Ultra-Performance Liquid Chromatography with Tandem Mass Spectrometry for Simultaneous Analysis of 22 Analytes of Oncheong-Eum, a Traditional Korean Herbal Formula
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Chemicals, and Reagents
2.2. Sample Preparation for Simultaneous Analysis by UPLC–MS/MS
2.3. Preparation of Sample and Standard Stock Solutions for Simultaneous Analysis by UPLC–MS/MS
2.4. UPLC–MS/MS Simultaneous Analysis Conditions
2.5. Validation of the UPLC–MS/MS MRM Simultaneous Analytical Method
2.6. Stability Test
3. Results and Discussion
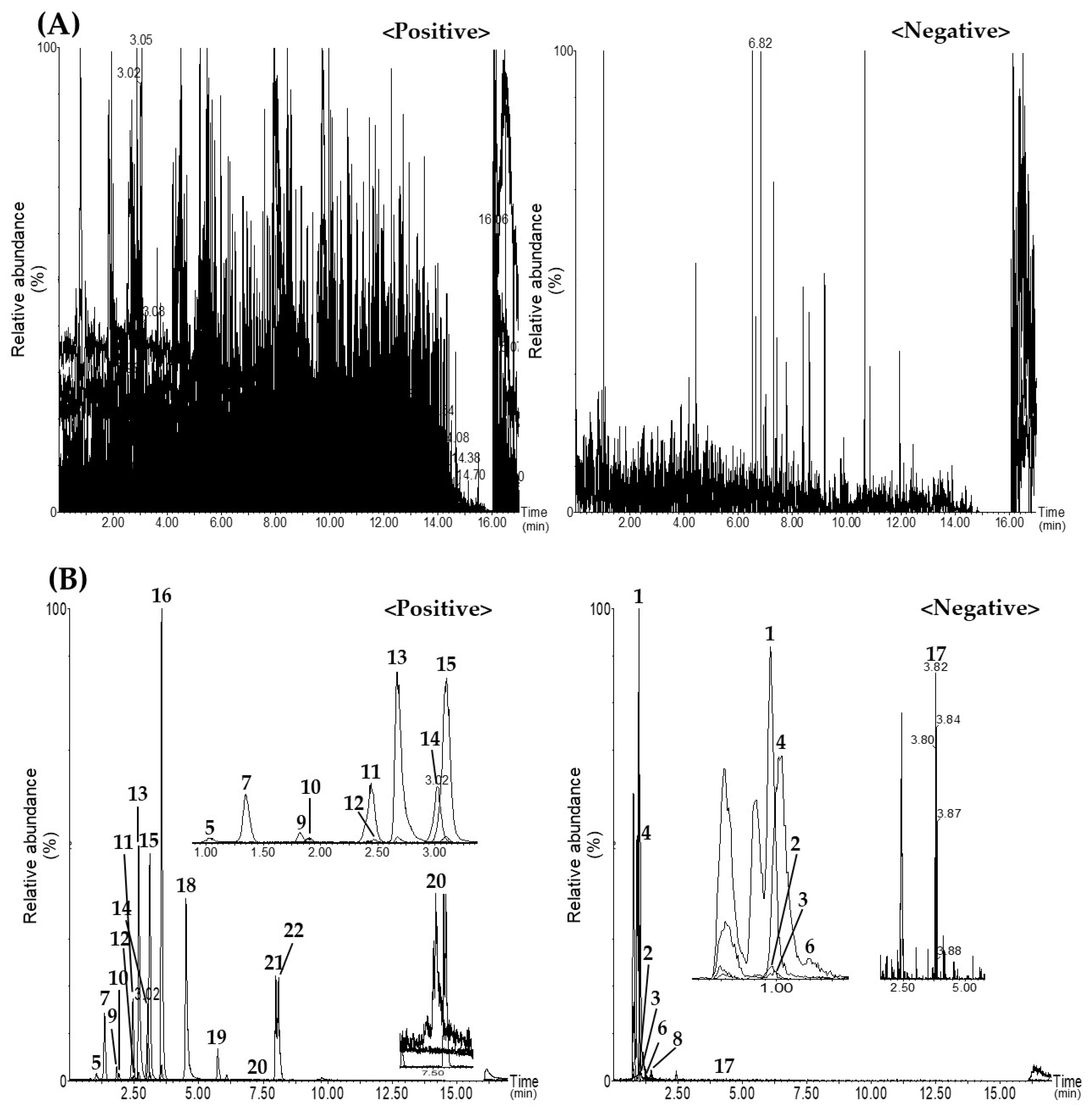
3.1. Optimization of Analytical Conditions for Simultaneous UPLC–MS/MS MRM Analysis
3.2. Setting of MRM Transition of Each Analyte for UPLC–MS/MS Analysis
3.3. Validation of the Optimized UPLC–MS/MS MRM Analytical Method
3.4. Stability of the Analytes
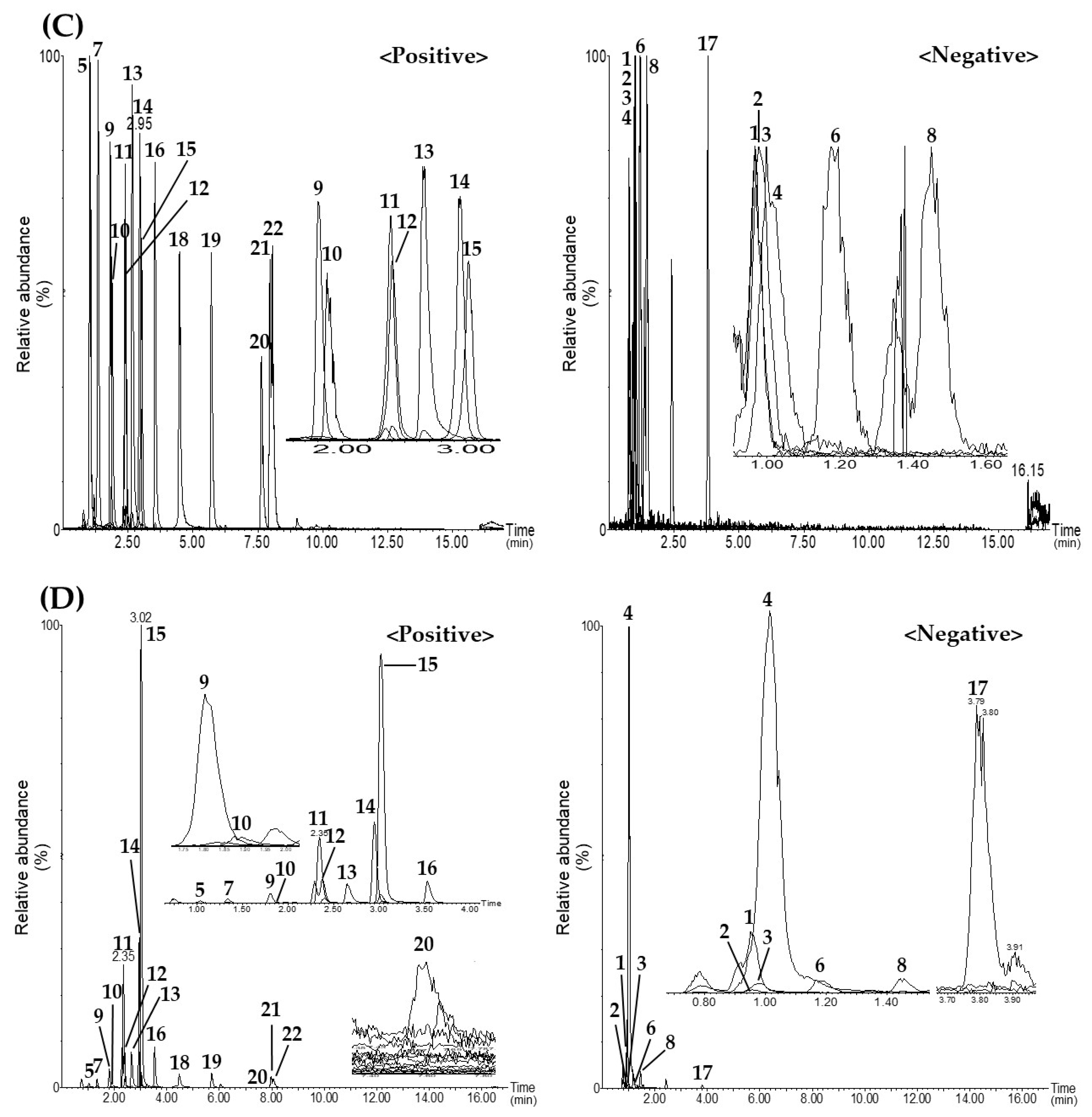
3.5. Simultaneous Analysis of the 22 Analytes in Various OCE Samples by the Optimized UPLC–MS/MS MRM Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Li, D.; Chem, Y.; Chen, W.; Xu, J.; Gao, L. Gut microbiota and aging: Traditional Chinese medicine and modern medicine. Clin. Interv. Aging 2023, 18, 963–986. [Google Scholar] [CrossRef]
- Seo, C.S.; Shin, H.K. Simultaneous analysis for quality control of traditional herbal medicine, Gungha-tang, using liquid chromatography-tandem mass spectrometry. Molecules 2022, 27, 1223. [Google Scholar] [CrossRef]
- Andoh, T.; Honda, Y.; Kawaharada, S.; Al-Akeel, A.; Nojima, H.; Kuraishi, Y. Inhibitory effect of the repeated treatment with Unsei-in on substance P-induced itch-associated responses through the downregulation of the expression of NK1 tachykinin receptor in mice. Biol. Pharm. Bull. 2003, 26, 896–898. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Andoh, T.; Al-Akeel, A.; Tsujii, K.; Nojima, H.; Kuraishi, Y. Repeated treatment with the traditional medicine Unsei-in inhibits substance P-induced itch-associated responses through downregulation of the expression of nitric oxide synthase 1 in mice. J. Pharmacol. Sci. 2004, 94, 207–210. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, J.M.; Lee, S.E.; Jung, H.J.; Choi, S.B.; Seo, H.S.; Jung, H.A.; Ko, W.S.; Yoon, H.J. Overseas clinical research trends of On Cheong Eum on skin disease. J. Korean Med. Ophthalmol. Otolaryngol. Dermatol. 2007, 30, 1–9. [Google Scholar]
- Wang, L.M.; Mineshita, S. Preventive effects of Unsei-in and Oren-gedoku-to, Chinese traditional medicines, against rat paw oedema and abdominal constriction in mice. J. Pharm. Pharmacol. 1996, 48, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Nose, M.; Sakushima, J.; Harada, D.; Ogihara, Y. Comparison of immunopharmacological actions of 8 kinds of Kampo-hozais clinically used in atopic dermatitis on delayed-type hypersensitivity in mice. Biol. Pharm. Bull. 1999, 22, 48–54. [Google Scholar] [CrossRef]
- Wang, L.M.; Yamamoto, T.; Wang, X.X.; Yang, L.; Koike, Y.; Shiba, K.; Mineshita, S. Effects of Oren-gedoku-to and Unsei-in, Chinese traditional medicines, on interleukin-8 and superoxide dismutase in rats. J. Pharm. Pharmacol. 1997, 49, 102–104. [Google Scholar] [CrossRef]
- An, T.E.B.; Lim, D.C. In vitro cytotoxicity, skin regeneration, anti-wrinkle, whitening and in vivo skin moisturizing effects of Oncheongeum. J. Korean Obstet. Gynecol. 2016, 29, 14–34. [Google Scholar] [CrossRef]
