Design of Experiment (DoE) for Optimization of HPLC Conditions for the Simultaneous Fractionation of Seven α-Amylase/Trypsin Inhibitors from Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Experimental Material
2.1.2. Standards and Chemicals
2.2. Extraction of Wheat α-Amylase/Trypsin Inhibitors
2.3. ATIs Fractionation and Purification
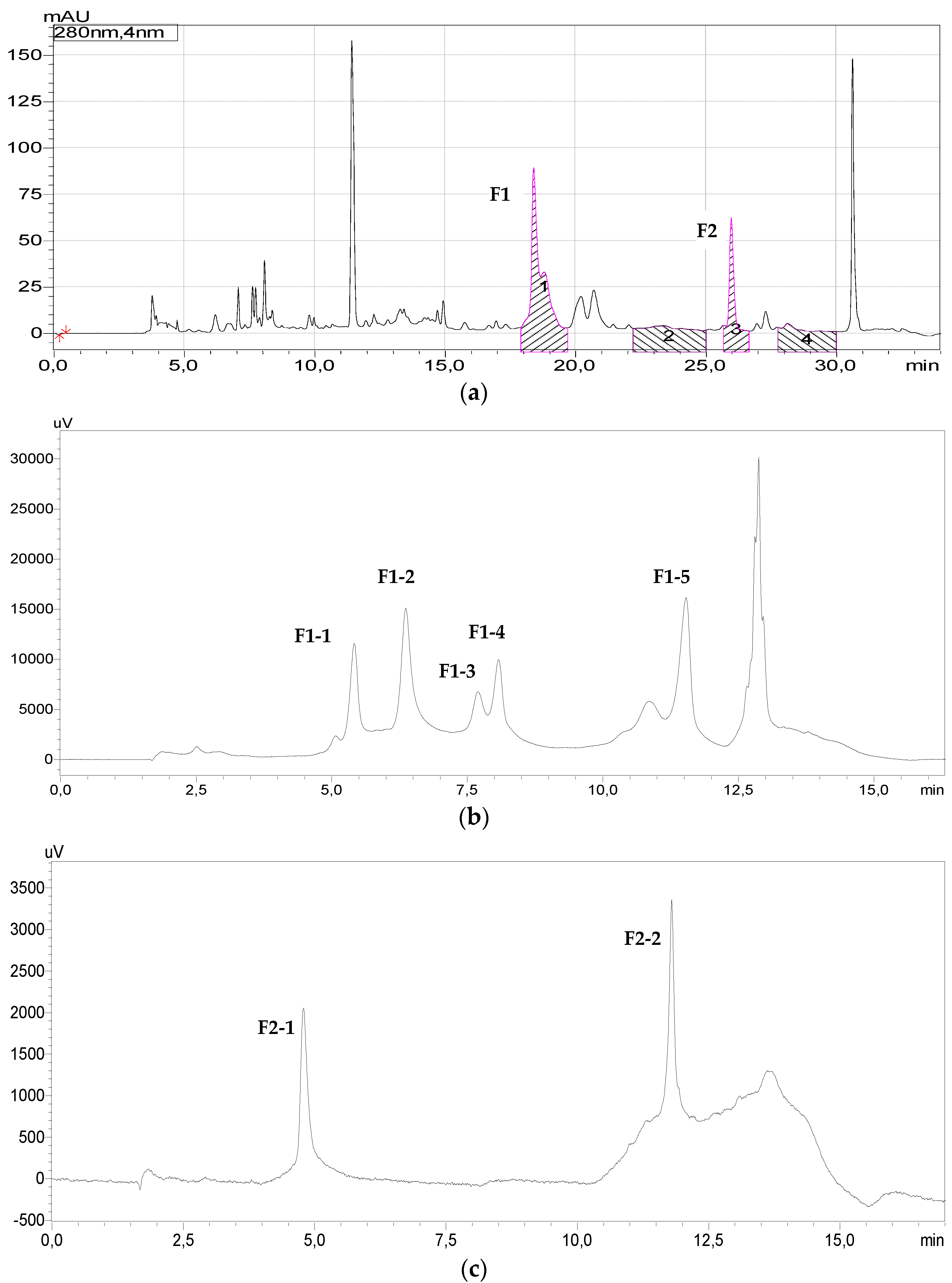
2.3.1. Reversed Phase Chromatography
2.3.2. Anion-Exchange Chromatography
2.4. Characterization of the Extracted and Purified Targeted Proteins
2.4.1. Protein Content
2.4.2. SDS PAGE
2.4.3. MALDI-TOF Mass Spectrometry Analysis
2.4.4. Targeted LC–MS/MS
2.5. Statistical Analysis
3. Results and Discussion
3.1. ATIs Extraction and Quantification
3.2. Reverse Phase Chromatography
3.3. Ion Exchange Chromatography
3.3.1. Ion Exchange Separation of Fraction of F2
3.3.2. Ion Exchange Chromatography of Fraction F1
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mundy, J.; Hejgaard, J.; Svendsen, I. Characterization of a bifunctional wheat inhibitor of endogenous α-amylase and subtilisin. FEBS Lett. 1984, 167, 210–214. [Google Scholar] [CrossRef] [Green Version]
- Altenbach, S.B.; Vensel, W.H.; Dupont, F.M. The spectrum of low molecular weight alpha-amylase/protease inhibitor genes expressed in the US bread wheat cultivar Butte 86. BMC Res. Notes 2011, 4, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junker, Y.; Zeissig, S.; Kim, S.J.; Barisani, D.; Wieser, H.; Leffler, D.A.; Zevallos, V.; Libermann, T.A.; Dillon, S.; Freitag, T.L.; et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J. Exp. Med. 2012, 209, 2395–2408. [Google Scholar] [CrossRef] [PubMed]
- Zevallos, V.F.; Raker, V.; Tenzer, S.; Jimenez-Calvente, C.; Ashfaq-Khan, M.; Russel, N.; Pickert, G.; Schild, H.; Steinbrink, K.; Schuppan, D. Nutritional Wheat Amylase-Trypsin Inhibitors Promote Intestinal Inflammation via Activation of Myeloid Cells. Gastroenterology 2017, 152, 1100–1113 e1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zevallos, V.F.; Raker, V.K.; Maxeiner, J.; Scholtes, P.; Steinbrink, K.; Schuppan, D. Dietary wheat amylase trypsin inhibitors exacerbate murine allergic airway inflammation. Eur. J. Nutr. 2019, 58, 1507–1514. [Google Scholar] [CrossRef]
