Abstract
Actinomycetes are natural architects of numerous secondary metabolites including antibiotics. With increased multidrug-resistant (MDR) pathogens, antibiotics that can combat such pathogens are urgently required to improve the health care system globally. The characterization of actinomycetes available in Nepal is still very much untouched which is the reason why this paper showcases the characterization of actinomycetes from Nepal based on their morphology, 16S rRNA gene sequencing, and metabolic profiling. Additionally, antimicrobial assays and liquid chromatography-high resolution mass spectrometry (LC-HRMS) of ethyl acetate extracts were performed. In this study, we employed a computational-based dereplication strategy for annotating molecules which is also time-efficient. Molecular annotation was performed through the GNPS server, the SIRIUS platform, and the available databases to predict the secondary metabolites. The sequencing of the 16S rRNA gene revealed that the isolates BN6 and BN14 are closely related to Streptomyces species. BN14 showed broad-spectrum antibacterial activity with the zone of inhibition up to 30 mm against Staphylococcus aureus (MIC: 0.3051 µg/mL and MBC: 9.7656 µg/mL) and Shigella sonnei (MIC: 0.3051 µg/mL and MBC: 4.882 µg/mL). Likewise, BN14 also displayed significant inhibition to Acinetobacter baumannii, Klebsiella pneumoniae, and Salmonella typhi. GNPS approach suggested that the extracts of BN6 and BN14 consisted of diketopiperazines ((cyclo(D-Trp-L-Pro), cyclo(L-Leu-L-4-hydroxy-Pro), cyclo(L-Phe-D-Pro), cyclo(L-Trp-L-Pro), cyclo(L-Val-L-Pro)), and polypeptide antibiotics (actinomycin D and X2). Additional chemical scaffolds such as bacterial alkaloids (bohemamine, venezueline B, and G), anthramycin-type antibiotics (abbeymycin), lipase inhibitor (ebelactone B), cytocidal (oxopropaline D), antifungal and antitumor antibiotics (reductiomycin, streptimidone, deoxynybomycin), alaremycin, fumaramidmycin, anisomycin, and others were also annotated, which were further confirmed by using the SIRIUS platform, and literature survey. Thus, the bioprospecting of natural products from Streptomyces species from Nepal could be a potential source for the discovery of clinically significant and new antimicrobial agents in the future.
1. Introduction
The unearthing and the furtherance of drugs for the treatment of infectious diseases, antibiotics, are considered one of the greatest leaps of the 20th century [1,2]. Between 1950 and 1960, microbiology and chemistry went side by side, and a large number of different classes of effective antibiotics were discovered; this decade is regarded as the ‘golden age’ of antibiotics [3]. However, microbes have continuously evolved and adapted with a wide range of metabolic mechanisms to overwhelm the effects of antimicrobial agents [4]. This causes non-susceptibility to the antimicrobial agents, and this phenomenon is known as antimicrobial resistance (AMR), which is mostly transferred among bacteria via mobile genetic elements such as bacteriophages, interferons, plasmids, and transposons [5].
The emergence of multidrug-resistant (MDR) pathogens poses a great threat to the global health system and a challenge to the effective use of antibiotics. The World Health Organization (WHO) has declared AMR among the top 10 global public health threats [6]. MDR pathogens have dented the existing antibiotic-based treatment era and raised the mortality rate [4]. As per a WHO report, AMR is currently accountable for the death of approximately 700,000 people globally each year and is expected to take the lives of 10 million each year by 2050 [7]. Another study showed AMR accounts for an economic loss of approximately USD 150 million to 30 billion annually [8]. The lack of significant research on new antibiotics is one of the contributing factors to the increased prevalence of AMR [9]. As a result, researchers are seeking alternative antibiotics that would be effective against MDR pathogens.
Natural products are the source of more than 75% of today’s antibiotics [10]. Actinomycetes are used as a potential natural source for the production of abundant classes of clinically and economically significant secondary metabolites including antibiotics [11]. Approximately two-thirds of today’s known antibiotics are derived from actinomycetes, of which the genus Streptomyces is the prolific producer [12]. The Streptomyces genome contains more than 20 biosynthetic gene clusters of higher clinical significance, including anti-infection agents that could handle the ascent of AMR [13]. No new class of antibiotics has passed the clinical phase in the last three decades [14]. The study of actinomycetes from diverse ecosystems could help to uncover new specific molecular targets and novel bioactive compounds because of their genetic variability [15].
Microbial screening explores uncultured bacterial lineages that could have novel biosynthetic pathways of secondary metabolites and offers new chemical scaffolds and pharmaceutical products [16,17]. However, present cultivation efforts to uncover new bacterial strains correspond to a minute fraction when compared to projected bacterial diversity globally [17,18]. Screening of natural-product-based drugs by pharmaceutical industries is losing momentum due to the rediscovery of known metabolites [19]. Having said that, metabolomics emerges as a new hope to detect metabolic pathways and metabolites of clinical importance [20,21]. Metabolomics involves the combination of high-throughput analytical techniques and bioinformatics intended for the extensive identification and quantification of biological samples [22,23]. Mass spectrometry (MS), nuclear magnetic resonance (NMR), and Fourier transform infrared (FT-IR) spectroscopy are common analytical platforms used in metabolomics [23]. Here, we used mass-spectrometry-based metabolomics. Liquid chromatography coupled with mass spectrometry (LC-MS) is a robust bio-analytical technique in metabolomics [24]. MS-based metabolomics finds its application in different fields including drug development, precision medicine, toxicology, environmental testing laboratories, and so on [22,24].
However, the microbial screening program in Nepal is still in the initial phase. We believe that the screening of actinomycetes from the untouched habitat of Nepal might lead to the isolation of new strains. Thus, the present study aims to isolate and characterize antibiotics producing actinomycetes from soils of Nepal with a focus on molecular annotation and molecular networking.
2. Materials and Methods
2.1. Soil Samples Collection and Isolation of Actinomycetes
Fourteen soil samples (BN1-BN14) were collected from various locations of altitudinal and geographic variations in Nepal (Supplementary Table S1). The samples BN6 and BN14 were collected from Rupa lake, Lekhnath (28°8′44.24″ N, 84°6′47.91″ E, 2683 ft) and Huti, Darchula (29°55′1.53″ N, 80°35′12.86″ E, 10,277 ft), respectively (Figure 1). The samples were air-dried for a week at room temperature and stored at 4 °C. Actinomycetes were isolated in the International Streptomyces Project 4 (ISP4) medium by serial dilution method [25]. The ISP4 medium was supplemented with 20 mg/mL nalidixic acid and 50 mg/mL cycloheximide to inhibit the growth of Gram-negative bacteria and fungus species, respectively [26]. For isolation of actinomycetes, 1 g of soil was suspended in 10 mL of distilled water using a vertex and serially diluted up to 1000-fold. Then, 100 μL of 1000-fold serially diluted suspension was spread over ISP4 plates. The plates were then incubated at 28 °C for 7–10 days. Based on the color, colony characteristics, and mycelium [27,28], the actinomycetes were sub-cultured and purified by the streak plate method over the ISP4 medium. Subsequently, the bacterial samples were stored in 20% (v/v) glycerol stocks at −20 °C for further use.
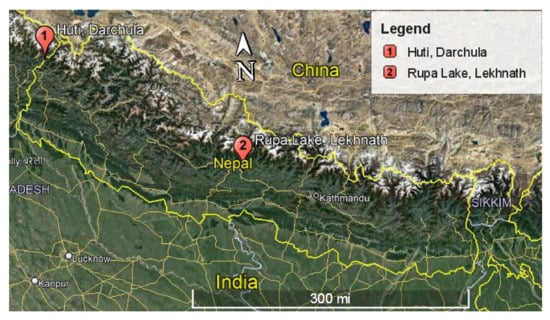
Figure 1.
Map of Nepal showing the sites for the collected soil samples.
