P.; Reina, R.; Badia-Fabregat, M.; Rosell, M.; Marco-Urrea, E.; García-Romera, I. Isolation of Ascomycota fungi with capability to transform PAHs: Insights into the biodegradation mechanisms of Penicillium oxalicum. Int. Biodeterior. Biodegrad. 2017, 122, 141–150. [Google Scholar] [CrossRef]
- Haritash, A.; Kaushik, C. Degradation of low molecular weight polycyclic aromatic hydrocarbons by microorganisms isolated from contaminated soil. Int. J. Environ. Sci. 2016, 6, 808–819. [Google Scholar]
- Birolli, W.G.; Santos, D.d.A.; Alvarenga, N.; Garcia, A.C.; Romão, L.P.; Porto, A.L. Biodegradation of anthracene and several PAHs by the marine-derived fungus Cladosporium sp. CBMAI 1237. Mar. Pollut. Bull. 2018, 129, 525–533. [Google Scholar] [CrossRef]
- Bharagava, R.N.; Chowdhary, P. Emerging and Eco-Friendly Approaches for Waste Management; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Lange, B.; Kremer, S.; Anke, H.; Sterner, O. Metabolism of pyrene by basidiomycetous fungi of the genera Crinipellis, Marasmius, and Marasmiellus. Can. J. Microbiol. 1996, 42, 1179–1183. [Google Scholar] [CrossRef]
- Li, P.; Li, H.; Stagnitti, F.; Wang, X.; Zhang, H.; Gong, Z.; Liu, W.; Xiong, X.; Li, L.; Austin, C. Biodegradation of pyrene and phenanthrene in soil using immobilized fungi Fusarium sp. Bull. Environ. Contam. Toxicol. 2005, 2, 75. [Google Scholar] [CrossRef] [PubMed]
- Khudhair, A.B.; Hadibarata, T.; Yusoff, A.R.M.; Teh, Z.C.; Adnan, L.A.; Kamyab, H. Pyrene metabolism by new species isolated from soil Rhizoctonia zeae SOL3. Water Air Soil Pollut. 2015, 226, 1–9. [Google Scholar] [CrossRef]
- Vieira, G.A.; Magrini, M.J.; Bonugli-Santos, R.C.; Rodrigues, M.V.; Sette, L.D. Polycyclic aromatic hydrocarbons degradation by marine-derived basidiomycetes: Optimization of the degradation process. Braz. J. Microbiol. 2018, 49, 749–756. [Google Scholar] [CrossRef]
- Lee, H.; Jang, Y.; Kim, J.M.; Kim, G.H.; Kim, J.J. White-rot fungus Merulius tremellosus KUC9161 identified as an effective degrader of polycyclic aromatic hydrocarbons. J. Basic Microbiol. 2013, 53, 195–199. [Google Scholar] [CrossRef]
- Hadibarata, T.; Teh, Z.C. Optimization of pyrene degradation by white-rot fungus Pleurotus pulmonarius F043 and characterization of its metabolites. Bioprocess Biosyst. Eng. 2014, 37, 1679–1684. [Google Scholar] [CrossRef]
- Wen, J.; Gao, D.; Zhang, B.; Liang, H. Co-metabolic degradation of pyrene by indigenous white-rot fungus Pseudotrametes gibbosa from the northeast China. Int. Biodeterior. Biodegrad. 2011, 65, 600–604. [Google Scholar] [CrossRef]
- Elyamine, A.M.; Kan, J.; Meng, S.; Tao, P.; Wang, H.; Hu, Z. Aerobic and anaerobic bacterial and fungal degradation of pyrene: Mechanism pathway including biochemical reaction and catabolic genes. Int. J. Mol. Sci. 2021, 22, 8202. [Google Scholar] [CrossRef]
- Mao, J.; Guan, W. Fungal degradation of polycyclic aromatic hydrocarbons (PAHs) by Scopulariopsis brevicaulis and its application in bioremediation of PAH-contaminated soil. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2016, 66, 399–405. [Google Scholar]
- Bishnoi, K.; Kumar, R.; Bishnoi, N.R. Biodegradation of Polycyclic Aromatic Hydrocarbons by White Rot Fungi Phanerochaete Chrysosporium in Sterile and Unsterile Soil; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Novotný, Č.; Svobodová, K.; Erbanová, P.; Cajthaml, T.; Kasinath, A.; Lang, E.; Šašek, V. Ligninolytic fungi in bioremediation: Extracellular enzyme production and degradation rate. Soil Biol. Biochem. 2004, 36, 1545–1551. [Google Scholar] [CrossRef]
- Aydin, S.; Karaçay, H.A.; Shahi, A.; Gökçe, S.; Ince, B.; Ince, O. Aerobic and anaerobic fungal metabolism and Omics insights for increasing polycyclic aromatic hydrocarbons biodegradation. Fungal Biol. Rev. 2017, 31, 61–72. [Google Scholar] [CrossRef]
- Ichinose, H. Cytochrome P 450 of wood-rotting basidiomycetes and biotechnological applications. Biotechnol. Appl. Biochem. 2013, 60, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Lladó, S.; Covino, S.; Solanas, A.; Viñas, M.; Petruccioli, M.; D’annibale, A. Comparative assessment of bioremediation approaches to highly recalcitrant PAH degradation in a real industrial polluted soil. J. Hazard. Mater. 2013, 248, 407–414. [Google Scholar] [CrossRef]