- Tatham, A.S.; Shewry, P.R. Allergens to wheat and related cereals. Clin. Exp. Allergy 2008, 38, 1712–1726. [Google Scholar] [CrossRef]
- Oda, S.; Schofield, J.D. Characterisation of Friabilin Polypeptides. J. Cereal Sci. 1997, 26, 29–36. [Google Scholar] [CrossRef]
- Sagu, S.T.; Zimmermann, L.; Landgraber, E.; Homann, T.; Huschek, G.; Ozpinar, H.; Schweigert, F.J.; Rawel, H.M. Comprehensive Characterization and Relative Quantification of alpha-Amylase/Trypsin Inhibitors from Wheat Cultivars by Targeted HPLC-MS/MS. Foods 2020, 9, 1448. [Google Scholar] [CrossRef]
- Kick, F.; Belitz, H.D.; Wieser, H.; Kieffer, R. Comparative studies of the Osborne protein fraction of wheat varieties with different dough and baking properties. Z. Lebensm. Unters. Forsch. 1992, 195, 437–442. [Google Scholar] [CrossRef]
- Kumar, A.; Nayak, R.; Purohit, S.R.; Rao, P.S. Impact of UV-C irradiation on solubility of Osborne protein fractions in wheat flour. Food Hydrocoll. 2021, 110, 105845. [Google Scholar] [CrossRef]
- Weegels, P.L.; Hamer, R.J.; Schofield, J.D. RP–HPLC and Capillary Electrophoresis of Subunits from Glutenin Isolated by SDS and Osborne Fractionation. J. Cereal Sci. 1995, 22, 211–224. [Google Scholar] [CrossRef]
- Zoccatelli, G.; Sega, M.; Bolla, M.; Cecconi, D.; Vaccino, P.; Rizzi, C.; Chignola, R.; Brandolini, A. Expression of alpha-amylase inhibitors in diploid Triticum species. Food Chem. 2012, 135, 2643–2649. [Google Scholar] [CrossRef] [PubMed]
- Sotkovsky, P.; Sklenar, J.; Halada, P.; Cinova, J.; Setinova, I.; Kainarova, A.; Golias, J.; Pavlaskova, K.; Honzova, S.; Tuckova, L. A new approach to the isolation and characterization of wheat flour allergens. Clin. Exp. Allergy 2011, 41, 1031–1043. [Google Scholar] [CrossRef]
- Carbonero, P.; García-Olmedo, F. A Multigene Family of Trypsin/α-Amylase Inhibitors from Cereals. In Seed Proteins; Springer: Dordrecht, The Netherlands, 1999; pp. 617–633. [Google Scholar] [CrossRef] [Green Version]
- Prandi, B.; Faccini, A.; Tedeschi, T.; Galaverna, G.; Sforza, S. LC/MS analysis of proteolytic peptides in wheat extracts for determining the content of the allergen amylase/trypsin inhibitor CM3: Influence of growing area and variety. Food Chem. 2013, 140, 141–146. [Google Scholar] [CrossRef]
