A New Theobromine-Based EGFRWT and EGFRT790M Inhibitor and Apoptosis Inducer: Design, Semi-Synthesis, Docking, DFT, MD Simulations, and In Vitro Studies
Abstract
:1. Introduction
Rationale
2. Results and Discussion
2.1. Molecular Docking
2.1.1. Molecular Docking Studies against the Wild EGFR Protein (EGFRWT)
2.1.2. Molecular Docking Studies against Mutant EGFR Protein (EGFRT790M)
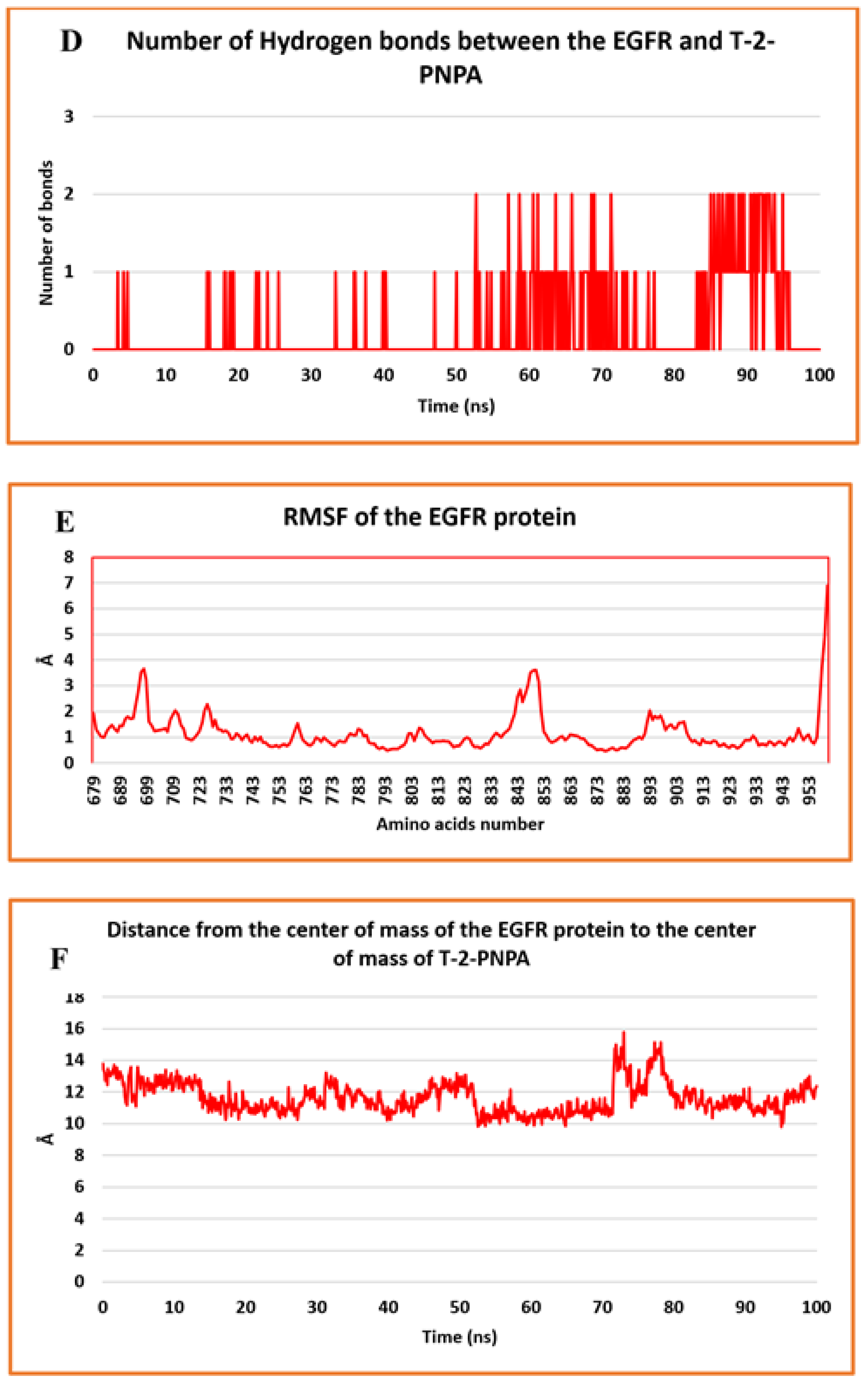
