Abstract
Infectious diseases are one of the core biological complications for public health. It is important to recognize the pathogen-specific mechanisms to improve our understanding of infectious diseases. Differentiations between bacterial- and viral-targeted human proteins are important for improving both prognosis and treatment for the patient. Here, we introduce machine learning-based classifiers to discriminate between the two groups of human proteins. We used the sequence, network, and gene ontology features of human proteins. Among different classifiers and features, the deep neural network (DNN) classifier with amino acid composition (AAC), dipeptide composition (DC), and pseudo-amino acid composition (PAAC) (445 features) achieved the best area under the curve (AUC) value (0.939), F1-score (94.9%), and Matthews correlation coefficient (MCC) value (0.81). We found that each of the selected top 100 of the bacteria- and virus-targeted human proteins from a candidate pool of 1618 and 3916 proteins, respectively, were part of distinct enriched biological processes and pathways. Our proposed method will help to differentiate between the bacterial and viral infections based on the targeted human proteins on a global scale. Furthermore, identification of the crucial pathogen targets in the human proteome would help us to better understand the pathogen-specific infection strategies and develop novel therapeutics.
1. Introduction
Despite the current improvements in antimicrobial therapy and vaccination, infectious diseases remain a major threat to public health worldwide. They cause significant morbidity across the nations, posing a major burden on the economy, and causing a substantial number of deaths in the less developed countries [1]. The majority of infectious diseases are caused by pathogenic bacteria and viruses. Pathogens interact with the host system right from the point of its entry into the host, primarily to evade the host immune response and create their own niche for survival and growth [2]. The identification of host proteins targeted by pathogens and pathogen–host protein–protein interactions (PPIs) is crucial to understand the mechanisms underlying the infectious diseases [3]. To differentiate between the bacterial- and viral-targeted host proteins is critical to delineate the specific infection strategies for these two groups of pathogens. While this may help in the diagnosis of the etiology, it is particularly important from the treatment perspective, which is distinct for bacterial and viral infections. Antibiotics kill bacterial pathogens but are ineffective against viruses. Finally, identification of the specific biological processes for the bacterial- and viral-targeted human proteins could improve disease prognosis and treatment.
Several studies attempted to explore the mechanisms underlying infectious diseases from the study of pathogen–host PPIs [4,5,6,7,8,9,10,11,12,13]. The availability of experimentally verified pathogen–host PPIs in the public domain significantly helped these efforts [14,15,16,17,18,19,20]. However, only one study compared pathogen–host PPIs for bacterial and viral infections [21]. This study addressed common as well as distinct infection strategies for bacterial and viral infections. To distinguish between bacterial- and viral-targeted human proteins, they only used the degree centrality, betweenness centrality, and gene ontology (GO) features of different proteins. They drew a general conclusion that viruses tend to interact with human proteins having much higher connectivity and centrality values than those for bacteria. They proposed that viral-targeted human proteins function in the cellular process to manipulate it, while bacteria-targeted human proteins interact with the immune system. Here, we used more rigorous techniques, such as machine learning algorithms, to differentiate the bacteria-targeted human proteins from the virus-targeted proteins. To this end, we used the sequence, network, and gene ontology features of human proteins extensively. We identified the best features set for the purpose of discriminating between bacterial- and viral-targeted proteins and listed the top predicted targets. Finally, the differences between the bacterial- and viral-targeted human proteins were validated by GO and pathway enrichment analysis.
2. Material and Methods
2.1. Data Collection
All the experimentally validated bacteria–human and virus–human protein–protein interaction (PPI) datasets were collected from PHISTO: a pathogen–host interaction search tool [22]. We found 8993 and 35,120 bacteria–human and virus–human PPIs, respectively, and detected 3673 bacterial- and 5887 viral-targeted human proteins. Out of these, 1780 proteins were common targets of both bacteria and viruses (shown in Figure 1) and were excluded from our analysis. We searched the remaining 1893 and 4107 respective bacterial- and viral-targeted human proteins, in UniProt, a worldwide hub of protein knowledge database [23]. We found 1618 and 3916 bacterial- and viral-targeted and reviewed human proteins, respectively, in UniProt (Supplementary Tables S1 and S2), which were considered for further analysis.
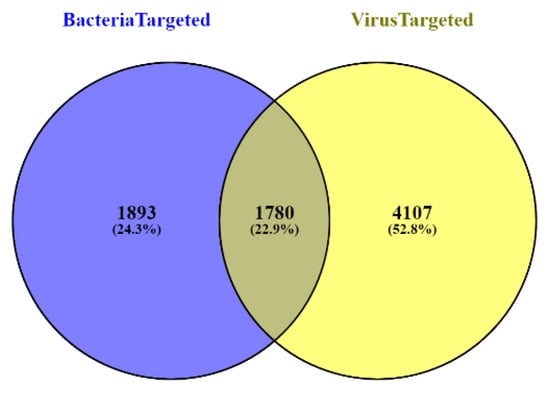
Figure 1.