- Rogniaux, H.; Pavlovic, M.; Lupi, R.; Lollier, V.; Joint, M.; Mameri, H.; Denery, S.; Larre, C. Allergen relative abundance in several wheat varieties as revealed via a targeted quantitative approach using MS. Proteomics 2015, 15, 1736–1745. [Google Scholar] [CrossRef]
- Geisslitz, S.; Ludwig, C.; Scherf, K.A.; Koehler, P. Targeted LC-MS/MS Reveals Similar Contents of alpha-Amylase/Trypsin-Inhibitors as Putative Triggers of Nonceliac Gluten Sensitivity in All Wheat Species except Einkorn. J. Agric. Food Chem. 2018, 66, 12395–12403. [Google Scholar] [CrossRef]
- Labrou, N.E. Protein purification: An overview. Methods Mol. Biol. 2014, 1129, 3–10. [Google Scholar] [CrossRef]
- Kallberg, K.; Johansson, H.O.; Bulow, L. Multimodal chromatography: An efficient tool in downstream processing of proteins. Biotechnol. J. 2012, 7, 1485–1495. [Google Scholar] [CrossRef]
- Wilken, L.R.; Nikolov, Z.L. Recovery and purification of plant-made recombinant proteins. Biotechnol. Adv. 2012, 30, 419–433. [Google Scholar] [CrossRef]
- Zuvela, P.; Skoczylas, M.; Jay Liu, J.; Ba Czek, T.; Kaliszan, R.; Wong, M.W.; Buszewski, B.; Heberger, K. Column Characterization and Selection Systems in Reversed-Phase High-Performance Liquid Chromatography. Chem. Rev. 2019, 119, 3674–3729. [Google Scholar] [CrossRef]
- Khatwani, S.L.; Pavlova, A.; Pirot, Z. Anion-exchange HPLC assay for separation and quantification of empty and full capsids in multiple adeno-associated virus serotypes. Mol. Ther. Methods Clin. Dev. 2021, 21, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Josic, D.; Kovac, S. Reversed-phase High Performance Liquid Chromatography of proteins. Curr. Protoc. Protein Sci. 2010, 61, 8.7.1–8.7.22. [Google Scholar] [CrossRef] [PubMed]
- Ostertag, F.; Schmidt, C.M.; Berensmeier, S.; Hinrichs, J. Development and validation of an RP-HPLC DAD method for the simultaneous quantification of minor and major whey proteins. Food Chem. 2021, 342, 128176. [Google Scholar] [CrossRef] [PubMed]
- Attimarad, M.; Venugopala, K.N.; Nair, A.B.; Sreeharsha, N.; Deb, P.K. Experimental Design Approach for Quantitative Expressions of Simultaneous Quantification of Two Binary Formulations Containing Remogliflozin and Gliptins by RP-HPLC. Separations 2022, 9, 23. [Google Scholar] [CrossRef]
- Snyder, L.R. Role of the solvent in liquid-solid chromatography. Review. Anal. Chem. 2002, 46, 1384–1393. [Google Scholar] [CrossRef]