- Ku, Y.R.; Chang, Y.S.; Wen, K.C.; Ho, L.K. Analysis and confirmation of synthetic anorexics in adulterated traditional Chinese medicines by high-performance capillary electrophoresis. J. Chromatogr. A 1999, 848, 537–543. [Google Scholar] [CrossRef]
- Xue, J.; Wang, R.; Chen, X.; Hu, S.; Bai, X. Three-phase hollow-fiber liquid-phase microextraction based on deep eutectic solvent as acceptor phase for extraction and preconcentration of main active compounds in a traditional Chinese medicinal formula. J. Sep. Sci. 2019, 42, 2239–2246. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Zhang, C.; Zhao, S.; Cheng, S.; Zhou, W.; Xu, J.; Zhan, P.; Zeper, A. Comprehensive chemical profiling and quantitative analysis of ethnic Yi medicine Miao-fu-Zhi-Tong granules using UHPLC–MS/MS. Chin. J. Nat. Med. 2023, 21, 214–225. [Google Scholar] [PubMed]
- Ge, N.; Li, Z.; Yang, L.; Yan, G.; Zhang, A.; Zhang, X.; Wu, X.; Sun, H.; Li, D.; Wang, X. Development and validation of a UPLC–MS/MS method for the quantification of components in the ancient classical Chinese medicine formula of Guyinjian. Molecules 2022, 27, 8611. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Cao, J.; Zhang, C.; Su, B.; Wang, S.; Ning, N.; Lei, T.; Li, P. A comprehensive strategy for quality evaluation of Wushe Zhiyang Pills by integrating UPLC-DAD fingerprint and multi-ingredients rapid quantitation with UPLC–MS/MS technology. J. Pharm. Biomed. Anal. 2022, 210, 114556. [Google Scholar] [CrossRef]
- Xie, M.; Yu, Y.; Zhu, Z.; Deng, L.; Ren, B.; Zhang, M. Simultaneous determination of six main components in Bushen Huoxue prescription by HPLC–CAD. J. Pharm. Biomed. Anal. 2021, 201, 114087. [Google Scholar] [CrossRef]
- Liu, L.; Chu, X.; Tian, C.; Xia, M.; Zhang, L.; Jiang, J.; Gui, S. Chemo proling and simultaneous analysis of different combinations of Sinomenii Caulis and Ramulus Cinnamomi using UHPLC–Q–TOF–MS, GC–MS and HPLC methods. J. Chromatogr. Sci. 2021, 59, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.C.; Huang, S.S.; Liu, P.Y.; Wang, B.C.; Tsai, C.F.; Wang, D.Y.; Cheng, H.F. Simultaneous quantification of six indicator compounds in Wen-Qing-Yin by high-performance liquid chromatography-diode array detection. J. Food Drug Anal. 2019, 27, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Seo, C.S.; Shin, H.K. Simultaneous analysis of 19 marker components for quality control of Oncheong-eum using HPLC–DAD. Molecules 2022, 27, 2992. [Google Scholar] [CrossRef]
- Oh, S.H. The Korean Herbal Pharmacopoeia, 5th ed.; Shinibooks: Seoul, Republic of Korea, 2016; p. 412. [Google Scholar]
- Cao, X.; You, G.; Li, H.; Li, D.; Wang, M.; Ren, X. Comparative investigation for rotten xylem (kuqin) and strip types (tiaoqin) of Scutellaria baicalensis Georgi base on fingerprinting and chemical pattern recognition. Molecules 2019, 24, 2431. [Google Scholar] [CrossRef]
- Xu, S.; Yang, L.; Tian, R.; Wang, Z.; Liu, Z.; Xie, P.; Feng, Q. Species differentiation and quality assessment of Radix Paeoniae Rubra (Chi-shao) by means of high-performance liquid chromatographic fingerprint. J. Chromatogr. A 2009, 1216, 2163–2168. [Google Scholar] [CrossRef]
- Ryuk, J.A.; Zheng, M.S.; Lee, M.Y.; Seo, C.S.; Li, Y.; Lee, S.H.; Moon, D.C.; Lee, H.W.; Lee, J.H.; Park, J.Y.; et al. Discrimination of Phellodendron amurense and P. chinense based on DNA analysis and the simultaneous analysis of alkaloids. Arch. Pharm. Res. 2012, 35, 1045–1054. [Google Scholar] [CrossRef]