2.2. MD Simulations
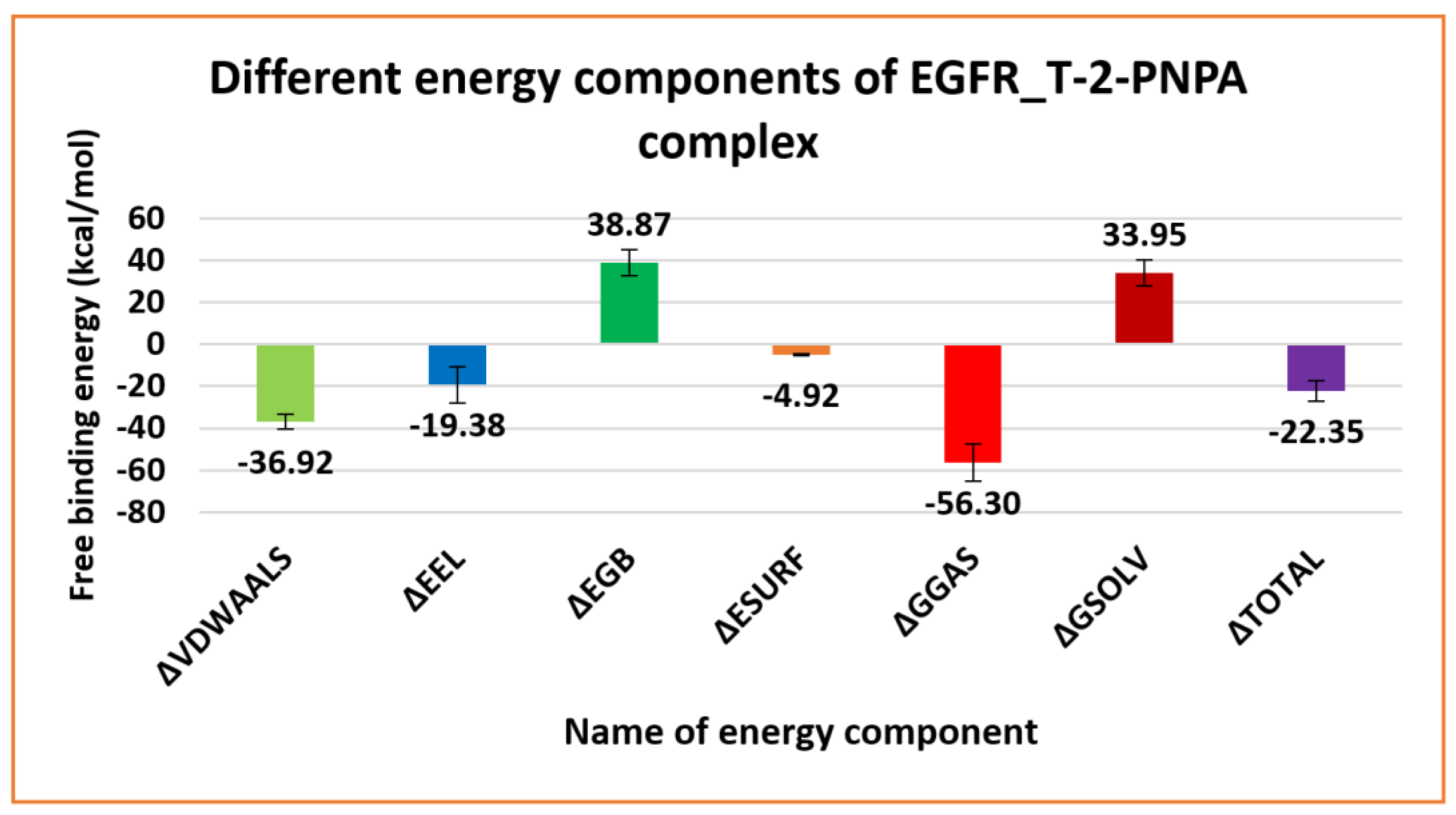
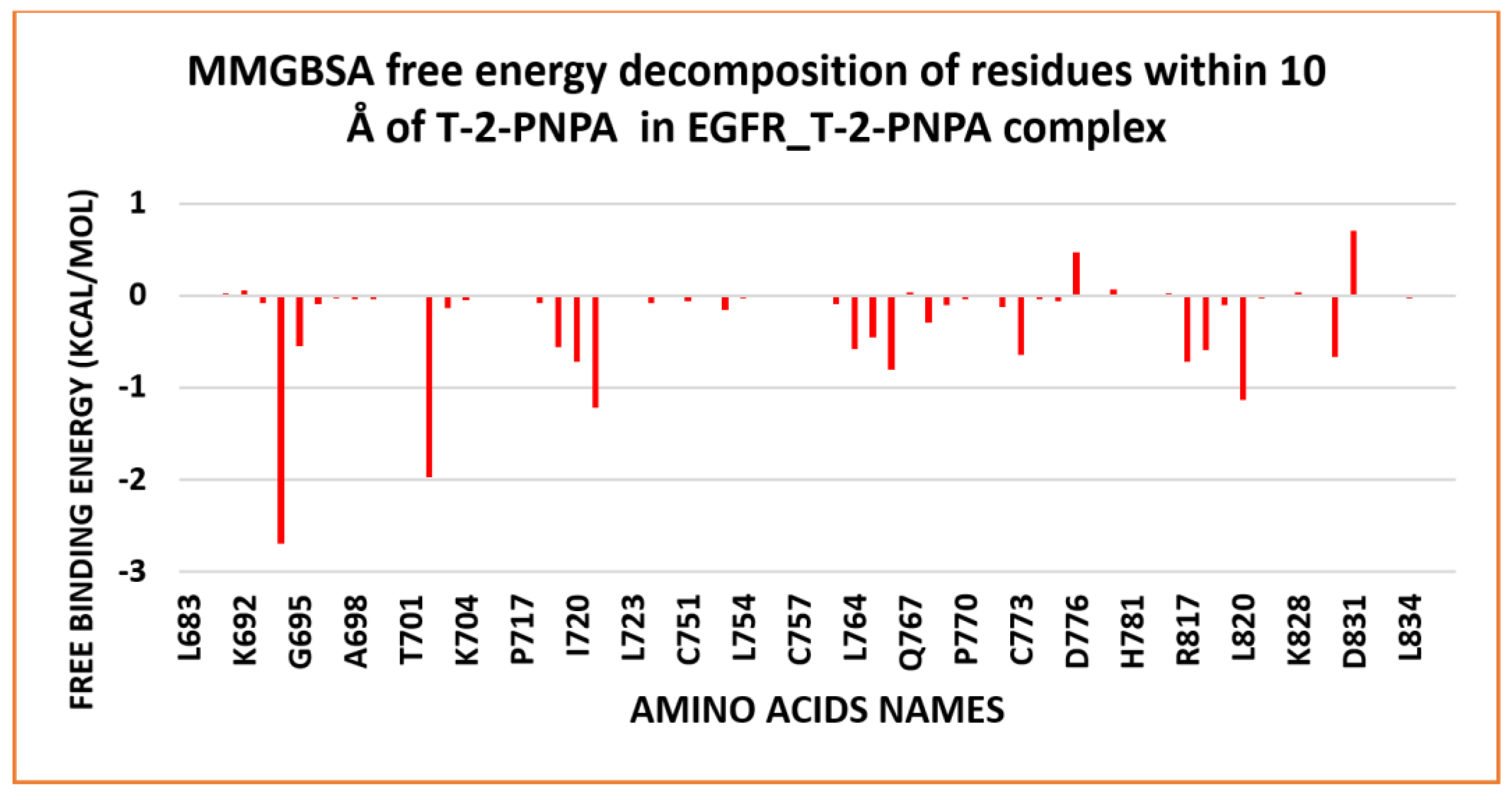
2.3. MM-GBSA
2.4. PLIP Study
2.5. DFT Studies