- Jandera, P.; Janas, P. Recent advances in stationary phases and understanding of retention in hydrophilic interaction chromatography. A review. Anal. Chim. Acta 2017, 967, 12–32. [Google Scholar] [CrossRef]
- Dousa, M.; Jires, J. HILIC-MS determination of dimethylamine in the active pharmaceutical ingredients and in the dosage forms of metformin. J. Pharm. Biomed. Anal. 2020, 191, 113573. [Google Scholar] [CrossRef]
- Wu, D.; Lucy, C.A. Study of the slope of the linear relationship between retention and mobile phase composition (Snyder-Soczewinski model) in normal phase liquid chromatography with bonded and charge-transfer phases. J. Chromatogr. A 2016, 1475, 31–40. [Google Scholar] [CrossRef]
- Pesek, J.J.; Matyska, M.T.; Boysen, R.I.; Yang, Y.; Hearn, M.T.W. Aqueous normal-phase chromatography using silica-hydride-based stationary phases. TrAC Trends Anal. Chem. 2013, 42, 64–73. [Google Scholar] [CrossRef]
- Gottschalk, U. 3.57—Overview of Downstream Processing in the Biomanufacturing Industry. Compr. Biotechnol. 2011, 3, 698–711. [Google Scholar] [CrossRef]
- Kumar, V.; Leweke, S.; Heymann, W.; von Lieres, E.; Schlegel, F.; Westerberg, K.; Lenhoff, A.M. Robust mechanistic modeling of protein ion-exchange chromatography. J. Chromatogr. A 2021, 1660, 462669. [Google Scholar] [CrossRef] [PubMed]
- Trappe, A.; Fussl, F.; Carillo, S.; Zaborowska, I.; Meleady, P.; Bones, J. Rapid charge variant analysis of monoclonal antibodies to support lead candidate biopharmaceutical development. J. Chromatogr. B 2018, 1095, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Murisier, A.; Duivelshof, B.L.; Fekete, S.; Bourquin, J.; Schmudlach, A.; Lauber, M.A.; Nguyen, J.M.; Beck, A.; Guillarme, D.; D’Atri, V. Towards a simple on-line coupling of ion exchange chromatography and native mass spectrometry for the detailed characterization of monoclonal antibodies. J. Chromatogr. A 2021, 1655, 462499. [Google Scholar] [CrossRef] [PubMed]
- Farsang, E.; Murisier, A.; Horvath, K.; Beck, A.; Kormany, R.; Guillarme, D.; Fekete, S. Tuning selectivity in cation-exchange chromatography applied for monoclonal antibody separations, part 1: Alternative mobile phases and fine tuning of the separation. J. Pharm. Biomed. Anal. 2019, 168, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Nesterenko, P.N.; Nesterenko, E.P. Hydrophobicity