- Lee, E.J.; Hong, J.K.; Whang, W.K. Simultaneous determination of bioactive marker compounds from Gardeniae fructus by high performance liquid chromatography. Arch. Pharm. Res. 2014, 37, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Seo, C.S.; Shin, H.K. Quantitative analysis of eight compounds in traditional Korean medicine, Gongjindan using HPLC, UPLC–MS/MS, and GC–MS/MS systems. Separations 2023, 10, 231. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Guidance for Industry, Q2B, Validation of Analytical Procedures: Methodology; Food and Drug Administration: Rockville, MD, USA, 1996. [Google Scholar]
- Jeong, S.Y.; Kim, H.M.; Lee, K.H.; Kim, K.Y.; Huang, D.S.; Kim, J.H.; Seong, R.S. Quantitative analysis of marker compounds in Angelica gigas, Angelica sinensis, and Angelica acutiloba by HPLC/DAD. Chem. Pharm. Bull. 2015, 63, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.E.; Seong, G.U.; Lee, Y.J.; Won, J.H. Quantitative analysis for the quality evaluation of active ingredients in Cnidium Rhizome. Yakhak Hoeji 2016, 60, 227–234. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, E.J.; Kim, J.S.; Lee, J.H.; Kang, S.S. Phytochemical studies on Rehmanniae Radix Preparata. Kor. J. Pharmacogn. 2011, 42, 117–126. [Google Scholar]
- Lv, X.; Li, Y.; Tang, C.; Zhang, Y.; Zhang, J.; Fan, G. Integration of HPLC-based fingerprint and quantitative analyses for differentiating botanical species and geographical growing origins of Rhizoma coptidis. Pharm. Bol. 2016, 54, 3264–3271. [Google Scholar] [CrossRef]
- Tong, L.; Wan, M.; Zhang, L.; Zhu, Y.; Sun, H.; Bi, K. Simultaneous determination of baicalin, wogonoside, baicalein, wogonin, oroxylin A and chrysin of Radix scutellariae extract in rat plasma by liquid chromatography tandem mass spectrometry. J. Pharm. Biomed. Anal. 2012, 70, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ji, L.; Song, S.; Wang, J.; Wei, N.; Jiang, M.; Bai, G.; Luo, G. Identification of the major constituents in Xuebijing injection by HPLC–ESI–MS. Phytochem. Anal. 2011, 22, 330–338. [Google Scholar] [CrossRef]
- Jaiswal, R.; Müller, H.; Müller, A.; Karar, M.G.E.; Kuhnert, N. Identification and characterization of chlorogenic acids, chlorogenic acid glycosides and flavonoids from Lonicera henryi L. (Caprifoliaceae) leaves by LC–MSn. Phytochem. 2014, 108, 252–263. [Google Scholar] [CrossRef]
- Zhou, T.; Liu, H.; Wen, J.; Fan, G.; Chai, Y.; Wu, Y. Fragmentation study of iridoid glycosides including epimers by liquid chromatography-diode array detection/electrospray ionization mass spectrometry and its application in metabolic fingerprint analysis of Gardenia jasminoides Ellis. Rapid Commun. Mass Spectrom. 2010, 24, 2520–2528. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, Y.; Shao, Y.; Chang, Z.; Guo, Y.; Feng, X.; Xu, D.; Zhang, J.; Song, Y.; Hou, R. Comparative pharmacokinetics and metabolites study of seven major bioactive components of Shaoyoa-Gancao decoction in normal and polycystic ovary syndrome rats by ultra high pressure liquid chromatography with tandem mass spectrometry. J. Sep. Sci. 2019, 42, 2467–2586. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Wang, X.; Li, H.; Shi, X.; Fan, W.; Zhao, S.; Liu, M.; Niu, L. Studies on metabolism of total glucosides of paeony from Paeoniae Radix Alba in rats by UPLC–Q–TOF–MS/MS. Biomed. Chromatogr. 