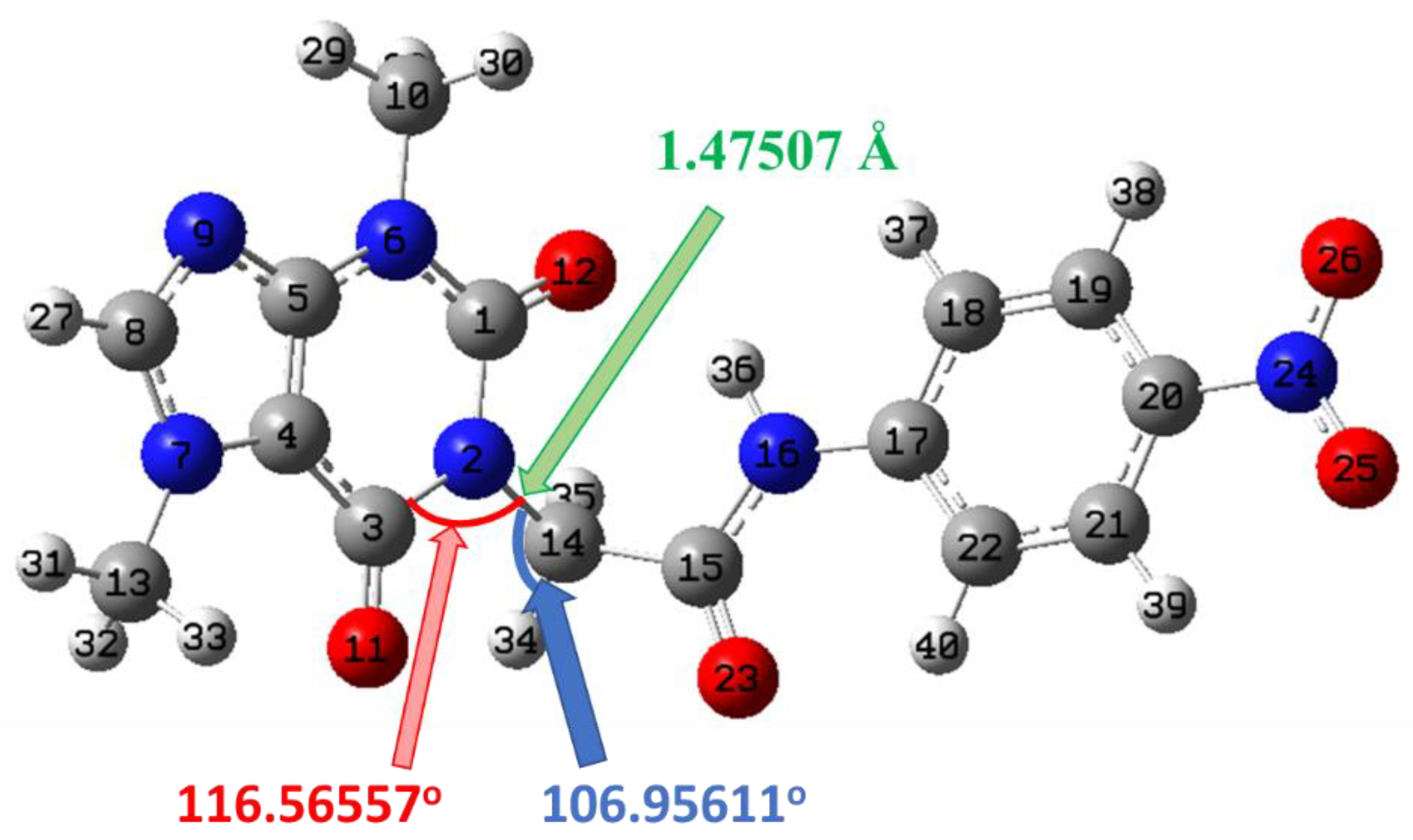
2.5.1. Geometry Optimization
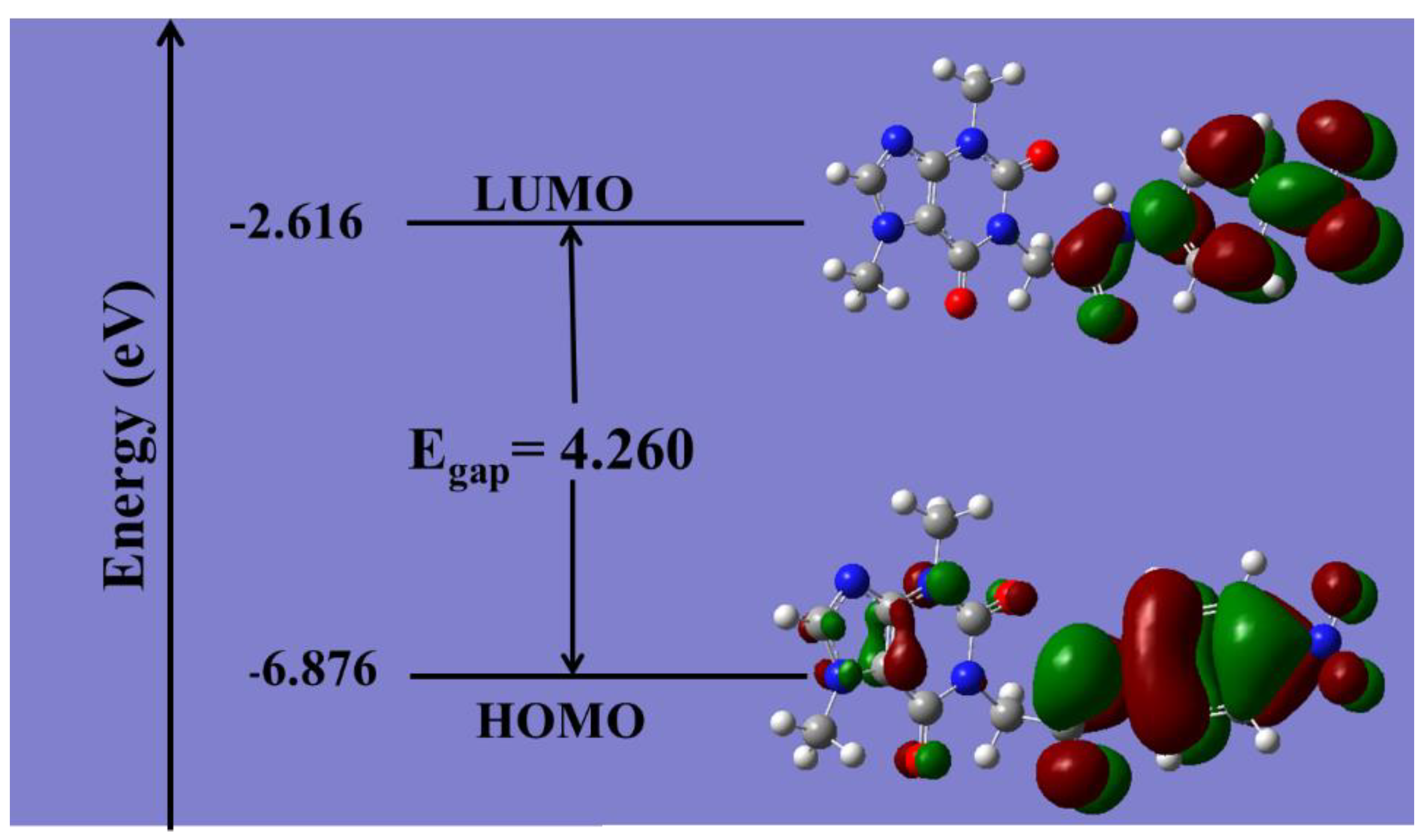
2.5.2. Frontier Molecular