Venn diagram of bacterial- and viral-targeted human proteins.
2.2. Sequence Features
All the above human protein sequences were downloaded from the UniProt database. For the prediction of proteins and PPIs, the sequence features, such as the amino acid composition (AAC), dipeptide composition (DC), pseudo-amino acid composition (PAAC), and composition-transition-distribution (CTD) were reported as important features [24,25,26]. We computed AAC, DC, PAAC, and CTD using PyDPI, a freely available python package for chemoinformatics, bioinformatics, and chemogenomics studies [27]. We used these sequence features to discriminate between the bacterial- from the viral-targeted human proteins.
2.3. Network Features
To compute network features for human proteins, we retrieved expert-curated human PPIs from the Human Protein Reference Database (HPRD) (Release 9) [28] and constructed a network using these PPIs. Network analyzer (cytoscape plugin) was used to compute the network properties, such as degree, closeness centrality, neighborhood connectivity, average shortest path length, betweenness centrality, clustering coefficient, topological coefficient, eccentricity, and radiality [29].
2.4. Gene Ontology (GO) Features
All the GO identifiers (IDs) for the respective 1618 and 3916 bacterial- and viral-targeted human proteins were downloaded from UniProt. We found a total of 23,737 GO IDs for 1618 bacteria-targeted human proteins, while the number of GO IDs for the viral-targeted human proteins was 67,035. The occurrence of each GO ID was counted separately for the above two groups, followed by sorting based on the occurrence value. The top 100 and 280 GO IDs for the bacterial- and viral-targeted human proteins were extracted for GO features. However, only 282 were unique among the top 380 GO IDs (Supplementary Table S3). Therefore, we considered the unique IDs for GO features (Supplementary Figure S1). For each human protein, the presence or absence of the top GO ID was considered as 1 or 0, respectively.
2.5. Classification
The distinction between the bacterial- and viral-targeted human proteins may be viewed as a binary (two-class) classification problem. To differentiate between the proteins, we used well-known classifiers, such as SVM, RF, and DNN.
2.5.1. Support Vector Machines (SVM)
The SVM classifier explicitly maps the data over a vector space to find a decision surface that maximizes the margin between data points of two classes. For the SVM classifier, we used the scikit-learn python package [30]. To find the best performance of the SVM classifier, we tested different combinations of cost and gamma parameters of radial basis function (RBF).
2.5.2. Random Forest (RF)
Several decision trees (DTs) grow simultaneously using a random subset of features in RF. In the RF classifier, each tree is a new object and “votes” for that class. Based on a majority vote, the forest elects the classification. We also used the scikit-learn python package for the RF classifier. Optimal parameters were utilized to find the best performance.
2.5.3. Deep Neural Networks (DNN)
The DNN method was shown to perform well with diverse problems. DNN is more robust and useful than other methods for complex classification problems and is becoming a popular algorithm in the field of modern computational biology. We used TensorFlow DNN, which is a widely-used deep learning package for classification, to discriminate between the bacterial- and viral-targeted human proteins [31].
2.6. 10-Fold Cross-Validation
To avoid the performance bias of the prediction methods, we used the 10-fold cross-validation technique. In 10-fold cross-validation, the whole dataset is divided into 10 sets (folds) of equal or nearly equal sizes. Training and testing are repeated 10 times so that each time, a different set (fold) goes out for testing, while the remaining 9 sets (folds) are used for training. The average performance measures over the 10 folds are considered for the overall performance of the model.
2.7. Feature Selection
We used several feature selection methods, such as univariate feature selection (UFS), recursive feature elimination (RFE), feature selection using SelectFromModel (SFM), and tree-based feature selection (TBFS). In UFS, the K best features were selected based on the univariate statistical tests. We used all the univariate statistical test methods available in scikit-learn for the purpose of classification. In RFE, the least important features are excluded in each recursive step, until the desired number of features is reached. The important features are selected from the model in SFM. In TBFS, a tree-based estimator computes the importance of the features and irrelevant features are discarded.
2.8. Performance Measures
The performance measures of the classification problem, such as sensitivity, specificity, accuracy, positive predictive value (PPV or precision), Mathews correlation coefficient (MCC), and F1-score were calculated using the following equations:
where
True Positive (TP): Bacterial-targeted human proteins are correctly identified as bacterial-targeted human proteins.
False Positive (FP): Viral-targeted human proteins are incorrectly identified as bacterial-targeted human proteins.