- Mouhamadou, B.; Faure, M.; Sage, L.; Marçais, J.; Souard, F.; Geremia, R.A. Potential of autochthonous fungal strains isolated from contaminated soils for degradation of polychlorinated biphenyls. Fungal Biol. 2013, 117, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Dell’Anno, F.; Rastelli, E.; Sansone, C.; Brunet, C.; Ianora, A.; Dell’Anno, A. Bacteria, fungi and microalgae for the bioremediation of marine sediments contaminated by petroleum hydrocarbons in the omics era. Microorganisms 2021, 9, 1695. [Google Scholar] [CrossRef] [PubMed]
- Ugya, Y.A.; Hasan, D.u.B.; Tahir, S.M.; Imam, T.S.; Ari, H.A.; Hua, X. Microalgae biofilm cultured in nutrient-rich water as a tool for the phycoremediation of petroleum-contaminated water. Int. J. Phytoremediat. 2021, 23, 1175–1183. [Google Scholar] [CrossRef]
- Özhan, K.; Miles, S.M.; Gao, H.; Bargu, S. Relative Phytoplankton growth responses to physically and chemically dispersed South Louisiana sweet crude oil. Environ. Monit. Assess. 2014, 186, 3941–3956. [Google Scholar] [CrossRef]
- Znad, H.; Al Ketife, A.M.D.; Judd, S.; AlMomani, F.; Vuthaluru, H.B. Bioremediation and nutrient removal from wastewater by Chlorella vulgaris. Ecol. Eng. 2018, 110, 1–7. [Google Scholar] [CrossRef]
- Xaaldi Kalhor, A.; Movafeghi, A.; Mohammadi-Nassab, A.D.; Abedi, E.; Bahrami, A. Potential of the green alga Chlorella vulgaris for biodegradation of crude oil hydrocarbons. Mar. Pollut. Bull. 2017, 123, 286–290. [Google Scholar] [CrossRef]
- El-Sheekh, M.M.; Hamouda, R.A.; Nizam, A.A. Biodegradation of crude oil by Scenedesmus obliquus and Chlorella vulgaris growing under heterotrophic conditions. Int. Biodeterior. Biodegrad. 2013, 82, 67–72. [Google Scholar] [CrossRef]
- Das, P.; AbdulQuadir, M.; Thaher, M.; Khan, S.; Chaudhary, A.K.; Alghasal, G.; Al-Jabri, H.M.S.J. Microalgal bioremediation of petroleum-derived low salinity and low pH produced water. J. Appl. Phycol. 2018, 31, 435–444. [Google Scholar] [CrossRef]
- Lei, A.; Wong, Y.; Tam, N. Removal of pyrene by different microalgal species. Water Sci. Technol. 2002, 46, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.-W.; Yuan, D.-X.; Lin, Q.-M.; Yang, T.-L. Accumulation and biodegradation of phenanthrene and fluoranthene by the algae enriched from a mangrove aquatic ecosystem. Mar. Pollut. Bull. 2008, 56, 1400–1405. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, Y.; Rasoul-Amini, S.; Fotooh-Abadi, E. The biotransformation, biodegradation, and bioremediation of organic compounds by microalgae 1. J. Phycol. 2011, 47, 969–980. [Google Scholar] [CrossRef]
- Morales, A.R.; Paniagua-Michel, J. Bioremediation of hexadecane and diesel oil is enhanced by photosynthetically produced marine biosurfactants. J. Bioremediat. Biodegrad 2013, 1, 1–5. [Google Scholar] [CrossRef]
- Ammar, S.H.; Khadim, H.J.; Mohamed, A.I. Cultivation of Nannochloropsis oculata and Isochrysis galbana microalgae in produced water for bioremediation and biomass production. Environ. Technol. Innov. 2018, 10, 132–142. [Google Scholar] [CrossRef]
- Radice, R.P.; De Fabrizio, V.; Donadoni, A.; Scopa, A.; Martelli, G. Crude Oil Bioremediation: From Bacteria to Microalgae. Processes 2023, 11, 442. [Google Scholar] [CrossRef]
- Gutierrez, T.; Rhodes, G.; Mishamandani, S.; Berry, D.; Whitman, W.B.; Nichols, P.D.; Semple, K.T.; Aitken, M.D. Polycyclic aromatic hydrocarbon degradation of phytoplankton-associated Arenibacter spp. and description of Arenibacter algicola sp. nov., an aromatic hydrocarbon-degrading bacterium. Appl. Environ. Microbiol. 2014, 80, 618–628. [Google Scholar] [CrossRef]
- Chernikova, T.N.; Bargiela, R.; Toshchakov, S.V.; Shivaraman, V.; Lunev, E.A.; Yakimov, M.M.; Thomas, D.N.; Golyshin, P.N. Hydrocarbon-degrading bacteria Alcanivorax and Marinobacter associated with microalgae Pavlova lutheri and Nannochloropsis oculata. Front. Microbiol. 2020, 11, 572931. [Google Scholar] [CrossRef]
- Ghodrati, M.; Kosari-Nasab, M.; Zarrini, G.; Movafeghi, A. Crude oil contamination enhances the lipoxygenase gene expression in the green microalga scenedesmus dimorphus. Biointerface Res. Appl. Chem. 2021, 11, 11431–11439. [Google Scholar]
- SureshKumar, P.; Thomas, J.; Poornima, V. Structural insights on bioremediation of polycyclic aromatic hydrocarbons using microalgae: A modelling-based computational study. Environ. Monit. Assess. 2018, 190, 92. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Shukla, P. Futuristic avenues of metabolic engineering techniques in bioremediation. Biotechnol. Appl. Biochem. 2020, 69, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, B.S.; Esmaeili, H.; Foroutan, R. Cadmium(II) Removal from Aqueous Solution Using Microporous Eggshell: Kinetic and Equilibrium Studies. Indones. J. Chem. 2018, 18, 265. [Google Scholar] [CrossRef]