2.2. Morphological and Molecular Characterization of Actinomycetes
2.2.1. Macroscopic and Microscopic Characterization
The aerial and submerged mycelium along with diffused pigments of isolates were studied following Bergey’s Manual of Systematic Bacteriology [28]. Microscopic characterization was performed by the Gram staining method and observed under 100× magnification [29].
2.2.2. Extraction of Genomic DNA and 16S rRNA Amplification
Bacterial genomic DNA was isolated through the phenol-chloroform method and it was examined on 0.4% agarose gel electrophoresis (Cleaver, Scientific, Rugby, UK) and viewed under Gel Doc (UVITEC, Cambridge, UK). The amplification of the 16S rRNA gene was carried out by polymerase chain reaction (PCR) (Thermo Fisher Scientific, Waltham, MA, USA) using universal primers; 27F: 5′-AGAGTTTGATCCTGGCTCAG-3′, and 1492R: 5′-GGTTACCTTGTTACGACTT-3′ (Merck). The primers were diluted to a working concentration of 10 μM using nuclease-free water. The amplification was carried out in a 50 μL reaction mixture using 5X premix (Solis Biodyne) containing Taq-polymerase within 29 cycles. A cycle consisted of an initial denaturation temperature of 95 °C, an annealing temperature of 51.4 °C, and an extension at 95 °C [30]. The amplified products were purified by using a New England Biolabs purification kit. The purified 16S rRNA products were sequenced by Macrogen Inc., Seoul, South Korea.
2.2.3. Sequence Analysis
The 16S rRNA sequences were obtained by Sanger dideoxy sequencing method, and the nucleotide sequences were compared with the NCBI database by using the BLAST sequence similarity search program [31]. Multiple sequence alignment was performed by using ClustalW software and Molecular Evolutionary Genetics Analysis 11 (MEGA11) [32]. A phylogenetic tree was constructed by the neighbor-joining method using MEGA11 software [32]. The 16S rRNA sequence of the BN6 was deposited in the NCBI and its accession number was obtained.
2.3. Flask Fermentation and Extraction of Bioactive Metabolites
Individual actinomycetes were cultured in a conical flask containing 20 mL Tryptic Soy Broth (TSB) with glass beads at 140 rpm and 28 °C in a shaking incubator for 3 to 4 days. After proper growth of bacteria, 1 mL of bacterial inoculum was transferred to 100 mL TSB and kept for 7 to 8 days on a shaking incubator until the bacterial growth attained the stationary phase. To pull out secondary metabolites, flask fermentation culture was shaken overnight with an equal volume of ethyl acetate. The mixture was separated using a separating funnel, and the ethyl acetate fraction was dried in the water bath at 40 °C and stored at 4 °C for further use.
2.4. Antibacterial Susceptibility Test
The primary screening of actinomycetes was carried out by the perpendicular streaking method [33] in the MHA medium. The tested pathogens were Staphylococcus aureus ATCC 43300, Escherichia coli ATCC 2591, Klebsiella pneumoniae ATCC 700603, Salmonella typhi ATCC 14028, Shigella sonnei ATCC 25931, and Acinetobacter baumannii ATCC 19606. The antibacterial activity of the actinomycetes extracts was screened against the aforementioned ATCC strains by the agar well diffusion method using neomycin (1 mg/mL) and DMSO (50%) as a positive and negative control, respectively, as described previously [34]. Each well contained 5 µg of bacterial extract. The assays were performed in triplicate for method validation and reproducibility testing. Based on the zone of inhibition, the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of the BN14 extract were further determined as discussed previously [34].
2.5. GNPS-Based Molecular Networking
All the positively ionized HRMS/MS data were piled up using Bruker MetaboScape 3.0 software. Extracted ion chromatogram (EIC) correlation of 0.8 and mass range from 150 to 1500 Da were optimized during piling up. While processing in the software, the adduct ions were kept as [M+H]+ (primary ion), [M+Na]+, [M+K]+, [2M+H]+, [2M+Na]+, [M+2H]2+, [M+2Na]2+, [M-H2O+H]+, [M+H2O+H]+, [2M+H2O+H]+, and [2M-H2O+H]+ [35,36]. The data were transferred to the Global Natural Product Social Molecular Networking (GNPS) server after bucketing and examined for spectral database matching and adjusting a cosine score parameter of 0.7. The best hits compounds in the GNPS library were enlisted in a table along with their GNPS accession IDs [35,36].
2.6. Metabolic Profiling
A low resolution-liquid chromatography–mass spectrometry (LR-LC-MS) profile was achieved using the HPLC system (1100 Series, Agilent Technologies, Waldbronn, Germany) coupled with a mass spectrometer (ABSCIEX 3200 Q TRAP LC-MS/MS, Germany GmbH, Darmstadt, Germany). HR-LC-ESI-MS/MS (Bruker TOF-MS MaXis Impact ESI-HR-MS) profiling was also performed for the ethyl acetate extracts as previously explained [37]. The data were acquired in both positive and negative ionization modes. The LR-LC-MS data were analyzed using Analyst® 1.6.3 software (ABSCIEX). Molecular annotation was performed using the Dictionary of Natural Products 30.2 Chemical Search (https://dnp.chemnetbase.com/ (accessed on 7 April 2022)) and the Natural Products Atlas 2.0 databases (https://www.npatlas.org/ (accessed on 7 April 2022)) [38,39]. The HR-LC-ESI-MS/MS data were read in Bruker Compass DataAnalysis 4.4 (Bruker Daltonics GmbH, Billerica, MA, USA). Further, the molecular annotation was also carried out in SIRIUS 4.9.12 using CSI: FingerID interface [40,41].
3. Results
3.1. Macroscopic and Microscopic Characterization of Actinomycetes
A total of 47 actinomycetes were isolated from 14 different samples (Supplementary Tables S2 and S3). Out of 47 isolates, seven actinomycetes (4 from BN6 and 3 from BN14) were grown in a culture plate. The isolates (one from each sample) showing potential antibacterial activity in primary screening were considered for further studies. The colors of aerial and substrate mycelium for BN6 were white and greyish white, respectively, with no diffusible pigment. The cultural physiognomies of BN14 were greyish-white aerial mycelium, reddish-yellow substrate mycelium, and yellow diffusible pigmentation. The Gram staining showed that they were flagellated Gram-positive bacteria with hair-like mycelium. Figure 2 shows the macroscopic and microscopic characteristics of BN6 and BN14.
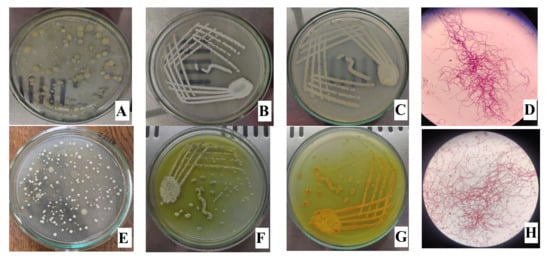
Figure 2.
Isolation plate (A,E), aerial mycelium (B,F), substrate mycelium (C,G), and Gram-staining (D,H) of BN6 (first row) and BN14 (second row).
3.2. Extraction of Genomic DNA and PCR
The genomic DNA was visualized using Gel Doc and 16S rRNA was amplified using universal primers. The size of amplified PCR products was approximately 1.5 kb as compared to the ladder (Supplementary Figure S1).
3.3. Sequence Analysis
The BLAST sequence search revealed that 16S rRNA gene sequencing of BN6 (Accession: ON024786) and BN14 was found to resemble the genus Streptomyces. The analysis showed that BN6 has the highest sequence similarity to Streptomyces galilaeus (99.63%) and Streptomyces variabilies (99.54%). Similarly, BN14 displayed the highest sequence similarity to Streptomyces griseoflavus (96.35%) and Kitasatospora purpeofusca (92.76%). The relation of BN6 and BN14 with other Streptomyces species is presented in the phylogenetic tree (Figure 3).