True Negative (TN): Viral-targeted human proteins are correctly identified as viral-targeted human proteins.
False Negative (FN): Bacterial-targeted human proteins are incorrectly identified as viral-targeted human proteins.
The area under the receiver operating characteristic curve (AUC), for all the cases, was also computed.
2.9. GO Enrichment Analysis
The top 100 bacterial-targeted and the same number of viral-targeted human proteins predicted by our method were considered for GO enrichment analysis. To this end, we used Enrichr, a comprehensive gene set enrichment analysis web server, 2016 update [32]. We considered only the biological process terms with p-values < 0.05 for the GO enrichment analysis.
2.10. Pathway Enrichment Analysis
The above mentioned 200 human proteins (100 each of the bacterial- and viral-targeted proteins) were also considered for pathway enrichment analysis. We used the Reactome Pathway Knowledgebase for this purpose [33]. Pathways with p-value < 0.05 were treated as enriched pathways.
3. Results
3.1. Selection of Features
Important features of human proteins, such as the sequence, GO, and networks were considered to discriminate between the bacteria- and virus-targeted human proteins. For individual sequence features, dipeptide composition (DC) achieved the highest AUC of 0.931, with an F1-score of 90.3%, and MCC of 0.67 (Table 1 and Supplementary Table S4). However, the sequence features AAC, PAAC, and CTD showed poor performances with CTD being the poorest. We tested different combinations of the above features to achieve a high performance. We observed that a combination of AAC, DC, and PAAC achieved the best AUC of 0.939, F1-score of 94.9% and MCC of 0.81.

Table 1.
Features-wise performance measures on bacterial- and viral-targeted human proteins.
Of the other features, the GO feature attained the maximum AUC of 0.886, F1-score of 86.4% and MCC of 0.51. On the other hand, the network feature was unable to distinguish between the bacteria- and virus-targeted human proteins. We also tested mixed features set to measure the performance. We found that the combination of AAC, DC, PAAC, and GO features achieved the highest AUC of 0.914, F1-score of 88.3% and MCC of 0.60. Together, the above results suggested that the combination of the AAC, DC, and PAAC features attained the highest level of performance.
We applied multiple feature selection methods, such as UFS, RFE, SFM, and TBFS for the combination of AAC, DC, and PAAC features. We observed that TBFS achieved the highest AUC of 0.805, F1-score of 84% and MCC of 0.44 (Table 2 and Supplementary Table S5). However, features selected by these methods were unable to attain a similar performance as the original features set. This result suggested that several features selection methods were unable to perform better than the primary features. As a result, we selected a combination of AAC, DC, and PAAC (445 features) as the best features set.

Table 2.
Selected feature-wise performance measures of bacterial- and viral-targeted human proteins.
3.2. Performance Comparison of Different Classifiers
To find the best classifier for our dataset, we compared the performance of SVM, RF, and DNN classifiers. Different parameter-based performances were calculated for these classifiers and only the best result was reported here. In the majority of cases, we observed that the DNN classifier achieved the best performance (Table 1 and Table 2). As shown in Figure 2 and Figure 3, the performance of the DNN classifier is far superior to SVM and RF. Together, the results suggested that DNN performed better than other conventional MLT.
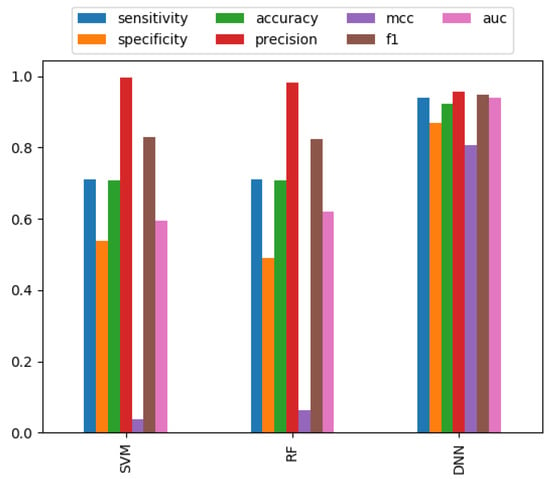
Figure 2.
Performance measures of different classifiers for the combination of AAC, DC, and PAAC features set.
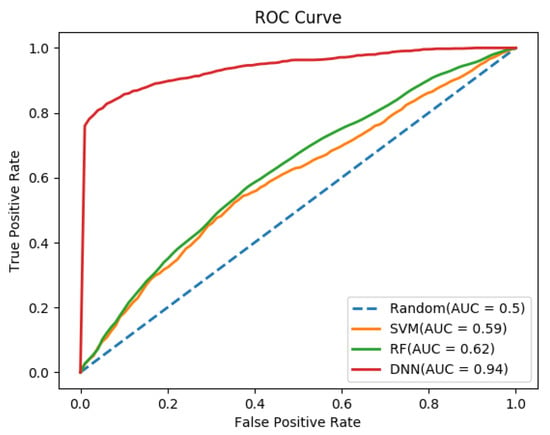
Figure 3.