- Li, X.; Li, H.; Qu, C. A Review of the Mechanism of Microbial Degradation of Petroleum Pollution. IOP Conf. Ser. Mater. Sci. Eng. 2019, 484, 012060. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Bahrani, S.; Sadrmousavi-Dizaj, A.; Arjmand, O.; Omidifar, N.; Lai, C.W.; Chiang, W.-H.; Gholami, A. Bioinorganic Synthesis of Sodium Polytungstate/Polyoxometalate in Microbial Kombucha Media for Precise Detection of Doxorubicin. Bioinorg. Chem. Appl. 2022, 2022, 2265108. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Pi, Y.; Bao, M.; Xu, N.; Li, Y.; Lu, J. Biodegradation of different petroleum hydrocarbons by free and immobilized microbial consortia. Environ. Sci. Process. Impacts 2015, 17, 2022–2033. [Google Scholar] [CrossRef]
- Pinheiro Pires, A.P.; Arauzo, J.; Fonts, I.; Domine, M.E.; Fernández Arroyo, A.; Garcia-Perez, M.E.; Montoya, J.; Chejne, F.; Pfromm, P.; Garcia-Perez, M. Challenges and Opportunities for Bio-oil Refining: A Review. Energy Fuels 2019, 33, 4683–4720. [Google Scholar] [CrossRef]
- Hou, D.; Al-Tabbaa, A.; O’Connor, D.; Hu, Q.; Zhu, Y.-G.; Wang, L.; Kirkwood, N.; Ok, Y.S.; Tsang, D.C.; Bolan, N.S. Sustainable remediation and redevelopment of brownfield sites. Nat. Rev. Earth Environ. 2023, 4, 271–286. [Google Scholar] [CrossRef]
- Saxena, G.; Kumar, V.; Shah, M.P. Bioremediation for Environmental Sustainability: Toxicity, Mechanisms of Contaminants Degradation, Detoxification and Challenges; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Kafle, A.; Timilsina, A.; Gautam, A.; Adhikari, K.; Bhattarai, A.; Aryal, N. Phytoremediation: Mechanisms, plant selection and enhancement by natural and synthetic agents. Environ. Adv. 2022, 8, 100203. [Google Scholar] [CrossRef]
- Lim, M.W.; Von Lau, E.; Poh, P.E. A comprehensive guide of remediation technologies for oil contaminated soil—Present works and future directions. Mar. Pollut. Bull. 2016, 109, 14–45. [Google Scholar] [CrossRef]
- Xiang, L.; Harindintwali, J.D.; Wang, F.; Redmile-Gordon, M.; Chang, S.X.; Fu, Y.; He, C.; Muhoza, B.; Brahushi, F.; Bolan, N. Integrating biochar, bacteria, and plants for sustainable remediation of soils contaminated with organic pollutants. Environ. Sci. Technol. 2022, 56, 16546–16566. [Google Scholar] [CrossRef] [PubMed]
- Meckenstock, R.U.; Boll, M.; Mouttaki, H.; Koelschbach, J.S.; Cunha Tarouco, P.; Weyrauch, P.; Dong, X.; Himmelberg, A.M. Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons. Microb. Physiol. 2016, 26, 92–118. [Google Scholar] [CrossRef] [PubMed]
- Okolafor, F.; Ekhaise, F.O. Microbial enzyme remediation of poly-aromatic hydrocarbon (PAH’s): A review. J. Int. Environ. Appl. Sci. 2022, 17, 10–21. [Google Scholar]
- Iranzo, M.; Sainz-Pardo, I.; Boluda, R.; Sanchez, J.; Mormeneo, S. The use of microorganisms in environmental remediation. Ann. Microbiol. 2001, 51, 135–144. [Google Scholar]
- Graj, W.; Lisiecki, P.; Szulc, A.; Chrzanowski, Ł.; Wojtera-Kwiczor, J. Bioaugmentation with petroleum-degrading consortia has a selective growth-promoting impact on crop plants germinated in diesel oil-contaminated soil. Water Air Soil Pollut. 2013, 224, 1–15. [Google Scholar] [CrossRef]
- El-Naas, M.H.; Acio, J.A.; El Telib, A.E. Aerobic biodegradation of BTEX: Progresses and prospects. J. Environ. Chem. Eng. 2014, 2, 1104–1122. [Google Scholar] [CrossRef]
- Grover, R.; Burse, S.A.; Shankrit, S.; Aggarwal, A.; Kirty, K.; Narta, K.; Srivastav, R.; Ray, A.K.; Malik, G.; Vats, A. Myg1 exonuclease couples the nuclear and mitochondrial translational programs through RNA processing. Nucleic Acids Res. 2019, 47, 5852–5866. [Google Scholar] [CrossRef]
- Li, J.; Yang, H. Polycyclic Aromatic Hydrocarbon Improves the Anaerobic Biodegradation of Benz [α] Anthracene in Sludge Via Boosting the Microbial Activity and Bioavailability. Pak. J. Zool. 2021, 53, 1–6. [Google Scholar] [CrossRef]
- Oliveira, L.G.; Araujo, K.C.; Barreto, M.C.; Bastos, M.E.P.; Lemos, S.G.; Fragoso, W.D. Applications of chemometrics in oil spill studies. Microchem. J. 2021, 166, 106216. [Google Scholar] [CrossRef]
- Tavares, T.S.; da Rocha, E.P.; Esteves Nogueira, F.G.; Torres, J.A.; Silva, M.C.; Kuca, K.; Ramalho, T.C. Δ-FeOOH as support for immobilization peroxidase: Optimization via a chemometric approach. Molecules 2020, 25, 259. [Google Scholar] [CrossRef]
- Vasudevan, M.; Nambi, I.M.; Kumar, G.S. Scenario-based modelling of mass transfer mechanisms at a petroleum contaminated field site-numerical implications. J. Environ. Manag. 2016, 175, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Bezza, F.A.; Chirwa, E.M.N. Production and applications of lipopeptide biosurfactant for bioremediation and oil recovery by Bacillus subtilis CN2. Biochem. Eng. J. 2015, 101, 168–178. [Google Scholar] [CrossRef]