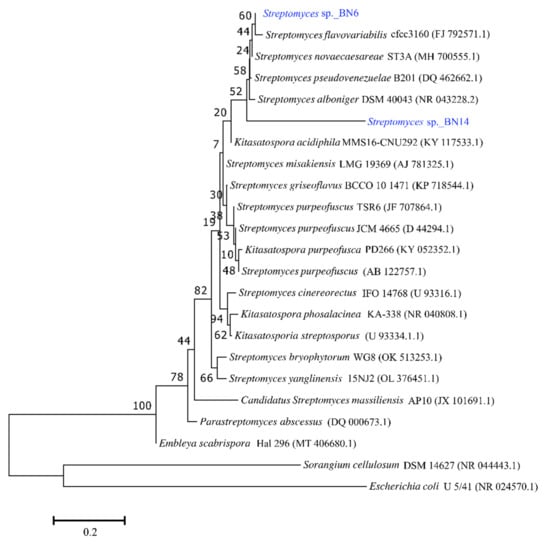
Figure 3.
Neighbor-joining phylogenetic tree of 16S rRNA of isolates BN6 and BN14. Horizontal branch lengths proportional to the estimated number of nucleotide substitutions, and bootstrap probabilities (as percentages), as determined for 1000 resamplings, are given above and beside the internal branches. The bar in the lower-left corner indicates 0.20 substitutions per nucleotide position. Genebank accession numbers are shown in parentheses. Sorangium cellulosum DSM 14627 (NR 044443.1) and Escherichia coli U 5/41 (NR 024570.1) were used as an outgroup to root the tree.
3.4. Antibacterial Susceptibility Test
The antibacterial activity of potent isolates was provided in Supplementary Table S4. The antibacterial activity of BN14 was highest for S. aureus and S. sonnei with a zone of inhibition of 30 mm for each bacterium. Thus, MIC and MBC of BN14 extract were determined for these test organisms. BN14 showed broad-spectrum antibacterial activity. The zones of inhibition exhibited by BN14 were higher than those displayed by BN6 (Supplementary Figure S2). Thus, BN6 was less potent than BN14 in the antimicrobial screening assays. The zones of inhibition shown by BN6 and BN14 against different test organisms were presented in the bar diagram (Figure 4). MIC of BN14 against S. aureus and S. sonnei was 0.3051 µg/mL each while that of neomycin (positive control) was 1.56 µg/mL for each bacterium. The MBC of BN14 against S. aureus and S. sonnei was 9.7656 µg/mL and 4.882 µg/mL, respectively, whereas 12.5 and 6.25 µg/mL, respectively, for neomycin.
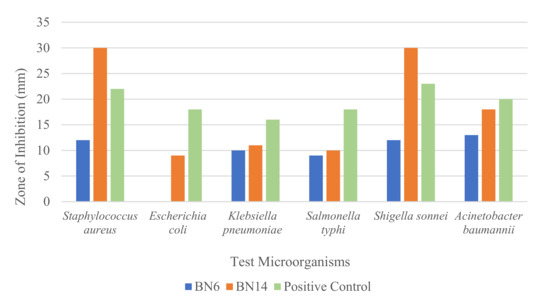
Figure 4.
A bar chart showing the zone of inhibition exhibited by extracts of BN6 and BN14 isolates (5 µg extract in each well and 1 mg/mL neomycin).
3.5. GNPS-Based Molecular Networking
A total of 1442 molecular ions were observed to have MS2 spectra of BN6 and BN14 represented by nodes that were connected by 1809 edges in the molecular network (Supplementary Figure S3). Out of 1442, 713 molecu
lar ions were self-looped, 96 ion pairs formed two-node clusters, and the remaining 537 ions were connected by variable node counts in 60 different big clusters. The GNPS-based molecular network provided the hits of 20 known compounds and 352 unknown molecular ion peaks in the GNPS library based on their relative abundance (greater than 100,000) (Supplementary Tables S5 and S6). The GNPS approach suggested that the crude extracts of BN6 and BN14 were found to consist of diketopiperazines ((cyclo(D-Trp-L-Pro), cyclo(L-Leu-L-4-hydroxy-Pro), cyclo(L-Phe-D-Pro), cyclo(L-Trp-L-Pro), cyclo(L-Val-L-Pro)), and polypeptide antibiotics (actinomycin D and X2). These metabolites were further verified by the SIRIUS platform, literature survey, and database libraries.
3.6. Metabolic Profiling
Metabolic profiling of the extracts of samples BN6 and BN14 was carried out using the SIRIUS platform and literature survey. The analysis accessed at least 54 secondary metabolites from the extracts. The LC-HR-MS spectra of BN14 showed the molecular ion peaks of [M+H]+ at m/z 1255.6333 and [M+Na]+ at m/z 1277.6153 that corresponded to actinomycin D (Supplementary Figure S4) [42]. The other key fragments such as 956.5417 [(MH+-(Pro-Sar-MeVal)], 875.4499 [(MH+-(Val-Pro-MeVal)], 659.3151, 629.3241, 558.0932, 459.1363, 399.2378, 300.1931, 203.1803, and 168.9069 in BN14 matched with standard fragmentation pattern of actinomycin D [42,43]. Similarly, the spectra of BN14 displayed the molecular ion peaks of [M+H]+ at m/z 1269.6165 and [M+Na]+ at m/z 1291.5987 which corresponded to actinomycin X2 (Supplementary Figure S5) [42]. The presence of [M+2H]2+ at m/z 635.3125 further confirmed the presence of actinomycin X2 [42]. Likewise, a list of annotated compounds from BN6 and BN14 is tabulated in Table 1 and Table 2 respectively along with ppm error less than 5, key fragments, and SIRIUS score. Some of the chemical scaffolds annotated from BN6 and BN14 are represented in Figure 5.

Table 1.
A list of annotated compounds using SIRIUS in Streptomyces sp._BN6.

Table 2.
A list of annotated compounds using SIRIUS in Streptomyces sp._BN14.
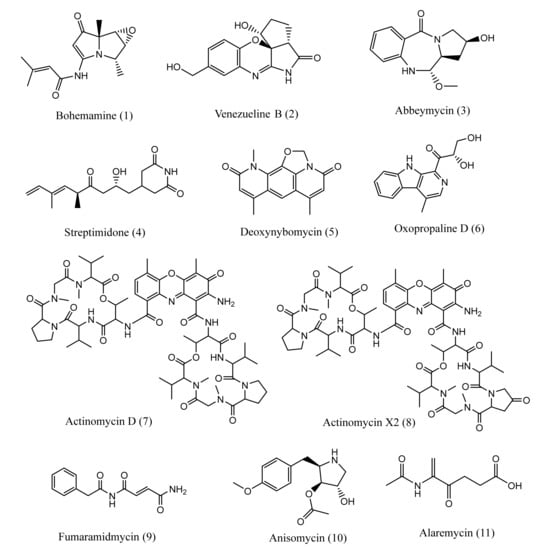
Figure 5.
Chemical structures of some annotated metabolites from BN6 (1–6) and BN14 (7–11).
4. Discussion
Natural products have captivated the scientific world ceaselessly. Due to evolution over millions of years, natural products have attained unique chemical heterogeneity that has resulted in drug-like properties and diversified biological activities in them [92]. They have provided several lead compounds as therapeutic agents. Among such lead compounds, a substantial number were isolated from the microbial communities [93]. Approximately 80% of the antibiotics currently in medical use are produced by actinobacteria of which 50% is derived from Streptomyces [94,95]. More than 7600 bioactive metabolites are already derived from Streptomyces with a possibility of discovering more than 100,000 new metabolites [96,97]. It has been suggested to explore and expand the dimension of microbial natural products research [93]. Thus, in this study, we explored the antibiotics producing Streptomyces from Nepalese biodiversity.
In Nepal, much focus has been given to studying floral biodiversity and plant-based metabolites. On the other hand, Nepalese microbial diversity has been already proven to contain potential Streptomyces to inhibit various pathogenic microorganisms [98,99,100,101]. The crude extracts derived from Nepalese Streptomyces species were found to inhibit methicillin-resistant S. aureus [100,101], ESBL-producing E. coli [30], and other Gram-negative and Gram-positive bacteria such as S. typhi, S. paratyphi, Proteus mirabilis, P. vulgaris, S. sonnei, K. pneumoniae, K. oxytoca, E. coli, Bacillus subtilis, and S. aureus [98,101,102]. However, these research works lack molecule annotation and metabolic profiling.