AUC of different classifiers (SVM, RF, and DNN) for the combination of AAC, DC, and PAAC features set.
3.3. Gene Ontology Enrichment Analysis
Prediction probability scores of all the bacteria- and virus-targeted human proteins were sorted (Supplementary Tables S6 and S7). Prediction scores for the top 100 bacteria-targeted and the same number of virus-targeted human proteins were investigated further to understand the specific infection strategies. GO enrichment analysis of the predicted bacteria-targeted proteins displayed negative regulation for catalytic activity, cellular response to hypoxia, cellular catabolic process, nitric oxide biosynthetic process, nitric oxide metabolic process, calcium ion import, RIG-I signaling pathway, cell adhesion mediated by integrin, and heart rate, etc. (Table 3). In contrast, virus-targeted human proteins showed biological processes, such as the peptide biosynthetic process, translation, mitochondrial ATP synthesis-coupled electron transport, mitochondrial translation elongation, cellular macromolecule biosynthetic process, mitochondrial translational termination, respiratory electron transport chain, and translational termination upon GO enrichment analysis (Table 4). Overall, the top bacteria- and virus-targeted human proteins were related to 48 and 96 enriched biological processes, respectively. We found that most of the enriched biological processes were distinct for bacteria- and virus-targeted human proteins (Figure 4).

Table 3.
Top 20 GO biological processes for bacterial-targeted human proteins.

Table 4.
Top 20 GO biological processes for viral-targeted human proteins.
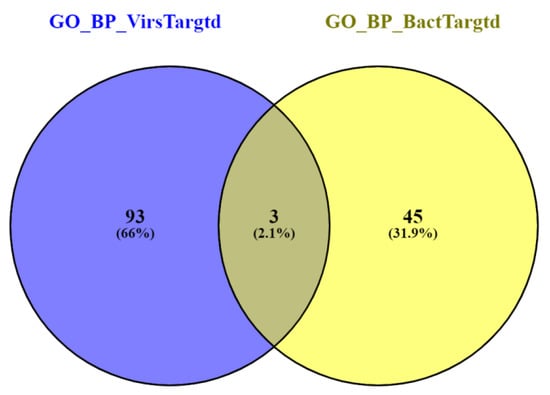
Figure 4.
Venn diagram of enriched biological processes for bacterial- and viral-targeted human proteins.
Pathway enrichment analysis showed the uptake and function of anthrax toxins, defective NEU1 causing sialidosis, and Vitamin B1 (thiamin) metabolism pathways for the top 100 bacteria-targeted human proteins (Table 5). Likewise, the top predicted virus-targeted human proteins showed the enrichment of pathways, including the formation of the cornified envelope, keratinization, translation, and mitochondrial translation termination, etc. (Table 6). We found that the enriched pathways for bacteria- and virus-targeted human proteins were different (Figure 5). The above results suggested that bacterial-targeted human proteins enriched gene ontology (GO) and pathways distinct from viral-targeted human protein.

Table 5.
Top 5 pathways for bacterial-targeted human proteins.

Table 6.
Top 5 pathways for viral-targeted human proteins.
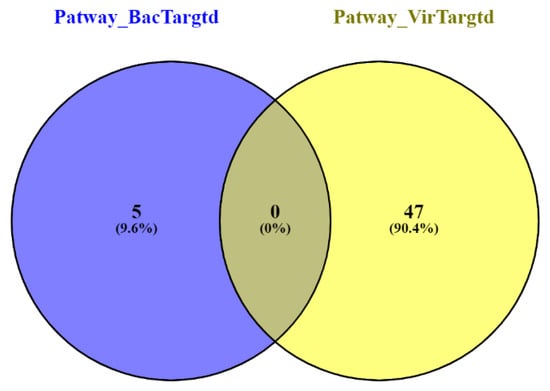
Figure 5.
Venn diagram of enriched pathways for bacterial- and viral-targeted human proteins.