- Chandankere, R.; Yao, J.; Cai, M.; Masakorala, K.; Jain, A.K.; Choi, M.M. Properties and characterization of biosurfactant in crude oil biodegradation by bacterium Bacillus methylotrophicus USTBa. Fuel 2014, 122, 140–148. [Google Scholar] [CrossRef]
- Chirwa, E.M.; Mampholo, T.; Fayemiwo, O. Biosurfactants as demulsifying agents for oil recovery from oily sludge–performance evaluation. Water Sci. Technol. 2013, 67, 2875–2881. [Google Scholar] [CrossRef]
- Bhanse, P.; Kumar, M.; Singh, L.; Awasthi, M.K.; Qureshi, A. Role of plant growth-promoting rhizobacteria in boosting the phytoremediation of stressed soils: Opportunities, challenges, and prospects. Chemosphere 2022, 303, 134954. [Google Scholar] [CrossRef]
- Zabed, H.M.; Akter, S.; Yun, J.; Zhang, G.; Awad, F.N.; Qi, X.; Sahu, J. Recent advances in biological pretreatment of microalgae and lignocellulosic biomass for biofuel production. Renew. Sustain. Energy Rev. 2019, 105, 105–128. [Google Scholar] [CrossRef]
- Alegbeleye, O.O.; Opeolu, B.O.; Jackson, V.A. Polycyclic aromatic hydrocarbons: A critical review of environmental occurrence and bioremediation. Environ. Manag. 2017, 60, 758–783. [Google Scholar] [CrossRef]
- Kalashgrani, M.Y.; Harzand, F.V.; Javanmardi, N.; Nejad, F.F.; Rahmanian, V. Recent Advances in Multifunctional magnetic nano platform for Biomedical Applications: A mini review. Adv. Appl. NanoBio-Technol. 2022, 3, 31–37. [Google Scholar]
- Lee, D.W.; Lee, H.; Lee, A.H.; Kwon, B.-O.; Khim, J.S.; Yim, U.H.; Kim, B.S.; Kim, J.-J. Microbial community composition and PAHs removal potential of indigenous bacteria in oil contaminated sediment of Taean coast, Korea. Environ. Pollut. 2018, 234, 503–512. [Google Scholar] [CrossRef]
- Davidson, S.-L.; Niepa, T.H. Micro-technologies for assessing microbial dynamics in controlled environments. Front. Microbiol. 2022, 12, 745835. [Google Scholar] [CrossRef]
- Amiard-Triquet, C.; Cossu-Leguille, C.; Mouneyrac, C. Biomarkers of defense, tolerance, and ecological consequences. Ecol. Biomark. Indic. Ecotoxicol. Eff. 2013, 2, 45–74. [Google Scholar]
- Steinberg, C.E.; Steinberg, C.E. Activation of oxygen: Multipurpose tool. In Stress Ecology: Environmental Stress as Ecological Driving Force and Key Player in Evolution; Springer: Berlin/Heidelberg, Germany, 2012; pp. 7–45. [Google Scholar]
- Fan, L.; Reynolds, D.; Liu, M.; Stark, M.; Kjelleberg, S.; Webster, N.S.; Thomas, T. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. USA 2012, 109, E1878–E1887. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, W.; Wu, Y.; Zhou, Z.; Lai, Q.; Shao, Z. Multiple alkane hydroxylase systems in a marine alkane degrader, Alcanivorax dieselolei B-5. Environ. Microbiol. 2011, 13, 1168–1178. [Google Scholar] [CrossRef]
- Kokol, V.; Doliška, A.; Eichlerová, I.; Baldrian, P.; Nerud, F. Decolorization of textile dyes by whole cultures of Ischnoderma resinosum and by purified laccase and Mn-peroxidase. Enzym. Microb. Technol. 2007, 40, 1673–1677. [Google Scholar] [CrossRef]
- Gajić, G.; Mitrović, M.; Pavlović, P. Phytobial remediation by bacteria and fungi. In Assisted Phytoremediation; Elsevier: Amsterdam, The Netherlands, 2022; pp. 285–344. [Google Scholar]
- Cabrera, M.Á.; Márquez, S.L.; Pérez-Donoso, J.M. Comparative genomic analysis of Antarctic pseudomonas isolates with 2, 4, 6-trinitrotoluene transformation capabilities reveals their unique features for xenobiotics degradation. Genes 2022, 13, 1354. [Google Scholar] [CrossRef] [PubMed]
- Tapadar, S.; Tripathi, D.; Pandey, S.; Goswami, K.; Bhattacharjee, A.; Das, K.; Palwan, E.; Rani, M.; Kumar, A. Role of extremophiles and extremophilic proteins in industrial waste treatment. In Removal of Emerging Contaminants through Microbial Processes; Springer: Berlin/Heidelberg, Germany, 2021; pp. 217–235. [Google Scholar]
- Kahng, H.-Y. Cellular responses of Pseudomonas sp. KK1 to two-ring polycyclic aromatic hydrocarbon, naphthalene. J. Microbiol. 2002, 40, 38–42. [Google Scholar]
- Pham, M.D.; Lin, Y.-P.; Van Vuong, Q.; Nagababu, P.; Chang, B.T.-A.; Ng, K.Y.; Chen, C.-H.; Han, C.-C.; Chen, C.-H.; Li, M.S. Inactivation of the particulate methane monooxygenase (pMMO) in Methylococcus capsulatus (Bath) by acetylene. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2015, 1854, 1842–1852. [Google Scholar] [CrossRef]