2015, 29, 1769–1779. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, X. Rapid determination of isomeric benzoylpaeoniflorin and benzoylalbiflorin in rat plasma by LC–MS/MS method. Int. J. Anal. Chem. 2017, 2017, 1693464. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, X.; Dai, W.; Yan, S.; Huang, H.; Liang, X.; Li, Y.; Zhang, W. Chemical fingerprinting of Liuwei Dihuang Pill and simultaneous determination of its major bioactive constituents by HPLC coupled with multiple detections of DAD, ELSD and ESI–MS. J. Pharm. Biomed. Anal. 2009, 49, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Yang, D.H.; Jung, M.; Jung, E.H.; Eom, H.Y.; Suh, J.H.; Min, J.W.; Kim, U.; Min, H.; Kim, J.; et al. Simultaneous determination of chromones and coumarins in Radix Saposhnikoviae by high performance liquid chromatography with diode array and tandem mass detectors. J. Chromatogr. A 2011, 1218, 6319–6330. [Google Scholar] [CrossRef]
- Ahn, M.J.; Lee, M.K.; Kim, Y.C.; Sung, S.H. The simultaneous determination of coumarins in Angelica gigas root by high performance liquid chromatography–diode array detector coupled with electrospray ionization/mass spectrometry. J. Pharm. Biomed. Anal. 2008, 46, 258–266. [Google Scholar] [CrossRef]
- Wang, D.; Liu, Z.; Guo, M.; Liu, S. Structural elucidation and identification of alkaloids in Rhizoma Coptidis by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2004, 39, 1356–1365. [Google Scholar] [CrossRef]
- Chen, L.; Qi, J.; Chang, Y.; Zhu, D.; Yu, B. Identification and determination of the major constituents in traditional Chinese medicinal formula Danggui-Shaoyao-San by HPLC–DAD–ESI–MS/MS. J. Pharm. Biomed. Anal. 2009, 50, 127–137. [Google Scholar] [CrossRef]
- Tao, S.; Li, H.; Liu, J. Metabolic profiling of ligustilide and identification of the metabolite in rat and human hepatocytes by liquid chromatography combined with high-resolution mass spectrometry. J. Sep. Sci. 2020, 43, 4405–4413. [Google Scholar]
| Analyte 1 | Ion Mode | Molecular Weight | Precursor Ion (Q1) | Product Ion (Q3) | Cone Voltage (V) | Collision Energy (eV) | Retention Time (min) |
|---|---|---|---|---|---|---|---|
| 1 | − | 170.1 | 169.0 | 125.0 | 25 | 15 | 0.97 |
| 2 | − | 404.4 | 403.2 | 241.0 | 35 | 10 | 0.97 |
| 3 | − | 496.5 | 495.4 | 137.0 | 40 | 25 | 0.98 |
| 4 | − | 354.3 | 353.2 | 191.0 | 20 | 20 | 1.02 |
| 5 | + | 126.1 | 127.0 | 109.0 | 20 | 10 | 1.02 |
| 6 | − | 388.4 | 387.2 | 123.0 | 25 | 15 | 1.20 |
| 7 | + | 480.5 | 481.4 | 197.1 | 20 | 15 | 1.33 |
| 8 | − | 480.5 | 479.2 | 121.0 | 32 | 25 | 1.44 |
| 9 | + | 408.4 | 409.4 | 247.2 | 30 | 15 | 1.81 |
| 10 | + | 184.2 | 195.0 | 177.0 | 15 | 10 | 1.89 |
| 11 | + | 373.8 | 338.4 | 322.3 | 30 | 30 | 2.38 |
| 12 | + | 355.8 | 320.1 | 290.0 | 45 | 25 | 2.39 |
| 13 | + | 446.4 | 447.3 | 271.0 | 25 | 15 | 2.65 |
| 14 | + | 387.9 | 352.1 | 336.0 | 40 | 30 | 2.95 |
| 15 | + | 371.8 | 336.1 | 320.0 | 35 | 30 | 3.02 |
| 16 | + | 460.4 | 461.3 | 285.1 | 30 | 20 | 3.54 |
| 17 | − | 584.6 | 583.4 | 121.0 | 40 | 25 | 3.80 |
| 18 | + | 270.2 | 271.1 | 123.0 | 40 | 30 | 4.47 |
| 19 | + | 284.3 | 285.1 | 270.0 | 40 | 20 | 5.70 |
| 20 | + | 190.1 | 191.0 | 91.0 | 30 | 25 | 7.66 |
| 21 | + | 328.4 | 329.2 | 229.0 | 35 | 20 | 7.96 |
| 22 | + | 328.4 | 329.2 | 229.0 | 35 | 20 | 8.07 |
| Analyte 1 | Linear Range (μg/L) | Regression Equation 2 | r2 | LOD (μg/L) | LOQ (μg/L) |
|---|---|---|---|---|---|