of polymer based anion-exchange columns for ion chromatography. Heliyon 2021, 7, e07290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, X. Mixed-mode chromatography in pharmaceutical and biopharmaceutical applications. J. Pharm. Biomed. Anal. 2016, 128, 73–88. [Google Scholar] [CrossRef] [Green Version]
- Maria, S.; Joucla, G.; Garbay, B.; Dieryck, W.; Lomenech, A.M.; Santarelli, X.; Cabanne, C. Purification process of recombinant monoclonal antibodies with mixed mode chromatography. J. Chromatogr. A 2015, 1393, 57–64. [Google Scholar] [CrossRef]
- Kazarian, A.A.; Taylor, M.R.; Haddad, P.R.; Nesterenko, P.N.; Paull, B. Single column comprehensive analysis of pharmaceutical preparations using dual-injection mixed-mode (ion-exchange and reversed-phase) and hydrophilic interaction liquid chromatography. J. Pharm. Biomed. Anal. 2013, 86, 174–181. [Google Scholar] [CrossRef]
- Santarelli, X.; Cabanne, C. Mixed Mode Chromatography: A Novel Way Toward New Selectivity. Curr. Protein Pept. Sci. 2019, 20, 14–21. [Google Scholar] [CrossRef]
- Kreusser, J.; Jirasek, F.; Hasse, H. Influence of pH value and salts on the adsorption of lysozyme in mixed-mode chromatography. Eng. Life Sci. 2021, 21, 753–768. [Google Scholar] [CrossRef]
- Sagu, S.T.; Nso, E.J.; Karmakar, S.; De, S. Optimisation of low temperature extraction of banana juice using commercial pectinase. Food Chem. 2014, 151, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Sagu, S.T.; Landgraber, E.; Henkel, I.M.; Huschek, G.; Homann, T.; Bussler, S.; Schluter, O.K.; Rawel, H. Effect of Cereal alpha-Amylase/Trypsin Inhibitors on Developmental Characteristics and Abundance of Digestive Enzymes of Mealworm Larvae (Tenebrio molitor L.). Insects 2021, 12, 454. [Google Scholar] [CrossRef] [PubMed]
- Sagu, S.T.; Landgraber, E.; Rackiewicz, M.; Huschek, G.; Rawel, H. Relative Abundance of Alpha-Amylase/Trypsin Inhibitors in Selected Sorghum Cultivars. Molecules 2020, 25, 5982. [Google Scholar] [CrossRef]
- Sagu, S.T.; Huschek, G.; Bonick, J.; Homann, T.; Rawel, H.M. A New Approach of Extraction of alpha-Amylase/trypsin Inhibitors from Wheat (Triticum aestivum L.), Based on Optimization Using Plackett-Burman and Box-Behnken Designs. Molecules 2019, 24, 3589. [Google Scholar] [CrossRef] [Green Version]