- Patil, T.; Pendse, A.; Aruna, K. Optimization of Urease Production by Bacillus megaterium Tara26 Isolated from Marble Quarry Sample and Its Application in Reduction of Water Hardness. Indian J. Appl. Res. 2020, 10, 50–57. [Google Scholar]
- Goyal, M.; Kalra, K.; Sareen, V.; Soni, G. Xylanase production with xylan rich lignocellulosic wastes by a local soil isolate of Trichoderma viride. Braz. J. Microbiol. 2008, 39, 535–541. [Google Scholar] [CrossRef]
- Pedroso, M.M.; Ely, F.; Mitić, N.; Carpenter, M.C.; Gahan, L.R.; Wilcox, D.E.; Larrabee, J.L.; Ollis, D.L.; Schenk, G. Comparative investigation of the reaction mechanisms of the organophosphate-degrading phosphotriesterases from Agrobacterium radiobacter (OpdA) and Pseudomonas diminuta (OPH). JBIC J. Biol. Inorg. Chem. 2014, 19, 1263–1275. [Google Scholar] [CrossRef]
- Keong, C.Y.; Chin, T.C.; Umar, N.A.B.; Mustapha, N.M.; Mohamad, S. The Effects of a Powder of the Fruiting Body of Commercial Lingzhi or Reishi Medicinal Mushroom Ganoderma lucidum (W. Curt.: Fr.) P. Karts.,] on Hypercholesterolemic Rat Skin, Applied With a Topical Application of Benzo (a) pyrene. Int. J. Med. Mushrooms 2010, 12, 367–378. [Google Scholar] [CrossRef]
- Khatoon, N.; Jamal, A.; Ali, M.I. Polymeric pollutant biodegradation through microbial oxidoreductase: A better strategy to safe environment. Int. J. Biol. Macromol. 2017, 105, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.S.; Gummadi, S.N. Biotechnological approach to caffeine degradation: Current trends and perspectives. In Microorganisms in Sustainable Agriculture and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 435–451. [Google Scholar]
- Darwesh, O.M.; Matter, I.A.; Eida, M.F. Development of peroxidase enzyme immobilized magnetic nanoparticles for bioremediation of textile wastewater dye. J. Environ. Chem. Eng. 2019, 7, 102805. [Google Scholar] [CrossRef]
- Bhatt, P. Smart Bioremediation Technologies: Microbial Enzymes; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Van Beilen, J.B.; Funhoff, E.G. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 2007, 74, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Kweon, O.; Jones, R.C.; Freeman, J.P.; Edmondson, R.D.; Cerniglia, C.E. Complete and integrated pyrene degradation pathway in Mycobacterium vanbaalenii PYR-1 based on systems biology. J. Bacteriol. 2007, 189, 464–472. [Google Scholar] [CrossRef]
- Kweon, O.; Kim, S.-J.; Freeman, J.P.; Song, J.; Baek, S.; Cerniglia, C.E. Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. MBio 2010, 1, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Kadri, T.; Rouissi, T.; Brar, S.K.; Cledon, M.; Sarma, S.; Verma, M. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: A review. J. Environ. Sci. 2017, 51, 52–74. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Yang, W.; Xu, F.; Wang, W.; Feng, L.; Bartlam, M.; Wang, L.; Rao, Z. Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: Unveiling the long-chain alkane hydroxylase. J. Mol. Biol. 2008, 376, 453–465. [Google Scholar] [CrossRef]
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58. [Google Scholar] [CrossRef]
- Wang, M.; Garrido-Sanz, D.; Sansegundo-Lobato, P.; Redondo-Nieto, M.; Conlon, R.; Martin, M.; Mali, R.; Liu, X.; Dowling, D.; Rivilla, R. Soil microbiome structure and function in ecopiles used to remediate petroleum-contaminated soil. Front. Environ. Sci. 2021, 9, 39. [Google Scholar] [CrossRef]
- Kim, S.-J.; Kweon, O.; Cerniglia, C.E. Proteomic applications to elucidate bacterial aromatic hydrocarbon metabolic pathways. Curr. Opin. Microbiol. 2009, 12, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Mallick, S.; Chatterjee, S.; Dutta, T.K. A novel degradation pathway in the assimilation of phenanthrene by Staphylococcus sp. strain PN/Y via meta-cleavage of 2-hydroxy-1-naphthoic acid: Formation of trans-2, 3-dioxo-5-(2′-hydroxyphenyl)-pent-4-enoic acid. Microbiology 2007, 153, 2104–2115. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Xie, X.; Wang, X.; Lin, L.; Yang, L.; Luan, T.; Chen, B. Transcriptional response of Mycobacterium sp. strain A1-PYR to multiple polycyclic aromatic hydrocarbon contaminations. Environ. Pollut. 2018, 243, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Regar, R.K.; Bajaj, A.; Ch, R.; Satyanarayana, G.N.V.; Mudiam, M.K.R.; Manickam, N. Simultaneous biodegradation of polyaromatic hydrocarbons by a Stenotrophomonas sp: Characterization of nid genes and effect of surfactants on degradation. Indian J. Microbiol. 2017, 57, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yan, J.; Wang, L.; Zhang, Y.; Liu, D.; Geng, H.; Xiong, L. Characterization of the phthalate acid catabolic gene cluster in phthalate acid esters transforming bacterium-Gordonia sp. strain HS-NH1. Int. Biodeterior. Biodegrad. 2016, 106, 34–40. [Google Scholar] [CrossRef]