| 1 | 250.00–5000.00 | y = 366.26x + 35,869.50 | 0.9967 | 1.88 × 10−1 | 6.21 × 101 |
| 2 | 250.00–5000.00 | y = 2.96x + 418.22 | 0.9950 | 1.64 × 101 | 5.40 × 101 |
| 3 | 10.00–500.00 | y = 128.97x + 849.98 | 0.9986 | 4.93 × 10−1 | 1.63 |
| 4 | 100.00–2500.00 | y = 523.14x + 20,049.70 | 0.9996 | 2.96 × 10−1 | 9.78 × 10−1 |
| 5 | 10.00–500.00 | y = 5852.87x + 50,893.70 | 0.9992 | 3.85 × 10−1 | 1.27 |
| 6 | 250.00–5000.00 | y = 1.15x + 181.45 | 0.9976 | 2.12 × 101 | 6.99 × 101 |
| 7 | 250.00–5000.00 | y = 2936.14x + 106,224.00 | 0.9993 | 4.64 × 10−2 | 1.53 × 10−1 |
| 8 | 250.00–5000.00 | y = 4.65x + 398.67 | 0.9975 | 2.31 × 101 | 7.62 × 101 |
| 9 | 10.00–500.00 | y = 21,943.90x + 137,062.00 | 0.9992 | 9.09 × 10−3 | 3.00 × 10−3 |
| 10 | 10.00–500.00 | y = 6981.34x + 17,442.00 | 0.9975 | 9.52 × 10−2 | 3.14 × 10−1 |
| 11 | 10.00–500.00 | y = 80,723.40x + 1,592,260.00 | 0.9972 | 7.58 × 10−3 | 2.50 × 10−2 |
| 12 | 10.00–500.00 | y = 7396.96x + 158,880.00 | 0.9969 | 3.33 × 10−2 | 1.10 × 10−1 |
| 13 | 250.00–5000.00 | y = 13,141.30x + 668,576.00 | 0.9983 | 3.09 × 10−2 | 1.02 × 10−1 |
| 14 | 10.00–500.00 | y = 96,358.10x + 1,478,850.00 | 0.9987 | 1.82 × 10−3 | 6.00 × 10−3 |
| 15 | 100.00–2500.00 | y = 32,388.10x + 3,056,940.00 | 0.9969 | 2.42 × 10−3 | 8.00 × 10−3 |
| 16 | 250.00–5000.00 | y = 19,403.60x + 2,923,960.00 | 0.9952 | 2.73 × 10−3 | 9.00 × 10−3 |
| 17 | 10.00–500.00 | y = 20.86x + 83.16 | 0.9986 | 3.20 × 10−1 | 1.06 |
| 18 | 250.00–5000.00 | y = 9583.75x + 352,523.00 | 0.9992 | 1.26 × 10−1 | 4.15 × 10−1 |
| 19 | 10.00–500.00 | y = 36,776.20x + 294,755.00 | 0.9992 | 2.12 × 10−3 | 7.00 × 10−3 |
| 20 | 10.00–500.00 | y = 267.38x + 2420.96 | 0.9977 | 5.68 | 1.88 × 101 |
| 21 | 10.00–500.00 | y = 58,707.80x + 602,086.00 | 0.9994 | 4.55 × 10−3 | 1.50 × 10−2 |
| 22 | 10.00–500.00 | y = 83,487.50x + 830,806.00 | 0.9993 | 4.24 × 10−3 | 1.40 × 10−3 |
| Analyte 1 | Spiked Amount (μg/L) | Found Amount (μg/L) | Accuracy (%) | SD 2 | RSD (%) |
|---|---|---|---|---|---|
| 1 | 400.00 | 3.95 × 102 | 98.69 | 6.62 | 6.71 |
| 800.00 | 9.24 × 102 | 115.47 | 4.77 | 4.13 | |
| 1600.00 | 1.83 × 103 | 114.13 | 4.82 | 4.22 | |
| 2 | 600.00 | 5.15 × 102 | 85.75 | 4.83 | 5.63 |
| 1200.00 | 1.18 × 103 | 97.96 | 5.11 | 5.22 | |
| 2400.00 | 2.41 × 103 | 100.35 | 5.15 | 5.14 | |
| 3 | 8.00 | 6.74 × 101 | 84.23 | 5.27 | 6.25 |
| 160.00 | 1.50 × 102 | 93.99 | 5.79 | 6.16 | |
| 320.00 | 2.94 × 102 | 91.86 | 8.57 | 9.32 | |
| 4 | 600.00 | 5.24 × 102 | 87.38 | 5.32 | 6.09 |
| 1200.00 | 1.14 × 103 | 95.39 | 2.87 | 3.01 | |
| 2400.00 | 2.18 × 103 | 90.85 | 3.67 | 4.04 | |
| 5 | 20.00 | 2.09 × 101 | 104.32 | 8.13 | 7.80 |
| 40.00 | 3.93 × 101 | 98.17 | 7.53 | 7.67 | |
| 80.00 | 7.68 × 101 | 96.06 | 8.02 | 8.35 | |
| 6 | 400.00 | 3.63 × 102 | 90.69 | 4.59 | 5.06 |
| 800.00 | 7.56 × 102 | 94.55 | 2.87 | 3.04 | |
| 1600.00 | 1.52 × 103 | 94.76 | 5.72 | 6.03 | |
| 7 | 800.00 | 8.00 × 102 | 99.98 | 6.60 | 6.61 |
| 1600.00 | 1.60 × 103 | 100.27 | 2.93 | 2.92 | |
| 3200.00 | 3.05 × 103 | 95.38 | 4.19 | 4.39 | |
| 8 | 1000.00 | 1.01 × 103 | 100.73 | 5.55 | 5.51 |
| 2000.00 | 2.25 × 103 | 112.56 | 1.01 | 0.90 | |
| 4000.00 | 4.42 × 103 | 110.42 | 2.45 | 2.22 | |
| 9 | 20.00 | 2.02 × 101 | 101.21 | 2.91 | 2.87 |
| 40.00 | 4.08 × 101 | 101.89 | 1.34 | 1.31 | |
| 80.00 | 8.04 × 101 | 100.50 | 1.22 | 1.21 | |
| 10 | 80.00 | 7.83 × 101 | 94.16 | 3.13 | 3.33 |
| 160.00 | 1.57 × 102 | 97.99 | 2.97 | 3.04 | |