- Lowry, O.; Rosebrough, N.; Farr, A.L.; Randall, R. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Sagu, S.T.; Huschek, G.; Homann, T.; Rawel, H.M. Effect of Sample Preparation on the Detection and Quantification of Selected Nuts Allergenic Proteins by LC-MS/MS. Molecules 2021, 26, 4698. [Google Scholar] [CrossRef]
- Baranyi, J.; Pin, C.; Ross, T. Validating and comparing predictive models. Int. J. Food Microbiol. 1999, 48, 159–166. [Google Scholar] [CrossRef]
- Ross, T. Indices for performance evaluation of predictive models in food microbiology. J. Appl. Bacteriol. 1996, 81, 501–508. [Google Scholar] [CrossRef]
- Manadas, B.; Mendes, V.M.; English, J.; Dunn, M.J. Peptide fractionation in proteomics approaches. Expert Rev. Proteom. 2014, 7, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]


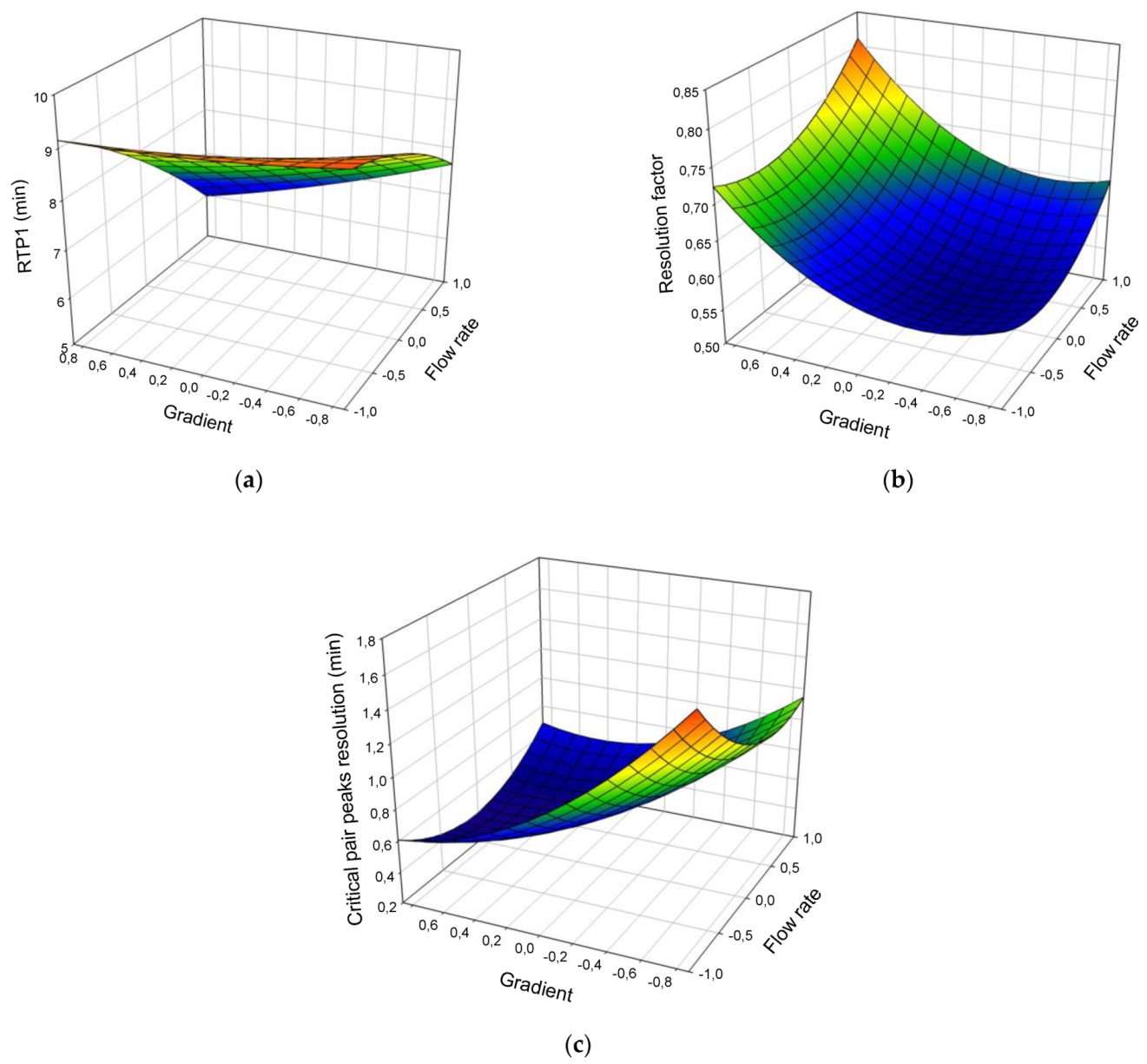
| Runs | Coded Values | Real Values | Experimental Responses | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Flow Rate (mL/min) | Gradient (%B/min) | Temp. (°C) | RTP1 (min) | ∆tCR (min) | RF - | ||||
| x1 | x2 | x3 | X1 | X2 | X3 | ||||
| 1 | 0 | 0 | 0 | 0.8 | 0.30 | 35 | 6.00 ± 0.12 | 0.55 ± 0.02 | 0.58 ± 0.01 |
| 2 | 1 | 0 | 0 | 1.2 | 0.30 | 35 | 5.08 ± 0.52 | 0.66 ± 0.01 | 0.67 ± 0.01 |
| 3 | −1 | 0 | 0 | 0.4 | 0.30 | 35 | 6.90 ± 0.42 | 0.96 ± 0.04 | 0.59 ± 0.03 |
| 4 | 0.5 | 0.866 | 0 | 1.0 | 0.50 | 35 | 5.03 ± 0.48 | 0.50 ± 0.01 | 0.73 ± 0.01 |
| 5 | −0.5 | −0.866 | 0 | 0.6 | 0.10 | 35 | 7.13 ± 0.14 | 1.35 ± 0.06 | 0.60 ± 0.00 |
| 6 | 0.5 | −0.866 | 0 | 1.0 | 0.10 | 35 | 6.32 ± 0.17 | 1.17 ± 0.05 | 0.59 ± 0.01 |
| 7 | −0.5 | 0.866 | 0 | 0.6 | 0.50 | 35 | 6.32 ± 0.09 | 0.37 ± 0.00 | 0.72 ± 0.01 |
| 8 | 0.5 | 0.289 | 0.816 | 1.0 | 0.37 | 40 | 5.66 ± 0.30 | 0.44 ± 0.01 | 0.72 ± 0.04 |
| 9 | −0.5 | −0.289 | −0.816 | 0.6 | 0.23 | 30 | 6.77 ± 0.26 | 0.94 ± 0.07 | 0.60 ± 0.01 |
| 10 | 0.5 | −0.289 | −0.816 | 1.0 | 0.23 | 30 | 5.78 ± 0.25 | 0.84 ± 0.04 | 0.59 ± 0.01 |
| 11 | 0 | 0.577 | −0.816 | 0.8 | 0.43 | 30 | 6.86 ± 0.41 | 0.55 ± 0.01 | 0.68 ± 0.02 |
| 12 | −0.5 | 0.289 | 0.816 | 0.6 | 0.37 | 40 | 6.49 ± 0.33 | 0.65 ± 0.02 | 0.60 ± 0.01 |
| 13 | 0 | −0.577 | 0.816 | 0.8 | 0.17 | 40 | 6.62 ± 0.45 | 0.84 ± 0.01 | 0.59 ± 0.01 |
| 14 | 0 | 0 | 0 | 0.8 | 0.30 | 35 | 6.11 ± 0.08 | 0.57 ± 0.01 | 0.59 ± 0.00 |