- Stingley, R.L.; Brezna, B.; Khan, A.A.; Cerniglia, C.E. Novel organization of genes in a phthalate degradation operon of Mycobacterium vanbaalenii PYR-1. Microbiology 2004, 150, 3749–3761. [Google Scholar] [CrossRef]
- Yunusa, Y.; Umar, Z. Effective microbial bioremediation via the multi-omics approach: An overview of trends, problems and prospects. UMYU J. Microbiol. Res. 2021, 6, 127–145. [Google Scholar] [CrossRef]
- Fischer, A.; Manefield, M.; Bombach, P. Application of stable isotope tools for evaluating natural and stimulated biodegradation of organic pollutants in field studies. Curr. Opin. Biotechnol. 2016, 41, 99–107. [Google Scholar] [CrossRef]
- Adelana, S. Summary of Isotopes in Contaminant Hydrogeology. In Water Encyclopedia. sl; John Wiley Sons Inc.: Hoboken, NJ, USA, 2005; pp. 1–12. [Google Scholar]
- Aelion, C.M.; Höhener, P.; Hunkeler, D.; Aravena, R. Environmental Isotopes in Biodegradation and Bioremediation; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, W.; Bhatt, P.; Chen, S. Recent advanced technologies for the characterization of xenobiotic-degrading microorganisms and microbial communities. Front. Bioeng. Biotechnol. 2021, 9, 632059. [Google Scholar] [CrossRef]
- Shahi, A.; Ince, B.; Aydin, S.; Ince, O. Assessment of the horizontal transfer of functional genes as a suitable approach for evaluation of the bioremediation potential of petroleum-contaminated sites: A mini-review. Appl. Microbiol. Biotechnol. 2017, 101, 4341–4348. [Google Scholar] [CrossRef]
- Bhatt, P.; Sethi, K.; Gangola, S.; Bhandari, G.; Verma, A.; Adnan, M.; Singh, Y.; Chaube, S. Modeling and simulation of atrazine biodegradation in bacteria and its effect in other living systems. J. Biomol. Struct. Dyn. 2022, 40, 3285–3295. [Google Scholar] [CrossRef] [PubMed]
- Chambon, J.C.; Bjerg, P.L.; Scheutz, C.; Bælum, J.; Jakobsen, R.; Binning, P.J. Review of reactive kinetic models describing reductive dechlorination of chlorinated ethenes in soil and groundwater. Biotechnol. Bioeng. 2013, 110, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bilal, M.; Iqbal, H.M.; Raj, A. Trends in predictive biodegradation for sustainable mitigation of environmental pollutants: Recent progress and future outlook. Sci. Total Environ. 2021, 770, 144561. [Google Scholar] [CrossRef] [PubMed]
- Jaskulak, M.; Grobelak, A.; Vandenbulcke, F. Modelling assisted phytoremediation of soils contaminated with heavy metals–main opportunities, limitations, decision making and future prospects. Chemosphere 2020, 249, 126196. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Yadav, S.; Kumar, A.; Hashem, A.; Abd_Allah, E.F. Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front. Microbiol. 2018, 9, 1132. [Google Scholar] [CrossRef]
- Hualpa-Cutipa, E.; Acosta, R.A.S.; Cariga, O.J.M.; Espinoza-Medina, M.A.; Hansen-Reyes, M.; Medina-Cerna, D.; Olanda, M.C.; Cortez-Lázaro, A.A. Omics Insights into Cold Environments: Cold-Tolerant Microorganisms and their Potential Use in Bioremediation. In Omics Insights in Environmental Bioremediation; Springer: Berlin/Heidelberg, Germany, 2022; pp. 437–453. [Google Scholar]
- Kotoky, R.; Rajkumari, J.; Pandey, P. The rhizosphere microbiome: Significance in rhizoremediation of polyaromatic hydrocarbon contaminated soil. J. Environ. Manag. 2018, 217, 858–870. [Google Scholar] [CrossRef]
- Suneja, G.; Srivastav, R. Impact of Microbial Genome Sequencing Advancements in Understanding Extremophiles. In Extreme Environments; CRC Press: Boca Raton, FL, USA, 2021; pp. 330–342. [Google Scholar]
- Staats, M.; Braster, M.; Röling, W.F. Molecular diversity and distribution of aromatic hydrocarbon-degrading anaerobes across a landfill leachate plume. Environ. Microbiol. 2011, 13, 1216–1227. [Google Scholar] [CrossRef]
- Yaohua, G.; Ping, X.; Feng, J.; Keren, S. Co-immobilization of laccase and ABTS onto novel dual-functionalized cellulose beads for highly improved biodegradation of indole. J. Hazard. Mater. 2018, 365, 118–124. [Google Scholar] [CrossRef]
- Li, Q.-S.; Ogawa, J.; Schmid, R.D.; Shimizu, S. Engineering cytochrome P450 BM-3 for oxidation of polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 2001, 67, 5735–5739. [Google Scholar] [CrossRef]