4. Discussion
Rapid, safe, cost-effective, and accurate tools for etiological diagnosis of suspected infections are of paramount importance for individual and public health. Particularly important is to discriminate between the bacterial and viral causes of infectious diseases given the alarming rise of antibiotic resistance, due to their indiscriminate and unnecessary use. An estimated 30–50% of antibiotics are prescribed in hospitalized patients of the United States for wrong indications, most commonly viral infections (https://www.cdc.gov/antibiotic-use/stewardship-report/outpatient.html, accessed on 21 October 2021) [34]. Traditional culture methods for bacterial infections are low throughput, time consuming, and labor intensive, in addition to the challenges of sample collection from some of the infected tissues, and the lack of wide availability of culture techniques for many pathogen species. On the other hand, the diagnosis of viral infections by serology may lack specificity, while nucleic acid detection methods require sophisticated equipment and technical expertise. However, no reliable methods or markers are currently available for the rapid diagnosis of bacterial and viral etiologies of infectious diseases.
Attempts have been made to develop complementary diagnostics for infectious diseases by focusing on specific host responses. In addition to being capable of discriminating between colonization and infection, this approach is not limited by the availability of infected tissue samples. Moreover, host response-based categorization of infections provides additional insights into the disease pathogenesis and immune response and may help to identify new targets for therapeutic intervention.
Multiple attempts have been made to diagnose infectious diseases based on host-specific biomarkers. Widely used parameters such as WBC counts and C-reactive protein (CRP), may aid to differentiate between bacterial and viral infections, but lack sensitivity and specificity, leading to frequent misdiagnosis. Newer bacterial infection markers, such as presepsin, procalcitonin, and CD64, are used for severe sepsis, while proADM may predict prognosis of the disease [35,36]. In contrast, cytokines, such as IL-2, IL-8, and IL-10 were suggested as early biomarkers for viral infection [37]. Several research groups reported that the antiviral host protein MxA is a clinically useful marker for acute viral infection and, combined with CRP and/or procalcitonin, may distinguish between bacterial and viral infections [38]. A double-blind, multicenter study found that a strategy to integrate CRP, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and interferon γ-induced protein-10 (IP-10) performed significantly better than the individual markers to identify acute viral infection in pediatric patients [39]. However, they did not validate their tools against reference diagnostic methods, limiting its utility. Other studies also suggested that a combination of markers may perform better than a single biomarker [40]. However, combining CRP with other markers did not improve the former’s ability to differentiate between bacterial and viral lower respiratory tract infections in a different study [41].
High throughput genomic and proteomic studies have been employed to identify infection-specific host gene sets. Although they were useful for novel biomarker discovery, the gene sets often contained a large number of candidates, making them difficult to apply clinically [42,43,44]. Through multi-cohort analysis of these large datasets, smaller gene sets optimized for the diagnosis of bacterial and viral infections were identified later on [45].
Machine learning techniques have been extensively used for disease biomarker discovery, including infectious diseases. However, they were mostly used for individual microbial species or groups of pathogens. The increasing availability of bacteria–human and virus–human PPIs now permits researchers to compare bacterial- and viral-specific infection strategies and identify host proteins that are differentially targeted by these two classes of pathogens. We employed well-known machine learning methods, such as SVM, RF, and DNN to the available PPI datasets to distinguish between bacteria- and virus-targeted human proteins.
We considered all the updated and comprehensive sets of experimentally validated bacteria–human and virus–human PPIs from PHISTO. We found 1780 human proteins that are common targets for bacteria and viruses. During the bacterial and viral infection, these common proteins might help to execute several commonalities, such as immune response patterns, acute onset, and response to antimicrobial agents in humans. The primary goal of the current study was to differentiate between bacterial- and viral-targeted human proteins. Therefore, we excluded these 1780 human proteins from our analysis. The proposed method used 1618 and 3917 bacterial- and viral-targeted human proteins. To ensure utilization of a larger dataset of two classes, we considered the complete dataset for building the model. For imbalance datasets, we found that performance measures, such as the AUC, MCC, and F1-score, were more important as opposed to sensitivity, specificity, and accuracy. Therefore, we compared the AUC, MCC, and F1-score for all the cases. We found that sequence and gene ontology features performed far better than network features. We witnessed that the network properties of human proteins was unable to distinguish between bacterial- and viral-targeted human proteins (Table 1), suggesting indistinguishable network feature patterns for bacterial and viral targeted human proteins. The majority of frequent GO IDs for bacterial- and viral-targeted human proteins are common (Supplementary Figure S1). Therefore, gene ontology features were unable to perform better than the sequence features. Among the sequence features, we found that DC achieved better performance than the others. A combination of AAC, DC, and PAAC features (445 features) achieved the best performance (Table 1). In addition to these, the feature set selected by different feature selection techniques also showed a poorer performance than the above features set. Therefore, we reported that the combination of AAC, DC, and PAAC (445 features) is the best feature set for discriminating between bacterial- and viral-targeted human proteins. If the two classes are distinct due to true biological reasons, then we can also get good performance results for conventional MLTs like SVM and RF (shown in Table 1, and Figure 2 and Figure 3). The DNN performed well due to a large number of data and features. Furthermore, we identified the top 100 human proteins targeted by bacteria and the top 100 human proteins targeted by viruses. The gene ontology enrichment analysis of these 200 proteins showed a greater number of enriched biological processes for viral-targeted human proteins rather than bacterial-targeted human proteins (Figure 4). Similarly, we observed a greater number of enriched pathways for viral-targeted human proteins than bacterial targeted human proteins. These results imply that viruses are influencing more biological processes and pathways than bacteria. As is known, viruses are totally dependent on the host. Therefore, they exploit more host machinery than bacteria. The above results indicate the same. In addition to this, we observed that the majority of the enriched biological processes and pathways were different for bacterial- and viral-targeted human proteins. These functional annotations also validated our method for discriminating between bacterial- and viral-targeted human proteins.