| 15 | 0 | 0 | 0 | 0.8 | 0.30 | 35 | 6.38 ± 0.41 | 0.55 ± 0.03 | 0.57 ± 0.01 |
| 16 | 0 | 0 | 0 | 0.8 | 0.30 | 35 | 6.15 ± 0.35 | 0.57 ± 0.01 | 0.57 ± 0.01 |
| 17 | 0 | 0 | 0 | 0.8 | 0.30 | 35 | 6.23 ± 0.09 | 0.58 ± 0.01 | 0.58 ± 0.01 |
| Proportion (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RP-HPLC | SAX-HPLC | |||||||||
| UniProt Access Nr. | Extract | F1-C18 | F2-C18 | F1-1 | F1-2 | F1-3 | F1-4 | F1-5 | F2-1 | F2-3 |
| P01083 | 23.7 ± 0.3 | 33.0 ± 1.7 | 3.0 ± 0.7 | 0.0 ± 0.0 | 0.9 ± 0.1 | 0.0 ± 0.0 | 0.4 ± 0.1 | 98.2 ± 0.7 | 0.8 ± 0.0 | 0.4 ± 0.0 |
| P17314 | 18.3 ± 0.5 | 0.7 ± 0.0 | 74.8 ± 1.3 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.4 ± 0.1 | 98.1 ± 0.8 |
| P16850 | 6.5 ± 0.0 | 9.5 ± 0.1 | 0.0 ± 0.4 | 2.6 ± 0.2 | 0.3 ± 0.0 | 0.5 ± 0.0 | 97.9 ± 0.5 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.0 |
| P01085 | 8.4 ± 0.1 | 12.3 ± 0.5 | 0.4 ± 0.0 | 95.1 ± 0.8 | 0.3 ± 0.0 | 0.1 ± 0.0 | 0.4 ± 0.1 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| P01084 | 1.3 ± 0.0 | 3.5 ± 0.4 | 0.0 ± 0.0 | 0.8 ± 0.0 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.2 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| P16851 | 13.4 ± 0.1 | 20.0 ± 0.1 | 0.2 ± 0.0 | 0.5 ± 0.0 | 0.6 ± 0.0 | 98.3 ± 0.4 | 0.4 ± 0.1 | 0.2 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 |
| P16159 | 12.1 ± 0.5 | 16.0 ± 0.3 | 1.7 ± 0.1 | 0.0 ± 0.0 | 96.9 ± 0.5 | 0.3 ± 0.0 | 0.1 ± 0.0 | 0.8 ± 0.1 | 0.6 ± 0.1 | 0.1 ± 0.0 |
| P81496 | 1.8 ± 0.0 | 0.1 ± 0.0 | 0.0 ± 0.0 | 1.0 ± 0.0 | 0.2 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.2 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Q43723 | 3.6 ± 0.0 | 3.8 ± 0.3 | 2.3 ± 0.1 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.9 ± 0.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.4 ± 0.0 | 0.3 ± 0.0 |
| P93602 | 1.8 ± 0.1 | 0.5 ± 0.0 | 1.1 ± 0.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.0 | 0.4 ± 0.0 | 0.1 ± 0.0 |
| P83207 | 6.7 ± 0.1 | 0.1 ± 0.0 | 15.9 ± 0.9 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 96.2 ± 0.4 | 0.8 ± 0.1 |
| Q4U199 | 1.2 ± 0.2 | 0.4 ± 0.0 | 0.5 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.0 ± 0.0 |
| Q41540 | 1.1 ± 0.1 | 0.2 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
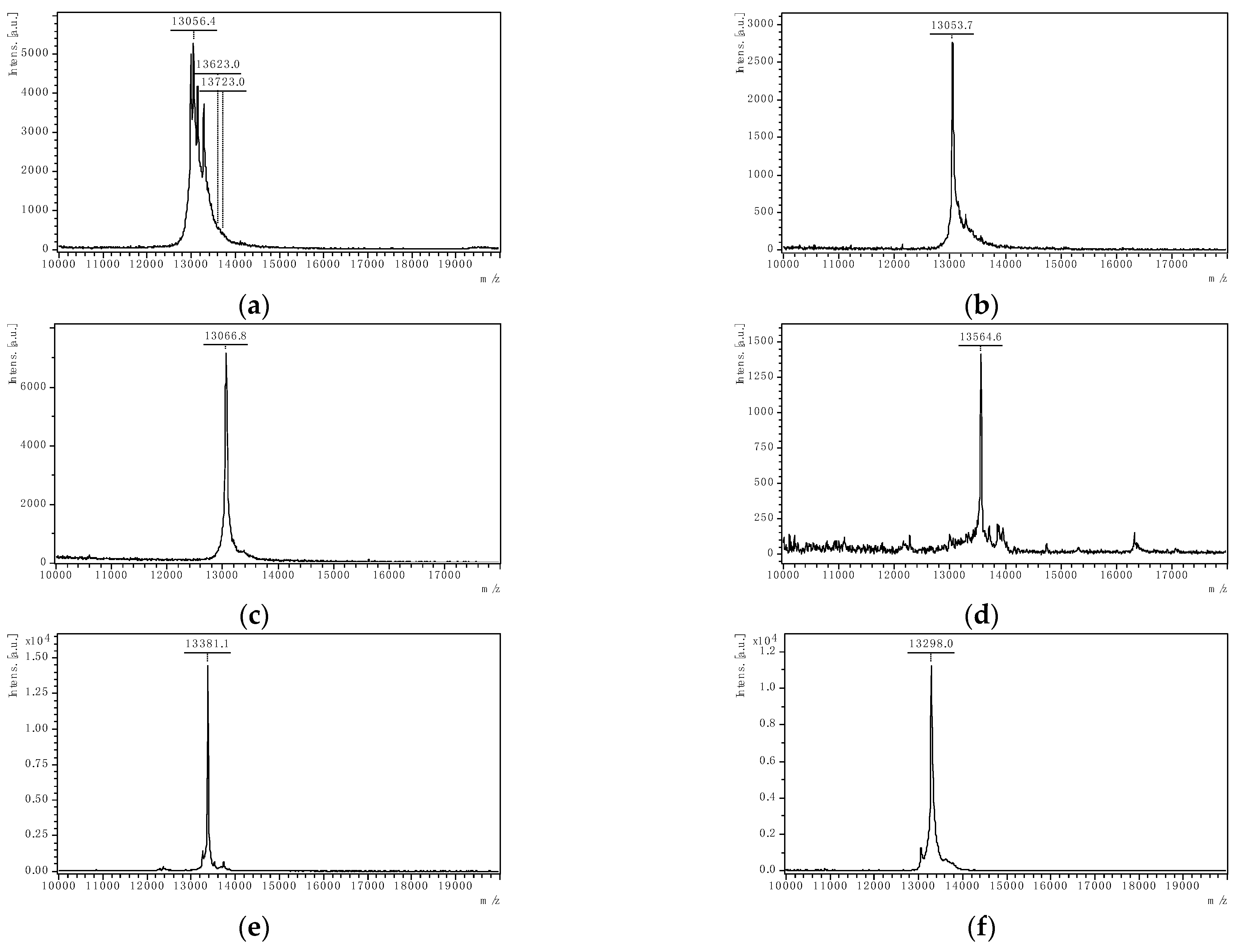