- Megharaj, M.; Ramakrishnan, B.; Venkateswarlu, K.; Sethunathan, N.; Naidu, R. Bioremediation approaches for organic pollutants: A critical perspective. Environ. Int. 2011, 37, 1362–1375. [Google Scholar] [CrossRef]
- Abtahi, H.; Parhamfar, M.; Saeedi, R.; Villasenor, J.; Sartaj, M.; Kumar, V.; Coulon, F.; Parhamfar, M.; Didehdar, M.; Koolivand, A. Effect of competition between petroleum-degrading bacteria and indigenous compost microorganisms on the efficiency of petroleum sludge bioremediation: Field application of mineral-based culture in the composting process. J. Environ. Manag. 2020, 258, 110013. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Iqbal, H.M.; Shah, S.Z.H.; Hu, H.; Wang, W.; Zhang, X. Horseradish peroxidase-assisted approach to decolorize and detoxify dye pollutants in a packed bed bioreactor. J. Environ. Manag. 2016, 183, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Singh, S.N. Biodegradation of benzo (a) pyrene mediated by catabolic enzymes of bacteria. Int. J. Environ. Sci. Technol. 2014, 11, 1571–1580. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, T.; Li, X.; Yao, J.; Liu, W.; Chang, S.; Chen, Y. The fate and enhanced removal of polycyclic aromatic hydrocarbons in wastewater and sludge treatment system: A review. Crit. Rev. Environ. Sci. Technol. 2019, 49, 1425–1475. [Google Scholar] [CrossRef]
- Sakshi; Singh, S.; Haritash, A. Polycyclic aromatic hydrocarbons: Soil pollution and remediation. Int. J. Environ. Sci. Technol. 2019, 16, 6489–6512. [Google Scholar] [CrossRef]
| PAH Name | Molecular Weight (g/mole) | Melting Point (°C) | Aqueous Solubility (mg/L) | Molecule Formula | Log KOW | Vapor Pressure (Pa, 25 °C) | Boiling Point (°C) |
|---|---|---|---|---|---|---|---|
| Pyrene | 22 | 150–156 | 0.135 | C16 H10 | 4.88 | 5.0 × 10−5 | 360–404 |
| Acenaphthene | 154 | 90–96 | 3.9 | C12H10 | 3.92 | 3.92 | 265–280 |
| Acenaphthylene | 152 | 92–93 | 16.1 | C12H8 | 3.94 | 3.87 | 265–280 |
| Fluorene | 166 | 116–118 | 1.9 | C13H10 | 4.18 | 1.66 | 293–295 |
| Chrysene | 228 | 252–256 | 0.002 | C18H12 | 5.81 | 4.0 × 10−6 | 441–448 |
| Naphthalene | 128 | 80.2 | 31.7 | C10H8 | 3.30 | 11.14 | 218 |
| Benzopyrene | 252 | 177–179 | 0.00162 | C20H12 | 6.13 | 6.0 × 10−8 | 493–496 |
| Benzofluoranthene | 252 | 167–168 | 0.0008 | C20H12 | 6.11 | 5.2 × 10−8 | 481 |
| Benzo(2)fluoranthene | 252 | 198–217 | 0.0015 | C20H12 | 5.78 | 5.0 × 10−7 | 480–471 |
| Phenanthrene | 178 | 96–101 | 1.15 | C14H10 | 4.46 | 1.06 × 10−1 | 339–340 |
| Bacterial Species | Hydrocarbon | Mechanism of Action | References |
|---|---|---|---|
| Pseudomonas putida | Pyrene | Cometabolism | [102] |
| Pseudomonas W10 | Phenanthrene | Biosurfactant production | [31] |
| Pseudomonas aeruginosa | n-alkanes (C16–C19), fluorene, phenanthrene, and pyrene | [96] | |
| Delftia sp. NL1 | diesel | [97] | |
| Rhodococcus sp. P14 | phenanthrene, pyrene, benzo(a)pyrene | Change in fatty acid composition of the cell membrane/biofilm formation/ | [101] |
| Acinetobacter | naphthalene, acenaphthene, and acenaphthylene | Biosurfactant production | [35] |
| Enterobacter cloacae | crude oil | [103] | |
| Stenotrophomonas sp. Pemsol | biphenyl, anthraquinone, phenanthrene, naphthalene, phenanthridine | Horizontal gene transfer | [104] |
| Achromobacter (AC15) | Pyrene | Biosurfactant production | [105] |
| Alcanivorax borkumensis | Alkanes | Alkane hydroxylase | [106] |
| Rhodococcus erythropolis | Alkanes | Cytochrome P450 | [107] |
| Rhodococcus sp. BCP1, and R. opacus R7 | Alkanes, fatty acids, aromatic compounds, and (PAHs) | Cytochrome P450 and dioxygenase systems | [108] |
| Sphingomonas sp. | (PAHs) | Dioxygenase | [109] |
| Sphingobium yanoikuyae B1 | (PAHs) and phenanthrene derivatives | Dioxygenase and monooxygenase systems | [110] |
| Mycobacterium sp. | (PAHs) | Dioxygenase | [111] |
| Gordonia sp. | Alkanes | Cytochrome P450 | [112] |
| Acinetobacter sp. | Alkanes | Alkane hydroxylase | [113] |
| Micrococcus sp., Bacillus sp., Corynebacterium sp., Flavobacterium sp., Pseudomonas sp., Acinetobacter sp., Moraxella sp. and flavobacterium sp. | xylene, benzene, hexane, crude oil, kerosene, gasoline, diesel and olive oil | Biosurfactant production | [114] |
| Moraxella sp., Pseudomonas sp., members of Enterobacteriaceae, Vibrionaceae | Resins | employ versatile enzymatic and metabolic processes, including biofilm formation and adaptation. | [115,116] |
| B. stereothermophilus, Pseudomonas sp., Corynebacterium sp., Vibrio sp., Nocardia sp., Bacillus sp., Achromobacter sp. | Monocyclic aromatic hydrocarbons | B. stereothermophilus thrives at high temperatures for thermophilic hydrocarbon degradation; the other strains employ a variety of enzymatic and metabolic mechanisms. | [115,116] |