5. Conclusions
We proposed a computational method to distinguish between the bacteria- and virus-targeted human proteins. We employed widely used and state-of-the-art machine learning techniques, such as SVM, RF, and DNN and integrated important biological information on human proteins, including the sequences, networks, and GO to achieve this goal. We found the best performance was with the sequence features and the DNN classifier. We developed a prediction model to maximize the performance measures and identify the best features to do the same. Therefore, we did not use the prediction for future data. However, the proposed model may be utilized for predicting and discriminating between the possible interactions of human proteins with bacterial and viral proteins. We identified distinct targets for bacterial and viral infections upon GO and pathway enrichment analysis of highly predicted human proteins. Bacterial targets predominantly included immune response-related genes and transcriptional machinery, while viruses targeted protein translation and mitochondrial energy metabolism. The distinction between bacteria- and virus-targeted human proteins might help to improve infection-specific diagnosis and treatment. In the future, we will look for the difference between RNA and DNA viruses, and Gram-positive and Gram-negative bacteria to understand the specific infection strategy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pr10020291/s1, Figure S1: Venn diagram of Gene Ontology (GO) IDs of bacteria- and virus-targeted human proteins., Table S1: Bacterial targeted reviewed human proteins, Table S2: Viral targeted reviewed human proteins, Table S3: Top GO IDs for bacterial and viral targeted human proteins, Table S4: Full table of features wise performance measures on bacterial and viral targeted human proteins, Table S5: Full table of selected feature-wise performance measures of bacterial and viral targeted human proteins, Table S6: Probability score of top 100 bacteria targeted human proteins, Table S7: Probability score of top 100 virus targeted human proteins.
Author Contributions
R.K.B., A.M., U.M. and S.D. conceived and designed experiments; R.K.B. executed experiments; R.K.B., A.M., U.M. and S.D. analyzed data and wrote manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
No separate funding was obtained for this study; intramural funds were utilized.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Source codes and supplementary information is available at https://github.com/ranjan1010/BacteriaVsVirusTargetedHumanProteinsWork (accessed on 5 January 2022).
Acknowledgments
R.K.B. acknowledges the Senior Research Fellowship of Indian Council of Medical Research [No. ISRM/11(39)/2017]. A.M. acknowledges the support received from the research project (Memo No: 355(Sanc.)/ST/P/S&T/6G-10/2018 dt.08/03/2019) of Dept. of Science & Technology and Biotechnology, Govt. of West Bengal, India at University of Kalyani.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
DNN: deep neural networks; AAC: amino acid composition; DC: dipeptide composition; PAAC: pseudo-amino acid composition; AUC: area under the curve; MCC: Matthews correlation coefficient; PPIs: protein-protein interactions; GO: gene ontology; CTD: composition-transition-distribution; HPRD: Human Protein Reference Database; SVM: support vector machines; RBF: radial basis function; RF: random forest; DTs: decision trees; UFS: univariate feature selection; RFE: recursive feature elimination; SFM: SelectFromModel; TBFS: tree-based feature selection; PPV: positive predictive value; TP: true positive; FP: false positive; TN: true negative; FN: false negative.