| 320.00 | 3.23 × 102 | 101.01 | 3.84 | 3.80 | |
| 11 | 20.00 | 2.04 × 101 | 102.02 | 9.84 | 9.65 |
| 40.00 | 4.26 × 101 | 106.60 | 5.33 | 5.00 | |
| 80.00 | 8.14 × 101 | 101.72 | 6.50 | 6.39 | |
| 12 | 40.00 | 3.64 × 101 | 90.98 | 4.16 | 4.57 |
| 80.00 | 8.31 × 101 | 103.88 | 0.86 | 0.83 | |
| 160.00 | 1.65 × 102 | 102.93 | 1.80 | 1.75 | |
| 13 | 1200.00 | 1.13 × 103 | 94.57 | 5.76 | 6.10 |
| 2400.00 | 2.46 × 103 | 102.38 | 4.53 | 4.43 | |
| 4800.00 | 4.80 × 103 | 99.90 | 4.91 | 4.92 | |
| 14 | 40.00 | 3.55 × 101 | 88.86 | 4.43 | 4.98 |
| 80.00 | 7.95 × 101 | 99.38 | 3.28 | 3.30 | |
| 160.00 | 1.56 × 102 | 97.69 | 2.04 | 2.09 | |
| 15 | 400.00 | 3.58 × 102 | 89.57 | 3.07 | 3.42 |
| 800.00 | 8.78 × 102 | 109.78 | 2.56 | 2.33 | |
| 1600.00 | 1.75 × 103 | 109.30 | 1.89 | 1.73 | |
| 16 | 600.00 | 5.31 × 102 | 88.50 | 3.51 | 3.96 |
| 1200.00 | 1.12 × 103 | 93.03 | 3.20 | 3.44 | |
| 2400.00 | 2.23 × 103 | 93.04 | 2.34 | 2.51 | |
| 17 | 40.00 | 3.66 × 101 | 91.39 | 7.46 | 8.16 |
| 80.00 | 8.35 × 101 | 104.35 | 7.22 | 6.92 | |
| 160.00 | 1.67 × 102 | 104.17 | 1.62 | 1.56 | |
| 18 | 600.00 | 5.28 × 102 | 87.99 | 6.20 | 7.05 |
| 1200.00 | 1.27 × 103 | 105.42 | 9.95 | 9.44 | |
| 2400.00 | 2.46 × 103 | 102.63 | 5.52 | 5.38 | |
| 19 | 20.00 | 2.01 × 101 | 100.33 | 2.23 | 2.22 |
| 40.00 | 4.15 × 101 | 103.71 | 2.83 | 2.73 | |
| 80.00 | 8.12 × 101 | 101.55 | 2.08 | 2.05 | |
| 20 | 20.00 | 1.98 × 101 | 99.06 | 3.87 | 3.91 |
| 40.00 | 3.85 × 101 | 96.16 | 4.19 | 4.36 | |
| 80.00 | 7.65 × 101 | 95.58 | 4.59 | 4.80 | |
| 21 | 40.00 | 3.95 × 101 | 98.68 | 7.90 | 8.01 |
| 80.00 | 7.79 × 101 | 97.32 | 4.24 | 4.35 | |
| 160.00 | 1.50 × 102 | 93.73 | 2.89 | 3.08 | |
| 22 | 40.00 | 3.99 × 101 | 99.72 | 3.54 | 3.55 |
| 80.00 | 8.19 × 101 | 102.43 | 4.49 | 4.38 | |
| 160.00 | 1.63 × 102 | 101.70 | 3.01 | 2.96 |
| Analyte 1 | Conc. (μg/L) | Intraday (n = 5) | Interday (n = 5) | Repeatability (n = 6) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Observed Conc. (μg/L) | Precision (RSD, %) | Accuracy (%) | Observed Conc. (μg/L) | Precision (RSD, %) | Accuracy (%) | Retention Time (RSD, %) | Peak Area (RSD, %) | ||
| 1 | 400.00 | 4.07 × 102 | 9.45 | 101.73 | 3.94 × 102 | 7.29 | 98.47 | 3.89 | 7.00 |
| 800.00 | 9.20 × 102 | 2.97 | 115.06 | 9.25 × 102 | 3.33 | 115.61 | |||
| 1600.00 | 1.80 × 103 | 3.22 | 112.34 | 1.83 × 103 | 3.45 | 114.38 | |||
| 2 | 600.00 | 5.48 × 102 | 9.09 | 91.39 | 5.20 × 102 | 7.69 | 86.60 | 3.87 | 5.48 |
| 1200.00 | 1.20 × 103 | 5.63 | 100.39 | 1.20 × 103 | 5.21 | 100.07 | |||
| 2400.00 | 2.37 × 103 | 4.50 | 98.56 | 2.46 × 103 | 4.40 | 102.52 | |||
| 3 | 80.00 | 7.83 × 101 | 3.99 | 97.91 | 6.96 × 101 | 4.74 | 87.02 | 4.07 | 7.44 |
| 160.00 | 1.68 × 102 | 5.09 | 104.72 | 1.53 × 102 | 5.89 | 95.65 | |||
| 320.00 | 3.33 × 102 | 3.89 | 104.21 | 3.02 × 102 | 6.16 | 94.25 | |||
| 4 | 600.00 | 5.97 × 102 | 3.78 | 99.55 | 5.35 × 102 | 4.74 | 89.22 | 3.64 | 7.21 |
| 1200.00 | 1.24 × 103 | 5.30 | 103.02 | 1.16 × 103 | 3.81 | 96.57 | |||
| 2400.00 | 2.32 × 103 | 4.13 | 96.55 | 2.23 × 103 | 3.85 | 92.88 | |||
| 5 | 20.00 | 1.71 × 101 | 6.10 | 85.32 | 1.97 × 101 | 6.70 | 98.62 | 3.89 | 8.31 |
| 40.00 | 3.95 × 101 | 3.66 | 98.84 | 4.06 × 101 | 5.75 | 101.46 | |||
| 80.00 | 7.66 × 101 | 2.45 | 95.79 | 8.12 × 101 | 4.17 | 101.48 | |||
| 6 | 400.00 | 3.83 × 102 | 5.96 | 95.78 | 3.60 × 102 | 5.16 | 90.00 | 3.41 | 3.90 |
| 800.00 | 7.75 × 102 | 5.09 | 96.82 | 7.64 × 102 | 4.13 | 95.55 | |||