| Alcaligenes sp., Arthrobacter sp., Xanthomonas sp., Pseudomonas sp., Mycobacterium sp., Bacillus sp., Burkholderia cepacia, Anabaena sp., | Polycyclic aromatic hydrocarbons | employ various enzymatic and metabolic processes to effectively break down hydrocarbons, contributing to bioremediation | [115,116] |
| Fungal Species | ID | Degradation Rate % | Hydrocarbons | References |
|---|---|---|---|---|
| Aspergillus ficuum A. fumigatus A. flavus | MB#5058 | 54.6 | Aromatic hydrocarbons | [134] |
| MB#352615 | 59.6 | Aromatic hydrocarbons | ||
| MB#347788 | 59.8 | Aromatic hydrocarbons | ||
| Cladosporium sp. | CBMAI 1237 | 62 | Aromatic hydrocarbons | [135] |
| Coriolopis byrsina | APC5 | 96.1 | (PAHs) | [136] |
| Crinipellis campanella C. perniciosa C. stipitaria | MB#285848 MB#500896 MB#100767 | 39 95 94 | Aromatic hydrocarbons | [137] |
| Fusarium sp. | FJ613115.1 | 18.2–74.6 | Aromatic hydrocarbons | [138] |
| Rhizoctonia zeae | SOL3 | 42 | Aromatic hydrocarbons | [139] |
| Marasmiellus sp. | CBMAI 1062 | 98.8 | (PAHs) | [140] |
| Merulius tremellosus | KUC9161 | 83.6 | (PAHs) | [141] |
| Polyporus sp. | S133 | 71 | (PAHs) | [142] |
| Armillaria sp. | FO22 | 63 | (PAHs) | [143] |
| Peniophora incarnata | KUC8836 | 82.6, 97.9 | (PAHs) | [144] |
| Trichoderma sp. | F03 | 78 | (PAHs) | [129] |
| Pleurotus pulmonarius | FO43 | 99 | (PAHs) | [142] |
| Scopulariopsis brevicaulis | PZ-4 | 64 | Aromatic hydrocarbons | [145] |
| Phlebia brevispora | KUC9045 | 63.3 | (PAHs) | [144] |
| Phanerochaete chrysosporium | 92.2 | (PAHs), chlorinated compounds | [146] | |
| Pseudotrametes gibbasa | 28.33 | (PAHs) | [143] |
| Enzyme Name | Organism | Degraded Pollutants | References |
|---|---|---|---|
| Peptidase, Hydrolase | Pseudoalteromonas | PAHs | [164,197] |
| Hydrolase, Peptidase | Colweillia | [164,198] | |
| Peptidase, Hydrolase | Cyclocasticus | [164] | |
| Cytochrome P450 | Haematococcus pluvialis | [164] | |
| Hydroperoxidase | Chlorophyceae (Dunaliella tertiolecta) | [199] | |
| Lipoxygenase | [164,200] | ||
| Methane monooxygenase | Methylomirabilis oxyfera | Methane | [201] |
| Cytochrome P-450 dependent alkane monooxygenase | Alcanivorax spp. | cycloalkanes | [166,202] |
| Hydrolase (AlkB1 and AlkB2) | n-alkanes | ||
| Laccase | Trametes versicolor, Pleurotus ostreatus | Phenolic compounds, dyes, and lignin | [203] |
| Dehalogenases | Burkholderia xenovorans, Trametes versicolor | Halogenated compounds, e.g., chlorinated solvents | [204] |
| Nitroreductase | Pseudomonas spp. | Nitroaromatic compounds and nitramine-type explosives | [205] |
| Alkaline Phosphatase | Pseudomonas putida | Phosphorous-containing pollutants, like organophosphates | [206] |
| Naphthalene Dioxygenase | Pseudomonas putida, Pseudomonas sp. KK1 | (PAHs) | [207] |
| Methane Monooxygenase | Methylosinus trichosporium, Methylococcus capsulatus | Methane and other hydrocarbons | [208] |
| Urease | Bacillus sp., Klebsiella sp. | Urea and related compounds | [209] |
| Xylanase | Aspergillus niger, Bacillus sp., Trichoderma viride | Xylan and lignocellulosic materials | [210] |
| Phosphotriesterase | Pseudomonas diminuta, Agrobacterium radiobacter | Organophosphates and pesticides | [211] |
| Glutathione S-Transferase | Ganoderma sp. | (PAHs) | [212] |
| Oxidoreductases | Pleurotus ostreatus | Organic compounds, including phenolic pollutants and lignin | [213] |
| Xanthine oxidase | Pseudomonas putida | Purine compounds | [214] |
| Monooxygenase binding flavin (AlmA) | Gammaproteobacteria | Long C22 and C36 n-alkanes | [164] |
| Chlorophyceae |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alaidaroos, B.A. Advancing Eco-Sustainable Bioremediation for Hydrocarbon Contaminants: Challenges and Solutions. Processes 2023, 11, 3036. https://doi.org/10.3390/pr11103036
Alaidaroos BA. Advancing Eco-Sustainable Bioremediation for Hydrocarbon Contaminants: Challenges and Solutions. Processes. 2023; 11(10):3036. https://doi.org/10.3390/pr11103036
Chicago/Turabian StyleAlaidaroos, Bothaina A. 2023. "Advancing Eco-Sustainable Bioremediation for Hydrocarbon Contaminants: Challenges and Solutions" Processes 11, no. 10: 3036. https://doi.org/10.3390/pr11103036
APA StyleAlaidaroos, B. A. (2023). Advancing Eco-Sustainable Bioremediation for Hydrocarbon Contaminants: Challenges and Solutions. Processes, 11(10), 3036. https://doi.org/10.3390/pr11103036