References
- WHO. Health in 2015: From MDGs to SDGs; WHO Press: Geneva, Switzerland, 2015; pp. 101–130. [Google Scholar]
- Nicholson, L.B. The immune system. Essays Biochem. 2016, 60, 275–301. [Google Scholar] [CrossRef] [Green Version]
- Nicod, C.; Banaei-Esfahani, A.; Collins, B.C. Elucidation of host-pathogen protein-protein interactions to uncover mechanisms of host cell rewiring. Curr. Opin. Microbiol. 2017, 39, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Gao, S.; Nguyen, N.N.; Fan, M.; Jin, J.; Liu, B.; Zhao, L.; Xiong, G.; Tan, M.; Li, S.; et al. Stringent homology-based prediction of H. sapiens-M. tuberculosis H37Rv protein-protein interactions. Biol. Direct 2014, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Kosesoy, I.; Gok, M.; Oz, C. A new sequence based encoding for prediction of host-pathogen protein interactions. Comput. Biol. Chem. 2019, 78, 170–177. [Google Scholar] [CrossRef]
- Alguwaizani, S.; Park, B.; Zhou, X.; Huang, D.S.; Han, K. Predicting Interactions between Virus and Host Proteins Using Repeat Patterns and Composition of Amino Acids. J. Healthc. Eng. 2018, 2018, 1391265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, X.; Yang, S.; Li, H.; Fu, C.; Zhang, Z. Machine-Learning-Based Predictor of Human-Bacteria Protein-Protein Interactions by Incorporating Comprehensive Host-Network Properties. J. Proteome Res. 2019, 18, 2195–2205. [Google Scholar] [CrossRef]
- Tyagi, N.; Krishnadev, O.; Srinivasan, N. Prediction of protein-protein interactions between Helicobacter pylori and a human host. Mol. Biosyst. 2009, 5, 1630–1635. [Google Scholar] [CrossRef]
- Penn, B.H.; Netter, Z.; Johnson, J.R.; Von Dollen, J.; Jang, G.M.; Johnson, T.; Ohol, Y.M.; Maher, C.; Bell, S.L.; Geiger, K.; et al. An Mtb-Human Protein-Protein Interaction Map Identifies a Switch between Host Antiviral and Antibacterial Responses. Mol. Cell 2018, 71, 637–648.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barman, R.K.; Saha, S.; Das, S. Prediction of interactions between viral and host proteins using supervised machine learning methods. PLoS ONE 2014, 9, e112034. [Google Scholar] [CrossRef]
- Wuchty, S. Computational prediction of host-parasite protein interactions between P. falciparum and H. sapiens. PLoS ONE 2011, 6, e26960. [Google Scholar] [CrossRef] [Green Version]
- Dyer, M.D.; Murali, T.M.; Sobral, B.W. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008, 4, e32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uetz, P.; Dong, Y.A.; Zeretzke, C.; Atzler, C.; Baiker, A.; Berger, B.; Rajagopala, S.V.; Roupelieva, M.; Rose, D.; Fossum, E.; et al. Herpesviral protein networks and their interaction with the human proteome. Science 2006, 311, 239–242. [Google Scholar] [CrossRef] [Green Version]
- Farooq, Q.U.A.; Khan, F.F. Construction and analysis of a comprehensive protein interaction network of HCV with its host Homo sapiens. BMC Infect. Dis. 2019, 19, 367. [Google Scholar] [CrossRef]
- Li, Y.; Liu, G.; Zhang, J.; Zhong, X.; He, Z. Identification of key genes in human airway epithelial cells in response to respiratory pathogens using microarray analysis. BMC Microbiol. 2018, 18, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Zhang, Y.; Li, Y.H.; Wang, S.; Zhang, J.J.; Zhang, C.X.; Zhang, Z.S. Investigating dysregulated pathways in Staphylococcus aureus (SA) exposed macrophages based on pathway interaction network. Comput. Biol. Chem. 2017, 66, 21–25. [Google Scholar] [CrossRef]
- Ehsani Ardakani, M.J.; Safaei, A.; Arefi Oskouie, A.; Haghparast, H.; Haghazali, M.; Mohaghegh Shalmani, H.; Peyvandi, H.; Naderi, N.; Zali, M.R. Evaluation of liver cirrhosis and hepatocellular carcinoma using Protein-Protein Interaction Networks. Gastroenterol. Hepatol. Bed Bench 2016, 9, S14–S22. [Google Scholar] [PubMed]
- Simos, T.; Georgopoulou, U.; Thyphronitis, G.; Koskinas, J.; Papaloukas, C. Analysis of protein interaction networks for the detection of candidate hepatitis B and C biomarkers. IEEE J. Biomed. Health Inform. 2015, 19, 181–189. [Google Scholar] [CrossRef]
- Wang, Q.; Lou, Z.; Zhai, L.; Zhao, H. Detection of Significant Pneumococcal Meningitis Biomarkers by Ego Network. Indian J. Pediatrics 2017, 84, 430–436. [Google Scholar] [CrossRef]
- Liu, J.; Ma, Z.; Liu, Y.; Wu, L.; Hou, Z.; Li, W. Screening of potential biomarkers in hepatitis C virus-induced hepatocellular carcinoma using bioinformatic analysis. Oncol. Lett. 2019, 18, 2500–2508. [Google Scholar] [CrossRef] [PubMed]