The emergence of MDR has posed a significant threat to global health and a challenge to the effective use of antibiotics. Some currently marketed antibiotics are ineffective against the resistance mechanism of such pathogens. This scenario calls urgent attention to exploring new antimicrobial agents with novel modes of action to combat MDR [103]. Soil is considered to be teeming with novel antibiotics, and soil microbes are regarded as the richest source of antibiotics [104]. The majority of antibiotics available in the market today are extracted from terrestrial microorganisms [103]. Thus, terrestrial Streptomyces species still could fulfill the quest for novel secondary metabolites against AMR [42]. In this study, we isolated 47 different actinomycetes from 14 soil samples (BN1 to BN14) collected from varying altitudes of Nepal (Supplementary Tables S2 and S3). Out of these, isolates identified from soil samples BN4, BN6, and BN14 showed significant antimicrobial activity (Supplementary Table S4). Additionally, each potent colony from BN4, BN6, and BN14 in primary screening was considered further. These selected colonies were transferred to TSB fermentation medium to obtain crude extracts. The sample BN14 showed significant inhibition against S. aureus, S. sonnei, S. typhi, A. baumanii, and K. pneumonia. These pathogens are enlisted on the WHO precedence list as antibiotic-resistant pathogens [3,105].
Being a creator of a wide range of bioactive compounds such as antibiotics, antitumor, antifungal, antihelminthic, and immunosuppressant drugs, actinomycetes are regarded as nature’s pharmacists [106]. The crude extracts of BN6 and BN14 were found to consist of diketopiperazines ((cyclo(D-Trp-L-Pro), cyclo(L-Leu-L-4-hydroxy-Pro), cyclo(L-Phe-D-Pro), cyclo(L-Trp-L-Pro), cyclo(L-Val-L-Pro)), and polypeptide antibiotics (actinomycin D and X2), bacterial alkaloids (bohemamine, venezueline B, and G), anthramycin-type antibiotics (abbeymycin), lipase inhibitor (ebelactone B), cytocidal (oxopropaline D), antifungal and antitumor antibiotic (reductiomycin, streptimidone, deoxynybomycin), antibiotics (alaremycin, fumaramidmycin, anisomycin), and other chemical scaffolds presented in Table 1. BN6 and BN14 represent the low- and high-altitude soil samples, respectively, synthesizing different metabolites in culture. The antimicrobial potential of molecules derived from isolates of BN14 is higher than those exhibited by isolates from BN6. Thus, this study also suggests that there would be a high possibility of getting potent metabolites in stress conditions (high altitude).
Diketopiperazines have drawn interest among researchers because of their wide array of biological properties that include antimicrobial, antiviral, antitumor, anti-Alzheimer, and haemosuppressor activities [107]. These cyclic peptides are also used in quorum-sensing signaling, inhibiting microtubule polymerization, and treating ischemic brain injury [107]. Diketopiperazines are believed to be undesired by-products during oligopeptides synthesis, or products of a nonribosomal pathway [108,109]. Actinomycin D and X2 are chemotherapeutic agents used to treat Wilm’s kidney tumors, rhabdomyosarcoma, breast cancer, and trophoblastic tumors by inhibiting the transcription processes and RNA activity in cancer cells [110,111]. Pyrrolobenzodiazepines like abbeymycin are selective DNA alkylating agents showing significant antineoplastic activity [112]. The antibiotic reductiomycin exhibits antitumor, antifungal, antibacterial, and antiviral activities [113,114]. Similarly, Deoxynybomycin displays cancer chemotherapy application by selectively inhibiting human lung carcinoma, osteoblastic sarcoma, gastric cancer, and monocytic leukemia [115].
Streptomyces galilaeus, closely related to BN6, is a producer of aclacinomycins and anthracyclines which are used in cancer chemotherapy [116,117]. However, the extract of BN6 did not exhibit a significant antimicrobial effect. The probable secondary metabolites annotated from BN6 are listed in Table 1. The culture medium, incubation condition, aeration, pH, and growth temperature could be the factors affecting the production of secondary metabolites in Streptomyces species [118,119,120]. The poorly expressed or cryptic biosynthetic gene clusters could be another factor in the non-production of more potent antimicrobial agents in BN6 [11]. On the other hand, BN14 is found to be more potent than BN6. The antibiotics including actinomycin D and actinomycin X2 might be the reason for the potency of BN14.
Different factors influence the production of secondary metabolites in bacteria and fungi. The type of culture medium affects the fabrication of metabolites. It is reported that the mass of metabolites synthesized by the same fungal species was one to two orders of magnitude greater in solid medium than in liquid medium [121]. Feeding of amino acids and the addition of other precursors (supplements) in fermentation also enhance the production of metabolites [122,123,124]. In the future, advances in the expression of cryptic gene clusters [125] in those Streptomyces strains could be beneficial to obtain novel and useful metabolites. The combined application of metabolomics, genomics, and heterologous expression plays a significant role to access novel natural products [125,126,127]. The metabolites, not detected in fermentation extract, are identified with the strategies inspired by functional genomics and bioinformatics [128].
5. Conclusions
In this study, we isolated 47 actinomycetes from 14 soil samples collected from different locations in Nepal. The strains in the soil samples BN6 and BN14 exhibited antibacterial activity against S. aureus, S. sonnei, S. typhi, A. baumanii, and K. pneumonia. Here, we employed a time-efficient computational-based dereplication strategy for annotating molecules. The GNPS and SIRIUS-based metabolomics of Streptomyces sp._BN6 and Streptomyces sp._BN14 extracts provided at least 54 different secondary metabolites. These metabolites include multiple chemical scaffolds such as diketopiperazines, polypeptide antibiotics, anthramycin-type antibiotics, antifungal and antitumor antibiotics, bacterial alkaloids, lipase inhibitors, cytocidal metabolites, and others. Thus, the bioprospecting of natural products from actinomycetes of Nepalese soils could be a potential source for the discovery of clinically significant and new antibiotics and antitumor agents in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pr10061173/s1, Table S1. Location of soil sample collection, Table S2. Soil profile, total isolated actinomycetes and characterization method of 14 samples, Table S3. Culture characteristic and gram staining of samples BN1-BN14, Table S4. Zone of inhibition exhibited by potent isolates from each sample, Table S5. Predicted known metabolites by GNPS, Table S6. Unknown molecular ion peaks suggested by GNPS; Figure S1. Purified 16S rRNA products of BN6 and BN14, Figure S2. Zone of inhibition exhibited by BN6 and BN14 (PC: Positive Control, neomycin; NC: Negative Control, DMSO), Figure S3. Molecular networking of MS2 data of BN6 and BN14, Figure S4. Mass spectrum of BN14 corresponding to Actinomycin D, Figure S5. Mass spectrum of BN14 corresponding Actinomycin X2.
Author Contributions
Conceptualization, N.P. and B.R.B.; methodology, N.P. and B.R.B.; validation, N.P. and B.R.B.; formal analysis, B.R.B., B.A., N.R., N.A. and K.B.; investigation, B.R.B., K.K. and U.L.; resources, N.P. and B.R.B.; data curation, B.R.B.; writing—original draft preparation, B.R.B.; writing—review and editing, N.P., B.P.R., A.A. and S.T.; visualization, B.R.B.; supervision, N.P.; project administration, N.P.; funding acquisition, N.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University Grants Commission, Nepal under grant number CRG-75/76-S&T-1 to Niranjan Parajuli.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
Authors would like to acknowledge Sobika Bhandari, Shreesti Shrestha, Ganesh BK, and Purnima Sharma for their help. We are very grateful to Gross lab, University of Tübingen, Germany for recording mass spectrometry.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic Resistance in Microbes: History, Mechanisms, Therapeutic Strategies and Future Prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Hug, J.J.; Bader, C.D.; Remškar, M.; Cirnski, K.; Müller, R. Concepts and Methods to Access Novel Antibiotics from Actinomycetes. Antibiotics 2018, 7, 44. [Google Scholar] [CrossRef]
- Lima, R.; Del Fiol, F.S.; Balcão, V.M. Prospects for the Use of New Technologies to Combat Multidrug-Resistant Bacteria. Front. Pharmacol. 2019, 10, 692. [Google Scholar] [CrossRef]
- Tavares, L.S.; Silva, C.S.F.; Souza, V.C.; Silva, V.L.; Diniz, C.G.; Santos, M.D.O. Strategies and Molecular Tools to Fight Antimicrobial Resistance: Resistome, Transcriptome, and Antimicrobial Peptides. Front. Microbiol. 2013, 4, 412. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 28 March 2021).