| Uni-Prot Access Nr. | Signal Peptids | Chain (Amino Acids) | Reference Mass (Da) | Determined Mass (m/z) | Error (%) |
|---|---|---|---|---|---|
| P01083 | 1-30 | 31-153 | 13,326 | 13,381 | 0.4 |
| P17314 | 1-25 | 26-168 | 15,832 | 15,524 | 1.9 |
| P16850 | 1-24 | 25-145 | 13,085 | 13,067 | 0.1 |
| P01085 | - | 1-124 | 13,328 | 13,298 | 0.2 |
| P16851 | 1-25 | 26-145 | 13,034 | 13,054 | 0.2 |
| P16159 | 1-24 | 25-143 | 13,437 | 13,565 | 0.9 |
| P83207 | - | 1-119 | 12,943 | 12,935 | 0.1 |
| Coefficients | δ0 | α1 | α2 | α 3 | β1 | β2 | β3 | γ12 | γ13 | γ23 | R2 | BF | AF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RTP1 | 8.23 | −1.26 *** | −0.59 ** | −0.18 | −0.25 | 0.13 | 0.41 | −0.38 | 0.26 | −0.94 | 0.93 | 1.00 | 1.02 |
| ∆tCR | 0.56 | −0.12 ** | −0.45 *** | −0.08 * | 0.25 ** | 0.29 ** | 0.08 | 0.18 * | −0.13 | 0.05 | 0.98 | 1.00 | 1.05 |
| RF | 0.58 | 0.03 ** | 0.07 *** | 0.01 | 0.05 ** | 0.09 ** | 0.04 * | 0.01 | 0.08 * | 0.00 | 0.96 | 1.00 | 1.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sagu, S.T.; Huschek, G.; Braga, T.W.; Rackiewicz, M.; Homann, T.; Rawel, H.M. Design of Experiment (DoE) for Optimization of HPLC Conditions for the Simultaneous Fractionation of Seven α-Amylase/Trypsin Inhibitors from Wheat (Triticum aestivum L.). Processes 2022, 10, 259. https://doi.org/10.3390/pr10020259
Sagu ST, Huschek G, Braga TW, Rackiewicz M, Homann T, Rawel HM. Design of Experiment (DoE) for Optimization of HPLC Conditions for the Simultaneous Fractionation of Seven α-Amylase/Trypsin Inhibitors from Wheat (Triticum aestivum L.). Processes. 2022; 10(2):259. https://doi.org/10.3390/pr10020259
Chicago/Turabian StyleSagu, Sorel Tchewonpi, Gerd Huschek, Tess Waldbach Braga, Michal Rackiewicz, Thomas Homann, and Harshadrai M. Rawel. 2022. "Design of Experiment (DoE) for Optimization of HPLC Conditions for the Simultaneous Fractionation of Seven α-Amylase/Trypsin Inhibitors from Wheat (Triticum aestivum L.)" Processes 10, no. 2: 259. https://doi.org/10.3390/pr10020259
APA StyleSagu, S. T., Huschek, G., Braga, T. W., Rackiewicz, M., Homann, T., & Rawel, H. M. (2022). Design of Experiment (DoE) for Optimization of HPLC Conditions for the Simultaneous Fractionation of Seven α-Amylase/Trypsin Inhibitors from Wheat (Triticum aestivum L.). Processes, 10(2), 259. https://doi.org/10.3390/pr10020259