- Durmus Tekir, S.; Cakir, T.; Ulgen, K.O. Infection Strategies of Bacterial and Viral Pathogens through Pathogen-Human Protein-Protein Interactions. Front. Microbiol. 2012, 3, 46. [Google Scholar] [CrossRef] [Green Version]
- Durmus Tekir, S.; Cakir, T.; Ardic, E.; Sayilirbas, A.S.; Konuk, G.; Konuk, M.; Sariyer, H.; Ugurlu, A.; Karadeniz, I.; Ozgur, A.; et al. PHISTO: Pathogen-host interaction search tool. Bioinformatics 2013, 29, 1357–1358. [Google Scholar] [CrossRef] [Green Version]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Meher, P.K.; Sahu, T.K.; Banchariya, A.; Rao, A.R. DIRProt: A computational approach for discriminating insecticide resistant proteins from non-resistant proteins. BMC Bioinform. 2017, 18, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meher, P.K.; Sahu, T.K.; Mohanty, J.; Gahoi, S.; Purru, S.; Grover, M.; Rao, A.R. nifPred: Proteome-Wide Identification and Categorization of Nitrogen-Fixation Proteins of Diaztrophs Based on Composition-Transition-Distribution Features Using Support Vector Machine. Front. Microbiol. 2018, 9, 1100. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, P.; Yan, J.; Li, J.; Fong, S.; Siu, S.W.I. AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci. Rep. 2018, 8, 1697. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.S.; Liang, Y.Z.; Yan, J.; Tan, G.S.; Xu, Q.S.; Liu, S. PyDPI: Freely available python package for chemoinformatics, bioinformatics, and chemogenomics studies. J. Chem. Inf. Model. 2013, 53, 3086–3096. [Google Scholar] [CrossRef] [PubMed]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assenov, Y.; Ramirez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Grijalva, C.G.; Nuorti, J.P.; Griffin, M.R. Antibiotic prescription rates for acute respiratory tract infections in US ambulatory settings. JAMA 2009, 302, 758–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drijkoningen, J.J.; Rohde, G.G. Pneumococcal infection in adults: Burden of disease. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2014, 20 (Suppl. S5), 45–51. [Google Scholar] [CrossRef] [Green Version]
- Mathew, B.; Roy, D.D.; Kumar, T.V. The use of procalcitonin as a marker of sepsis in children. J. Clin. Diagn. Res. JCDR 2013, 7, 305–307. [Google Scholar] [CrossRef]
- Yusa, T.; Tateda, K.; Ohara, A.; Miyazaki, S. New possible biomarkers for diagnosis of infections and diagnostic distinction between bacterial and viral infections in children. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2017, 23, 96–100. [Google Scholar] [CrossRef] [Green Version]
- Zav’yalov, V.P.; Hamalainen-Laanaya, H.; Korpela, T.K.; Wahlroos, T. Interferon-Inducible Myxovirus Resistance Proteins: Potential Biomarkers for Differentiating Viral from Bacterial Infections. Clin. Chem. 2019, 65, 739–750. [Google Scholar] [CrossRef] [Green Version]
- Srugo, I.; Klein, A.; Stein, M.; Golan-Shany, O.; Kerem, N.; Chistyakov, I.; Genizi, J.; Glazer, O.; Yaniv, L.; German, A.; et al. Validation of a Novel Assay to Distinguish Bacterial and Viral Infections. Pediatrics 2017, 140. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Zhu, J.; Song, L.; Cai, W.; Wang, J. Combined use of biomarkers for distinguishing between bacterial and viral etiologies in pediatric lower respiratory tract infections. Infect. Dis. 2015, 47, 289–293. [Google Scholar] [CrossRef] [PubMed]
- ten Oever, J.; Tromp, M.; Bleeker-Rovers, C.P.; Joosten, L.A.; Netea, M.G.; Pickkers, P.; van de Veerdonk, F.L. Combination of biomarkers for the discrimination between bacterial and viral lower respiratory tract infections. J. Infect. 2012, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Suarez, N.M.; Bunsow, E.; Falsey, A.R.; Walsh, E.E.; Mejias, A.; Ramilo, O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J. Infect. Dis. 2015, 212, 213–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Yu, J.; Crosby, S.D.; Storch, G.A. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc. Natl. Acad. Sci. USA 2013, 110, 12792–12797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsalik, E.L.; Henao, R.; Nichols, M.; Burke, T.; Ko, E.R.; McClain, M.T.; Hudson, L.L.; Mazur, A.; Freeman, D.H.; Veldman, T.; et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 2016, 8, 322ra11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweeney, T.E.; Wong, H.R.; Khatri, P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci. Transl. Med. 2016, 8, 346ra91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).