- WHO. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 23 January 2022).
- Levy, S.B.; Marshall, B. Antibacterial Resistance Worldwide: Causes, Challenges and Responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global Contributors to Antibiotic Resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef]
- Majer, H.M.; Ehrlich, R.L.; Ahmed, A.; Earl, J.P.; Ehrlich, G.D.; Beld, J. Whole Genome Sequencing of Streptomyces Actuosus ISP-5337, Streptomyces Sioyaensis B-5408, and Actinospica Acidiphila B-2296 Reveals Secondary Metabolomes with Antibiotic Potential. Biotechnol. Rep. 2021, 29, e00596. [Google Scholar] [CrossRef]
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical Ecology of Antibiotic Production by Actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Hopwood, D.A. Highlights of Streptomyces Genetics. Heredity 2019, 123, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tong, Z.; Shi, J.; Li, R.; Upton, M.; Wang, Z. Drug Repurposing for Next-Generation Combination Therapies against Multidrug-Resistant Bacteria. Theranostics 2021, 11, 4910–4928. [Google Scholar] [CrossRef] [PubMed]
- De Simeis, D.; Serra, S. Actinomycetes: A Never-Ending Source of Bioactive Compounds—An Overview on Antibiotics Production. Antibiotics 2021, 10, 483. [Google Scholar] [CrossRef]
- Colwell, R.R. Microbial Diversity: The Importance of Exploration and Conservation. J. Ind. Microbiol. Biotechnol. 1997, 18, 302–307. [Google Scholar] [CrossRef]
- Overmann, J.; Abt, B.; Sikorski, J. Present and Future of Culturing Bacteria. Annu. Rev. Microbiol. 2017, 71, 711–730. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna, F.H.; Reiter, K.C.; Fátima Almeida, P.; Pereira Passaglia, L.M. 2020 Systematic Review of Descriptions of Novel Bacterial Species: Evaluation of the Twenty-First Century Taxonomy through Text Mining. Int. J. Syst. Evol. Microbiol. 2020, 70, 2925–2936. [Google Scholar] [CrossRef]
- Onaka, H. Novel Antibiotic Screening Methods to Awaken Silent or Cryptic Secondary Metabolic Pathways in Actinomycetes. J. Antibiot. 2017, 70, 865–870. [Google Scholar] [CrossRef]
- Liu, R.; Bao, Z.-X.; Zhao, P.-J.; Li, G.-H. Advances in the Study of Metabolomics and Metabolites in Some Species Interactions. Molecules 2021, 26, 3311. [Google Scholar] [CrossRef]
- Idle, J.R.; Gonzalez, F.J. Metabolomics. Cell Metab. 2007, 6, 348–351. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Q.; Xu, Z.; Dou, J. Mass Spectrometry-Based Metabolomics in Health and Medical Science: A Systematic Review. RSC Adv. 2020, 10, 3092–3104. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite Identification and Quantitation in LC-MS/MS-Based Metabolomics. Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Hankemeier, T. Mass Spectrometry and Metabolomics: Past, Present and Future. Metabolomics 2013, 9, 1–3. [Google Scholar] [CrossRef]
- Basilio, A.; González, I.; Vicente, M.F.; Gorrochategui, J.; Cabello, A.; González, A.; Genilloud, O. Patterns of Antimicrobial Activities from Soil Actinomycetes Isolated under Different Conditions of PH and Salinity. J. Appl. Microbiol. 2003, 95, 814–823. [Google Scholar] [CrossRef]
- Kharel, M.K.; Shepherd, M.D.; Nybo, S.E.; Smith, M.L.; Bosserman, M.A.; Rohr, J. Isolation of Streptomyces Species from Soil. Curr. Protoc. Microbiol. 2010, 19, 10E.4.1–10E.4.5. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, X.; Jiang, Y.; Jiang, C. Morphological Identification of Actinobacteria. Actinobacteria Basics Biotechnol. Appl. 2016, 2016, 59–86. [Google Scholar] [CrossRef]
- Bergey’s Manual of Systematic Bacteriology: Volume 5: The Actinobacteria. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Whitman, W., Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M., Ludwig, W., Suzuki, K., Parte, A., Eds.; Springer: New York, NY, USA, 2012; ISBN 978-0-387-95043-3. [Google Scholar]
- Tripathi, N.; Sapra, A. Gram Staining. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Khadayat, K.; Sherpa, D.D.; Malla, K.P.; Shrestha, S.; Rana, N.; Marasini, B.P.; Khanal, S.; Rayamajhee, B.; Bhattarai, B.R.; Parajuli, N. Molecular Identification and Antimicrobial Potential of Streptomyces Species from Nepalese Soil. Int. J. Microbiol. 2020, 2020, 8817467. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Gislin, D.; Sudarsanam, D.; Antony Raj, G.; Baskar, K. Antibacterial Activity of Soil Bacteria Isolated from Kochi, India and Their Molecular Identification. J. Genet. Eng. Biotechnol. 2018, 16, 287–294. [Google Scholar] [CrossRef]
- Aryal, B.; Adhikari, B.; Aryal, N.; Bhattarai, B.R.; Khadayat, K.; Parajuli, N. LC-HRMS Profiling and Antidiabetic, Antioxidant, and Antibacterial Activities of Acacia Catechu (L.f.) Willd. BioMed Res. Int. 2021, 2021, 7588711. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.; Paudel, B.; Dahal, S.; Yadav, P.; Aryal, N.; Baral, B.; Bhattarai, H.D. Bioprospecting the Metabolome of Plant Urtica Dioica, L.: A Fast Dereplication and Annotation Workflow in Plant Metabolomics. Evid. Based Complementary Altern. Med. 2022, 2022, 3710791. [Google Scholar] [CrossRef] [PubMed]
- Handayani, I.; Saad, H.; Ratnakomala, S.; Lisdiyanti, P.; Kusharyoto, W.; Krause, J.; Kulik, A.; Wohlleben, W.; Aziz, S.; Gross, H.; et al. Mining Indonesian Microbial Biodiversity for Novel Natural Compounds by a Combined Genome Mining and Molecular Networking Approach. Mar. Drugs 2021, 19, 316. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, J.A.; Poynton, E.F.; Iskakova, D.; McMann, E.; Alsup, T.A.; Clark, T.N.; Fergusson, C.H.; Fewer, D.P.; Hughes, A.H.; McCadden, C.A.; et al. The Natural Products Atlas 2.0: A Database of Microbially-Derived Natural Products. Nucleic Acids Research 2022, 50, D1317–D1323. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching Molecular Structure Databases with Tandem Mass Spectra Using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef]
- Qureshi, K.A.; Bholay, A.D.; Rai, P.K.; Mohammed, H.A.; Khan, R.A.; Azam, F.; Jaremko, M.; Emwas, A.-H.; Stefanowicz, P.; Waliczek, M.; et al. Isolation, Characterization, Anti-MRSA Evaluation, and in-Silico Multi-Target Anti-Microbial Validations of Actinomycin X2 and Actinomycin D Produced by Novel Streptomyces Smyrnaeus UKAQ_23. Sci. Rep. 2021, 11, 14539. [Google Scholar] [CrossRef]