| 1600.00 | 1.60 × 103 | 6.97 | 100.28 | 1.59 × 103 | 6.24 | 98.04 | |||
| 7 | 800.00 | 7.50 × 102 | 1.30 | 93.78 | 7.76 × 102 | 5.18 | 97.06 | 2.90 | 7.91 |
| 1600.00 | 1.48 × 103 | 4.84 | 92.22 | 1.57 × 103 | 3.65 | 98.34 | |||
| 3200.00 | 2.94 × 103 | 4.12 | 91.76 | 3.10 × 103 | 4.42 | 96.95 | |||
| 8 | 1000.00 | 9.56 × 102 | 6.48 | 95.59 | 6.46 × 102 | 4.91 | 96.07 | 2.92 | 8.67 |
| 2000.00 | 2.14 × 103 | 2.69 | 107.17 | 1.44 × 103 | 2.60 | 104.27 | |||
| 4000.00 | 4.26 × 103 | 3.49 | 106.58 | 2.83 × 103 | 3.83 | 103.91 | |||
| 9 | 20.00 | 2.08 × 101 | 4.12 | 104.06 | 2.00 × 101 | 4.40 | 99.90 | 2.71 | 5.68 |
| 40.00 | 4.17 × 101 | 1.80 | 104.14 | 4.10 × 101 | 1.53 | 102.39 | |||
| 80.00 | 8.36 × 101 | 5.60 | 104.48 | 8.24 × 101 | 3.17 | 103.04 | |||
| 10 | 80.00 | 8.07 × 101 | 4.44 | 100.87 | 7.84 × 101 | 3.96 | 97.95 | 3.09 | 4.92 |
| 160.00 | 1.58 × 102 | 1.08 | 98.54 | 1.59 × 102 | 2.32 | 99.49 | |||
| 320.00 | 3.13 × 102 | 3.25 | 97.86 | 3.22 × 102 | 2.86 | 100.59 | |||
| 11 | 20.00 | 2.10 × 101 | 9.57 | 104.89 | 2.02 × 101 | 7.44 | 101.16 | 2.40 | 7.01 |
| 40.00 | 4.29 × 101 | 2.68 | 107.23 | 4.29 × 101 | 3.63 | 107.16 | |||
| 80.00 | 8.37 × 101 | 2.08 | 104.56 | 8.37 × 101 | 4.54 | 104.64 | |||
| 12 | 40.00 | 3.97 × 101 | 0.84 | 99.24 | 3.75 × 101 | 2.98 | 93.77 | 2.49 | 7.31 |
| 80.00 | 8.55 × 101 | 1.91 | 106.81 | 8.36 × 101 | 1.60 | 104.47 | |||
| 160.00 | 1.68 × 102 | 1.61 | 105.04 | 1.66 × 102 | 1.59 | 103.79 | |||
| 13 | 1200.00 | 1.26 × 103 | 3.12 | 104.62 | 1.17 × 103 | 4.85 | 97.30 | 2.20 | 6.45 |
| 2400.00 | 2.71 × 103 | 2.08 | 112.80 | 2.58 × 103 | 3.11 | 107.40 | |||
| 4800.00 | 5.29 × 103 | 2.34 | 110.12 | 5.09 × 103 | 3.37 | 105.96 | |||
| 14 | 40.00 | 3.30 × 101 | 2.01 | 82.38 | 4.92 × 101 | 2.95 | 90.11 | 1.98 | 7.05 |
| 80.00 | 7.58 × 101 | 1.56 | 94.71 | 1.06 × 102 | 2.11 | 98.98 | |||
| 160.00 | 1.53 × 102 | 2.96 | 95.53 | 2.14 × 102 | 1.99 | 99.52 | |||
| 15 | 400.00 | 3.62 × 102 | 2.45 | 90.40 | 3.58 × 102 | 3.45 | 89.37 | 2.07 | 8.55 |
| 800.00 | 8.45 × 102 | 2.62 | 105.57 | 8.65 × 102 | 2.23 | 108.09 | |||
| 1600.00 | 1.68 × 103 | 3.64 | 104.93 | 1.73 × 103 | 2.19 | 107.97 | |||
| 16 | 600.00 | 6.16 × 102 | 1.73 | 102.67 | 5.64 × 102 | 2.36 | 96.15 | 1.65 | 4.26 |
| 1200.00 | 1.22 × 103 | 0.97 | 101.63 | 1.16 × 103 | 2.10 | 104.31 | |||
| 2400.00 | 2.48 × 103 | 2.85 | 103.35 | 2.33 × 103 | 2.21 | 105.00 | |||
| 17 | 40.00 | 3.90 × 101 | 4.05 | 97.51 | 3.87 × 101 | 6.06 | 96.67 | 1.59 | 7.36 |
| 80.00 | 7.97 × 101 | 6.30 | 99.59 | 8.15 × 101 | 6.07 | 101.90 | |||
| 160.00 | 1.62 × 102 | 4.88 | 101.16 | 1.63 × 102 | 3.09 | 101.79 | |||
| 18 | 600.00 | 6.26 × 102 | 4.29 | 104.31 | 5.81 × 102 | 4.68 | 96.89 | 1.54 | 3.50 |
| 1200.00 | 1.28 × 103 | 6.56 | 106.75 | 1.27 × 103 | 6.56 | 105.73 | |||
| 2400.00 | 2.49 × 103 | 5.93 | 102.59 | 2.49 × 103 | 5.40 | 103.74 | |||
| 19 | 20.00 | 2.09 × 101 | 2.05 | 104.40 | 1.94 × 101 | 2.43 | 97.14 | 1.39 | 5.73 |
| 40.00 | 4.27 × 101 | 2.34 | 106.81 | 4.12 × 101 | 2.62 | 102.96 | |||
| 80.00 | 8.62 × 101 | 2.62 | 107.79 | 8.55 × 101 | 2.91 | 106.84 | |||
| 20 | 20.00 | 2.02 × 101 | 2.84 | 100.91 | 1.94 × 101 | 3.57 | 96.92 | 1.17 | 3.93 |
| 40.00 | 4.07 × 101 | 2.14 | 101.77 | 3.94 × 101 | 3.46 | 98.42 | |||
| 80.00 | 8.05 × 101 | 5.03 | 100.65 | 8.02 × 101 | 4.87 | 100.29 | |||
| 21 | 40.00 | 3.80 × 101 | 3.65 | 95.05 | 3.86 × 101 | 5.67 | 96.41 | 1.02 | 4.98 |