- Thomas, D.; Morris, M.; Curtis, J.M.; Boyd, R.K. Fragmentation Mechanisms of Protonated Actinomycins and Their Use in Structural Determination of Unknown Analogues. J. Mass Spectrom. 1995, 30, 1111–1125. [Google Scholar] [CrossRef]
- Paudel, B.; Maharjan, R.; Rajbhandari, P.; Aryal, N.; Aziz, S.; Bhattarai, K.; Baral, B.; Malla, R.; Bhattarai, H.D. Maculosin, a Non-Toxic Antioxidant Compound Isolated from Streptomyces Sp. KTM18. Pharm. Biol. 2021, 59, 931–934. [Google Scholar] [CrossRef]
- Wattana-Amorn, P.; Charoenwongsa, W.; Williams, C.; Crump, M.P.; Apichaisataienchote, B. Antibacterial Activity of Cyclo(L-Pro-L-Tyr) and Cyclo(D-Pro-L-Tyr) from Streptomyces Sp. Strain 22-4 against Phytopathogenic Bacteria. Nat. Prod. Res. 2016, 30, 1980–1983. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Gong, T.; Liu, F.; Zhang, P.-C.; Zhou, W.-Q.; Li, Y.; Zhu, P. A New Analogue of Echinomycin and a New Cyclic Dipeptide from a Marine-Derived Streptomyces Sp. LS298. Mar. Drugs 2015, 13, 6947–6961. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, G.; Bai, J.; Jing, Y.-K.; Pei, Y.-H. A Bisamide and Four Diketopiperazines from a Marine-Derived Streptomyces Sp. J. Asian Nat. Prod. Res. 2011, 13, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Pettit, G.R.; Du, J.; Pettit, R.K.; Richert, L.A.; Hogan, F.; Mukku, V.J.R.V.; Hoard, M.S. Antineoplastic Agents. 554. The Manitoba Bacterium Streptomyces Sp. J. Nat. Prod. 2006, 69, 804–806. [Google Scholar] [CrossRef]
- Le Goff, G.; Martin, M.-T.; Iorga, B.I.; Adelin, E.; Servy, C.; Cortial, S.; Ouazzani, J. Isolation and Characterization of Unusual Hydrazides from Streptomyces Sp. Impact of the Cultivation Support and Extraction Procedure. J. Nat. Prod. 2013, 76, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Liu, D.; Tian, L.; Wei, Y.; Proksch, P.; Zeng, J.; Lin, W. Venezuelines A-G, new Phenoxazine-Based Alkaloids and Aminophenols from Streptomyces Venezuelae and the Regulation of Gene Target Nur77. Bioorg. Med. Chem. Lett. 2013, 23, 301–304. [Google Scholar] [CrossRef]
- Bugni, T.S.; Woolery, M.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Bohemamines from a Marine-Derived Streptomyces Sp. J. Nat. Prod. 2006, 69, 1626–1628. [Google Scholar] [CrossRef]
- Vértesy, L.; Fehlhaber, H.-W.; Schulz, A. The Trehalase Inhibitor Salbostatin, a Novel Metabolite from Streptomyces Albus, ATCC21838. Angew. Chem. Int. Ed. 1994, 33, 1844–1846. [Google Scholar] [CrossRef]
- Shizuri, Y.; Ojika, M.; Yamada, K. Structure of an Antitumor Antibiotic, Reductiomycin. Tetrahedron Lett. 1981, 22, 4291–4294. [Google Scholar] [CrossRef]
- Lee, B.; Son, S.; Lee, J.K.; Jang, M.; Heo, K.T.; Ko, S.-K.; Park, D.-J.; Park, C.S.; Kim, C.-J.; Ahn, J.S.; et al. Isolation of New Streptimidone Derivatives, Glutarimide Antibiotics from Streptomyces Sp. W3002 Using LC-MS-Guided Screening. J. Antibiot. 2020, 73, 184–188. [Google Scholar] [CrossRef]
- Naganawa, H.; Wakashiro, T.; Yagi, A.; Kondo, S.; Takita, T. Deoxynybomycin from a Streptomyces. J. Antibiot. 1970, 23, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.; Gromyko, O.; Fedorenko, V.; Luzhetskyy, A.; Müller, R. Pimprinols A–C, from the Terrestrial Actinomycete, Streptomyces Sp. Tetrahedron Lett. 2012, 53, 3009–3011. [Google Scholar] [CrossRef]
- Hochlowski, J.E.; Andres, W.W.; Theriault, R.J.; Jackson, M.; McAlpine, J.B. Abbeymycin, a New Anthramycin-Type Antibiotic Produced by a Streptomycete. J. Antibiot. 1987, 40, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Uotani, K.; Naganawa, H.; Kondo, S.; Aoyagi, T.; Umezawa, H. Structural Studies on Ebelactone A and B, Esterase Inhibitors Produced by Actinomycetes. J. Antibiot. 1982, 35, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Abe, N.; Enoki, N.; Nakakita, Y.; Uchida, H.; Nakamura, T.; Munekata, M. Novel Cytocidal Compounds, Oxopropalines from Streptomyces Sp. G324 Producing Lavendamycin. II. Physico-Chemical Properties and Structure Elucidations. J. Antibiot. 1993, 46, 1678–1686. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Teramura, K.; Orita, M.; Matsumoto, H.; Yasumuro, K.; Abe, K. Effects of Ym-51084 and Ym-51085, New Inhibitors Produced by Streptomyces Sp. Q21705, on Cathepsin, L. J. Enzym. Inhib. 1996, 11, 115–121. [Google Scholar] [CrossRef]
- Geiger, A.; Keller-Schierlein, W.; Brandl, M.; Zähner, H. Metabolites of Microorganisms. 247. Phenazines from Streptomyces Antibioticus, Strain Tü 2706. J. Antibiot. 1988, 41, 1542–1551. [Google Scholar] [CrossRef]
- Comin, J.; Keller-Schierlein, W. Stoffwechselprodukte von Actinomyceten. 19. Mitteilung N-Acetyl-tyramin. Helv. Chim. Acta 1959, 42, 1730–1732. [Google Scholar] [CrossRef]
- Banskota, A.H.; McAlpine, J.B.; Sørensen, D.; Aouidate, M.; Piraee, M.; Alarco, A.-M.; Omura, S.; Shiomi, K.; Farnet, C.M.; Zazopoulos, E. Isolation and Identification of Three New 5-Alkenyl-3,3(2H)-Furanones from Two Streptomyces Species Using a Genomic Screening Approach. J. Antibiot. 2006, 59, 168–176. [Google Scholar] [CrossRef]
- Wang, F.; Xu, M.; Li, Q.; Sattler, I.; Lin, W. P-Aminoacetophenonic Acids Produced by a Mangrove Endophyte Streptomyces Sp. (Strain HK10552). Molecules 2010, 15, 2782–2790. [Google Scholar] [CrossRef]
- Tan, L.T.-H.; Chan, K.-G.; Pusparajah, P.; Yin, W.-F.; Khan, T.M.; Lee, L.-H.; Goh, B.-H. Mangrove Derived Streptomyces Sp. MUM265 as a Potential Source of Antioxidant and Anticolon-Cancer Agents. BMC Microbiol. 2019, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.W.; Cordell, G.A. Metabolism Studies of Indole Derivatives Using a Staurosporine Producer, Streptomyces Staurosporeus. J. Nat. Prod. 1997, 60, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Řezanka, T.; Spížek, J.; Přikrylová, V.; Prell, A.; Dembitsky, V.M. Five New Derivatives of Nonactic and Homo-Nonactic Acids from Streptomyces Globisporus. Tetrahedron 2004, 60, 4781–4787. [Google Scholar] [CrossRef]
- Hosny, M.; Rosazza, J.P.N. Microbial Hydroxylation and Methylation of Genistein by Streptomycetes. J. Nat. Prod. 1999, 62, 1609–1612. [Google Scholar] [CrossRef]
- Tchize Ndejouong, B.L.S.; Sattler, I.; Dahse, H.-M.; Kothe, E.; Hertweck, C. Isoflavones with Unusually Modified B-Rings and Their Evaluation as Antiproliferative Agents. Bioorganic Med. Chem. Lett. 2009, 19, 6473–6476. [Google Scholar] [CrossRef]
- Fyans, J.K.; Altowairish, M.S.; Li, Y.; Bignell, D.R.D. Characterization of the Coronatine-Like Phytotoxins Produced by the Common Scab Pathogen Streptomyces Scabies. MPMI 2015, 28, 443–454. [Google Scholar] [CrossRef]
- Pérez-Picaso, L.; Olivo, H.F.; Argotte-Ramos, R.; Rodríguez-Gutiérrez, M.; Rios, M.Y. Linear and Cyclic Dipeptides with Antimalarial Activity. Bioorg. Med. Chem. Lett. 2012, 22, 7048–7051. [Google Scholar] [CrossRef]
- Smith, K.; White, R.L.; Le, Y.; Vining, L. Isolation of N-Acetyl-3,4-Dihydroxy-L-Phenylalanine from Streptomyces Akiyoshiensis. J. Natural Prod. 1995, 58, 1274–1277. [Google Scholar] [CrossRef]
- Ou, Y.; Huang, J.; Li, X.; Kang, Q.; Pan, Y. Three New 2,5-Diketopiperazines from the Fish Intestinal Streptomyces Sp. MNU FJ-36. Nat. Prod. Res. 2016, 30, 1771–1775. [Google Scholar] [CrossRef]
- Shamim Hossain, M.; Aslam Hossain, M.; Mukhlesur Rahman, M.; Mojid Mondol, M.A.; Bhuiyan, M.S.A.; Gray, A.I.; Flores, M.E.; Rashid, M.A. Amides from the Fungus Streptomyces Hygroscopicus and Their Antimicrobial Activity. Phytochemistry 2004, 65, 2147–2151. [Google Scholar] [CrossRef]
- Awa, Y.; Iwai, N.; Ueda, T.; Suzuki, K.; Asano, S.; Yamagishi, J.; Nagai, K.; Wachi, M. Isolation of a New Antibiotic, Alaremycin, Structurally Related to 5-Aminolevulinic Acid from Streptomyces Sp. A012304. Biosci. Biotechnol. Biochem. 2005, 69, 1721–1725. [Google Scholar] [CrossRef] [PubMed]
- Suhara, Y.; Maruyama, H.B.; Koto, Y.; Miyasaka, Y.; Yokose, K. A New Antibiotic, Fumaramidmycin. II. Isolation, Structure and Syntheses. J. Antibiot. 1975, 28, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Sobin, B.A.; Tanner, F.W. Anisomycin,1 A New Anti-protozoab Antibiotics. J. Am. Chem. Soc. 1954, 76, 4053. [Google Scholar] [CrossRef]
- Sadovski, O.; Jaikaran, A.S.I.; Samanta, S.; Fabian, M.R.; Dowling, R.J.O.; Sonenberg, N.; Woolley, G.A. A Collection of Caged Compounds for Probing Roles of Local Translation in Neurobiology. Bioorg. Med. Chem. 2010, 18, 7746–7752. [Google Scholar] [CrossRef] [PubMed]
- Von Wittenau, M.S.; Els, H. The Structure of Indolmycin. J. Am. Chem. Soc. 1961, 83, 4678–4680. [Google Scholar] [CrossRef]
- Phay, N.; Yada, H.; Higashiyama, T.; Yokota, A.; Ichihara, A.; Tomita, F. NP-101A, Antifungal Antibiotic from Streptomyces Aurantiogriseus NPO-101. J. Antibiot. 1996, 49, 703–705. [Google Scholar] [CrossRef]
- Sakano, K.; Nakamura, S. New Antibiotics, Carbazomycins A and B. II. Structural Elucidation. J. Antibiot. 1980, 33, 961–966. [Google Scholar] [CrossRef]
- Fu, P.; Johnson, M.; Chen, H.; Posner, B.A.; MacMillan, J.B. Carpatamides A–C, Cytotoxic Arylamine Derivatives from a Marine-Derived Streptomyces Sp. J. Nat. Prod. 2014, 77, 1245–1248. [Google Scholar] [CrossRef]
- Shaaban, K.A.; Shepherd, M.D.; Ahmed, T.A.; Nybo, S.E.; Leggas, M.; Rohr, J. Pyramidamycins A-D and 3-Hydroxyquinoline-2-Carboxamide; Cytotoxic Benzamides from Streptomyces Sp. DGC1. J. Antibiot. 2012, 65, 615–622. [Google Scholar] [CrossRef]
- Jeong, S.-Y.; Shin, H.J.; Kim, T.S.; Lee, H.-S.; Park, S.; Kim, H.M. Streptokordin, a New Cytotoxic Compound of the Methylpyridine Class from a Marine-Derived Streptomyces Sp. KORDI-3238. J. Antibiot. 2006, 59, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, Y.; Yang, X.; Li, W.; Xiong, Z.; Zhao, L.; Xu, L.; Ding, Z. A New Cyclic Tetrapeptide from an Endophytic Streptomyces Sp. YIM67005. Nat. Prod. Res. 2014, 28, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Takagi, M.; Motohashi, K.; Shin-ya, K. Isolation of 2 New Metabolites, JBIR-74 and JBIR-75, from the Sponge-Derived Aspergillus Sp. FS14. J. Antibiot. 2010, 63, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, Y.; Kameyama, T.; Naganawa, H.; Okami, Y.; Takeuchi, T. Anisomycin and New Congeners Active against Human Tumor Cell Lines. J. Antibiot. 1993, 46, 1300–1302. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.; Wang, J.-D.; Liu, C.-X.; Yuan, J.-H.; Wang, X.-J.; Xiang, W.-S. A New Prenylated Indole Derivative from Endophytic Actinobacteria Streptomyces Sp. Neau-D50. Nat. Prod. Res. 2014, 28, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Lambalot, R.H.; Cane, D.E. Isolation and Characterization of 10-Deoxymethynolide Produced by Streptomyces Venezuelae. J. Antibiot. 1992, 45, 1981–1982. [Google Scholar] [CrossRef][Green Version]
- Hawas, U.W.; Shaaban, M.; Shaaban, K.A.; Speitling, M.; Maier, A.; Kelter, G.; Fiebig, H.H.; Meiners, M.; Helmke, E.; Laatsch, H. Mansouramycins A−D, Cytotoxic Isoquinolinequinones from a Marine Streptomycete. J. Nat. Prod. 2009, 72, 2120–2124. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, D.; Li, J.; Han, L.; Cui, X.; Lang, L.; Li, M.; Wang, Z.; Zhao, J.; Huang, X. Sannanine, a New Cytotoxic Alkaloid from Streptomyces Sannanensis. J. Antibiot. 2009, 62, 647–648. [Google Scholar] [CrossRef]
- Sorokina, M.; Steinbeck, C. Review on Natural Products Databases: Where to Find Data in 2020. J. Cheminform. 2020, 12, 20. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Jagannathan, S.V.; Manemann, E.M.; Rowe, S.E.; Callender, M.C.; Soto, W. Marine Actinomycetes, New Sources of Biotechnological Products. Mar. Drugs 2021, 19, 365. [Google Scholar] [CrossRef]
- Van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. Regulation of Antibiotic Production in Actinobacteria: New Perspectives from the Post-Genomic Era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef] [PubMed]
- Watve, M.G.; Tickoo, R.; Jog, M.M.; Bhole, B.D. How Many Antibiotics Are Produced by the Genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, A.; Thapa, A.; Budhathoki, A.; Sainju, M.; Shrestha, P.; Aryal, S. Isolation, Characterization, and Screening of Antimicrobial-Producing Actinomycetes from Soil Samples. Int. J. Microbiol. 2020, 2020, 2716584. [Google Scholar] [CrossRef]
- Shrestha, B.; Nath, D.K.; Maharjan, A.; Poudel, A.; Pradhan, R.N.; Aryal, S. Isolation and Characterization of Potential Antibiotic-Producing Actinomycetes from Water and Soil Sediments of Different Regions of Nepal. Int. J. Microbiol. 2021, 2021, 5586165. [Google Scholar] [CrossRef]