| 80.00 | 7.24 × 101 | 2.48 | 90.50 | 7.65 × 101 | 3.60 | 95.58 | |||
| 160.00 | 1.45 × 102 | 3.99 | 90.44 | 1.51 × 102 | 3.62 | 94.55 | |||
| 22 | 40.00 | 4.35 × 101 | 1.42 | 108.67 | 4.09 × 101 | 3.35 | 102.25 | 1.00 | 3.52 |
| 80.00 | 8.58 × 101 | 4.26 | 107.31 | 8.29 × 101 | 4.01 | 103.67 | |||
| 160.00 | 1.67 × 102 | 5.51 | 104.54 | 1.65 × 102 | 4.24 | 103.39 | |||
| Analyte 1 | Amount | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OCE–1 2 | OCE–2 | OCE–3 | OCE–4 | OCE–5 | ||||||
| Mean (mg/g) | RSD (%) | Mean (mg/g) | RSD (%) | Mean (mg/g) | RSD (%) | Mean (mg/g) | RSD (%) | Mean (mg/g) | RSD (%) | |
| 1 | 0.18 | 0.85 | 0.15 | 0.95 | 0.68 | 2.62 | 0.47 | 0.84 | 0.15 | 3.76 |
| 2 | 1.91 | 5.32 | 1.13 | 9.86 | 1.29 | 9.73 | 0.47 | 4.59 | 1.33 | 9.93 |
| 3 | 0.23 | 4.45 | 0.05 | 6.13 | 0.05 | 1.83 | 0.05 | 0.77 | 0.25 | 1.57 |
| 4 | 2.77 | 1.06 | 0.29 | 0.92 | 0.09 | 3.65 | 0.05 | 0.87 | 0.87 | 1.98 |
| 5 | 1.00 | 7.23 | 0.09 | 4.53 | 0.04 | 6.02 | 0.01 | 5.09 | 0.02 | 5.74 |
| 6 | 17.39 | 5.51 | 3.15 | 8.34 | 3.80 | 6.57 | 3.56 | 8.06 | 6.60 | 2.73 |
| 7 | 5.17 | 0.30 | 1.65 | 0.91 | 1.78 | 0.74 | 1.64 | 0.16 | 2.74 | 0.66 |
| 8 | 6.24 | 1.29 | 3.32 | 3.41 | 2.80 | 1.93 | 3.28 | 2.46 | 4.97 | 2.40 |
| 9 | 1.36 | 1.43 | 0.24 | 2.00 | 0.09 | 1.31 | 0.01 | 0.39 | <LOQ | – |
| 10 | 0.27 | 5.00 | 0.10 | 5.33 | 0.02 | 3.94 | 0.03 | 5.15 | 0.18 | 2.87 |
| 11 | 0.65 | 0.70 | 0.01 | 3.08 | 0.03 | 1.62 | 0.12 | 1.01 | 0.27 | 2.18 |
| 12 | 2.04 | 0.41 | <LOQ | – | 0.07 | 0.13 | 0.34 | 2.64 | 0.89 | 0.45 |
| 13 | 5.35 | 0.86 | 7.66 | 0.77 | 10.23 | 1.01 | 10.94 | 0.73 | 9.75 | 1.20 |
| 14 | 2.36 | 2.52 | 0.07 | 1.96 | 0.10 | 1.89 | 0.40 | 1.97 | 0.67 | 0.35 |
| 15 | 27.10 | 0.23 | 8.05 | 0.57 | 6.09 | 0.72 | 5.89 | 3.65 | 15.58 | 2.35 |
| 16 | 3.69 | 0.91 | 2.38 | 0.70 | 0.39 | 0.44 | 2.53 | 1.24 | 2.40 | 1.49 |
| 17 | 0.20 | 4.53 | 0.12 | 9.04 | 0.07 | 4.02 | 0.06 | 2.12 | 0.19 | 5.30 |
| 18 | 3.77 | 1.74 | 0.95 | 0.75 | 0.42 | 1.68 | 0.39 | 0.12 | 0.18 | 1.59 |
| 19 | 0.81 | 2.07 | 0.16 | 1.12 | 0.09 | 3.45 | 0.07 | 2.18 | 0.06 | 1.26 |
| 20 | 1.32 | 7.02 | 0.08 | 7.98 | 0.01 | 9.29 | 0.01 | 5.98 | 1.82 | 9.32 |
| 21 | 0.34 | 1.25 | 0.12 | 1.48 | 0.06 | 5.61 | 0.03 | 3.14 | <LOQ | – |
| 22 | 0.25 | 2.62 | 0.13 | 4.09 | 0.06 | 5.61 | 0.03 | 1.01 | <LOQ | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, C.-S.; Shin, H.-K. Ultra-Performance Liquid Chromatography with Tandem Mass Spectrometry for Simultaneous Analysis of 22 Analytes of Oncheong-Eum, a Traditional Korean Herbal Formula. Processes 2023, 11, 2906. https://doi.org/10.3390/pr11102906
Seo C-S, Shin H-K. Ultra-Performance Liquid Chromatography with Tandem Mass Spectrometry for Simultaneous Analysis of 22 Analytes of Oncheong-Eum, a Traditional Korean Herbal Formula. Processes. 2023; 11(10):2906. https://doi.org/10.3390/pr11102906
Chicago/Turabian StyleSeo, Chang-Seob, and Hyeun-Kyoo Shin. 2023. "Ultra-Performance Liquid Chromatography with Tandem Mass Spectrometry for Simultaneous Analysis of 22 Analytes of Oncheong-Eum, a Traditional Korean Herbal Formula" Processes 11, no. 10: 2906. https://doi.org/10.3390/pr11102906
APA StyleSeo, C.-S., & Shin, H.-K. (2023). Ultra-Performance Liquid Chromatography with Tandem Mass Spectrometry for Simultaneous Analysis of 22 Analytes of Oncheong-Eum, a Traditional Korean Herbal Formula. Processes, 11(10), 2906. https://doi.org/10.3390/pr11102906