- Baniya, A.; Singh, S.; Singh, M.; Nepal, P.; Adhikari, M.; Aryal, S.; Adhikari, A. Isolation and Screening of Antibiotics Producing Streptomyces Spp from the Soil Collected around the Root of Alnus Nepalensis from Godawari. Nepal J. Biotechnol. 2018, 6, 46–56. [Google Scholar] [CrossRef]
- Gurung, T.D.; Sherpa, C.; Agrawal, V.P.; Lekhak, B. Isolation and Characterization of Antibacterial Actinomycetes from Soil Samples of Kalapatthar, Mount Everest Region. Nepal J. Sci. Technol. 2009, 10, 173–182. [Google Scholar] [CrossRef]
- Yadav, J.; Shrestha, U.T.; Tiwari, K.B.; Sahukhal, G.S.; Agrawal, V.P. Streptomycin—Like Antibiotic from Streptomyces Spp. Isolated from Mount Everest Base Camp. Nepal J. Sci. Technol. 2008, 9, 73–77. [Google Scholar] [CrossRef][Green Version]
- Tortorella, E.; Tedesco, P.; Palma Esposito, F.; January, G.G.; Fani, R.; Jaspars, M.; De Pascale, D. Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef]
- Chandra, N.; Kumar, S. Antibiotics Producing Soil Microorganisms. In Antibiotics and Antibiotics Resistance Genes in Soils: Monitoring, Toxicity, Risk Assessment and Management; Hashmi, M.Z., Strezov, V., Varma, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–18. ISBN 978-3-319-66260-2. [Google Scholar]
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020; World Health Organization: Geneva, Switzerland, 2020; ISBN 978-92-4-000558-7. [Google Scholar]
- Fernández-Martínez, L.T. Microbiology Today, Natural Products and Drug Discovery; Microbiology Society: London, UK, 2019; pp. 158–161. ISSN 1464-0570. [Google Scholar]
- Song, Z.; Hou, Y.; Yang, Q.; Li, X.; Wu, S. Structures and Biological Activities of Diketopiperazines from Marine Organisms: A Review. Mar. Drugs 2021, 19, 403. [Google Scholar] [CrossRef]
- Sioud, S.; Karray-Rebai, I.; Aouissaoui, H.; Aigle, B.; Bejar, S.; Mellouli, L. Targeted Gene Disruption of the Cyclo (L-Phe, L-Pro) Biosynthetic Pathway in Streptomyces Sp. US24 Strain. J. Biomed. Biotechnol. 2007, 2007, 91409. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, R.; Zhou, X.; Xu, T.; Yang, X.; Liu, Y. Diketopiperazines from Marine Organisms. Chem. Biodivers. 2010, 7, 2809–2829. [Google Scholar] [CrossRef] [PubMed]
- Hollstein, U. Actinomycin. Chemistry and Mechanism of Action. Chem. Rev. 1974, 74, 625–652. [Google Scholar] [CrossRef]
- Liu, M.; Jia, Y.; Xie, Y.; Zhang, C.; Ma, J.; Sun, C.; Ju, J. Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces Costaricanus SCSIO ZS0073. Mar. Drugs 2019, 17, 240. [Google Scholar] [CrossRef]
- Gerratana, B. Biosynthesis, Synthesis and Biological Activities of Pyrrolobenzodiazepines. Med. Res. Rev. 2012, 32, 254–293. [Google Scholar] [CrossRef]
- Donohue, J.; Smith, A.B.; Carroll, P.J. Crystal and Molecular Structure of Reductiomycin. J. Crystallogr. Spectrosc. Res. 1984, 14, 35–43. [Google Scholar] [CrossRef]
- Shimizu, K.; Tamura, G. Reductiomycin, a New Antibiotic. I. Taxonomy, Fermentation, Isolation, Characterization and Biological Activities. J. Antibiot. 1981, 34, 649–653. [Google Scholar] [CrossRef]
- Egawa, K.; Yamori, T.; Nosaka, C.; Kunimoto, S.; Takeuchi, T.; Nose, K. Deoxynybomycin Is a Selective Anti-Tumor Agent Inducing Apoptosis and Inhibiting Topoisomerase I. Biol. Pharm. Bull. 2000, 23, 1036–1040. [Google Scholar] [CrossRef]
- Räty, K.; Kantola, J.; Hautala, A.; Hakala, J.; Ylihonko, K.; Mäntsälä, P. Cloning and Characterization of Streptomyces Galilaeus Aclacinomycins Polyketide Synthase (PKS) Cluster. Gene 2002, 293, 115–122. [Google Scholar] [CrossRef]
- Fujii, I.; Ebizuka, Y. Anthracycline Biosynthesis in Streptomyces Galilaeus. Chem. Rev. 1997, 97, 2511–2524. [Google Scholar] [CrossRef]
- Harir, M.; Bendif, H.; Bellahcene, M.; Pogni, Z.F.R. Streptomyces Secondary Metabolites; IntechOpen: London, UK, 2018; ISBN 978-1-78984-615-7. [Google Scholar]
- Al-Ansari, M.; Kalaiyarasi, M.; Almalki, M.A.; Vijayaraghavan, P. Optimization of Medium Components for the Production of Antimicrobial and Anticancer Secondary Metabolites from Streptomyces Sp. AS11 Isolated from the Marine Environment. J. King Saud Univ. Sci. 2020, 32, 1993–1998. [Google Scholar] [CrossRef]
- Thakur, D.; Bora, T.C.; Bordoloi, G.N.; Mazumdar, S. Influence of Nutrition and Culturing Conditions for Optimum Growth and Antimicrobial Metabolite Production by Streptomyces Sp. 201. J. De Mycol. Médicale 2009, 19, 161–167. [Google Scholar] [CrossRef]
- VanderMolen, K.M.; Raja, H.A.; El-Elimat, T.; Oberlies, N.H. Evaluation of Culture Media for the Production of Secondary Metabolites in a Natural Products Screening Program. AMB Express 2013, 3, 71. [Google Scholar] [CrossRef] [PubMed]
- Arend, K.I.; Bandow, J.E. Influence of Amino Acid Feeding on Production of Calcimycin and Analogs in Streptomyces Chartreusis. Int. J. Environ. Res. Public Health 2021, 18, 8740. [Google Scholar] [CrossRef]
- Barbuto Ferraiuolo, S.; Cammarota, M.; Schiraldi, C.; Restaino, O.F. Streptomycetes as Platform for Biotechnological Production Processes of Drugs. Appl. Microbiol. Biotechnol. 2021, 105, 551–568. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, Y.-H.; Chen, Y.; Chen, K.; Jiang, S.-X.; Huang, K.; Liu, Z.-Q.; Zheng, Y.-G. Enhanced AmB Production in Streptomyces Nodosus by Fermentation Regulation and Rational Combined Feeding Strategy. Front. Bioeng. Biotechnol. 2020, 8, 597. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Dhakal, D.; Pham, V.T.T.; Nguyen, H.T.; Sohng, J.-K. Recent Advances in Strategies for Activation and Discovery/Characterization of Cryptic Biosynthetic Gene Clusters in Streptomyces. Microorganisms 2020, 8, 616. [Google Scholar] [CrossRef]
- Wu, C.; Choi, Y.H.; van Wezel, G.P. Metabolic Profiling as a Tool for Prioritizing Antimicrobial Compounds. J. Ind. Microbiol. Biotechnol. 2016, 43, 299–312. [Google Scholar] [CrossRef]
- Ahmed, Y.; Rebets, Y.; Estévez, M.R.; Zapp, J.; Myronovskyi, M.; Luzhetskyy, A. Engineering of Streptomyces Lividans for Heterologous Expression of Secondary Metabolite Gene Clusters. Microb. Cell Factories 2020, 19, 5. [Google Scholar] [CrossRef]
- Winter, J.M.; Behnken, S.; Hertweck, C. Genomics-Inspired Discovery of Natural Products. Curr. Opin. Chem. Biol. 2011, 15, 22–31. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).