Orbital Analysis (FMO) Analysis
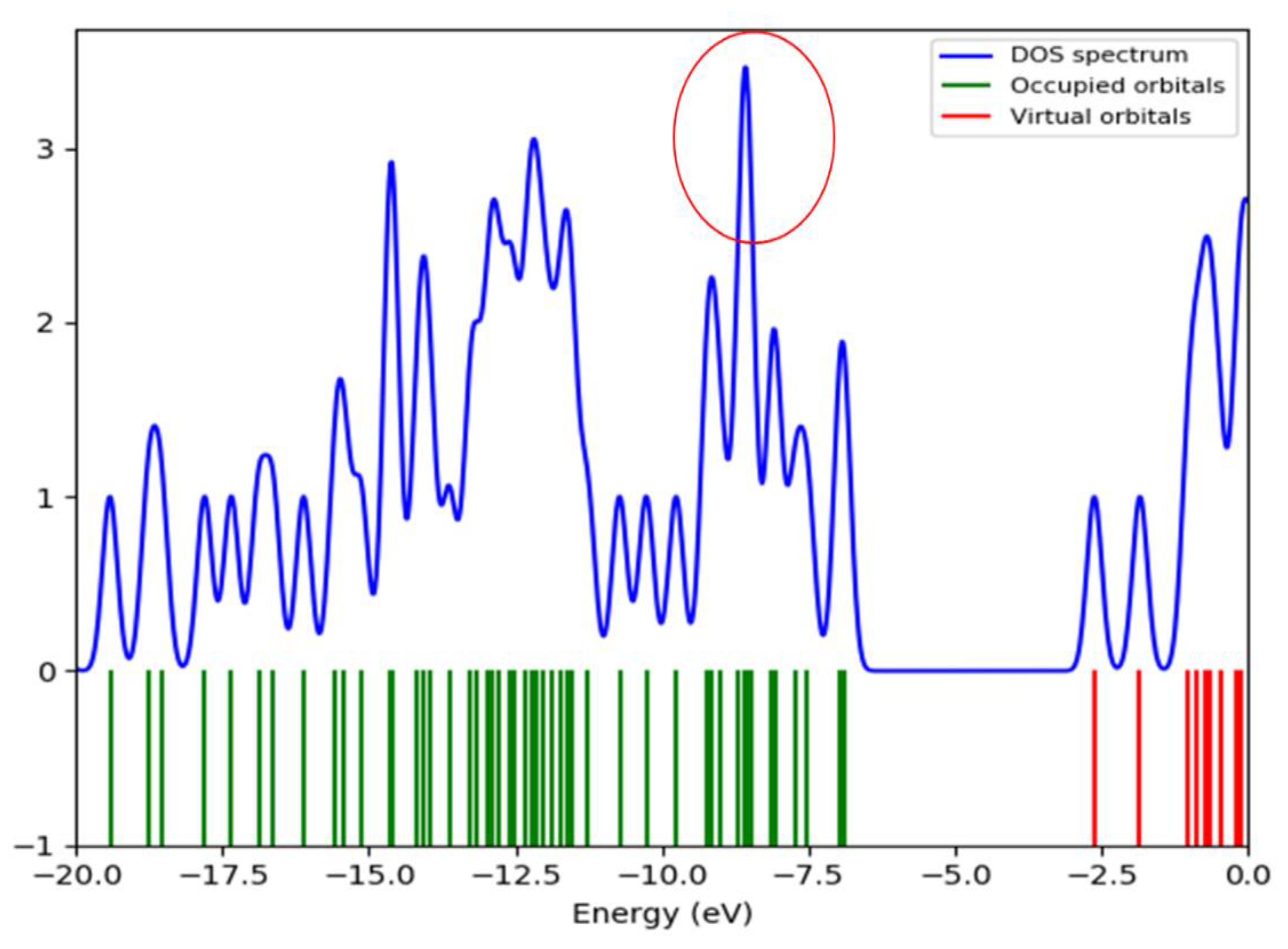
2.5.3. Global Reactive Indices and Total Density of State (TDOS)
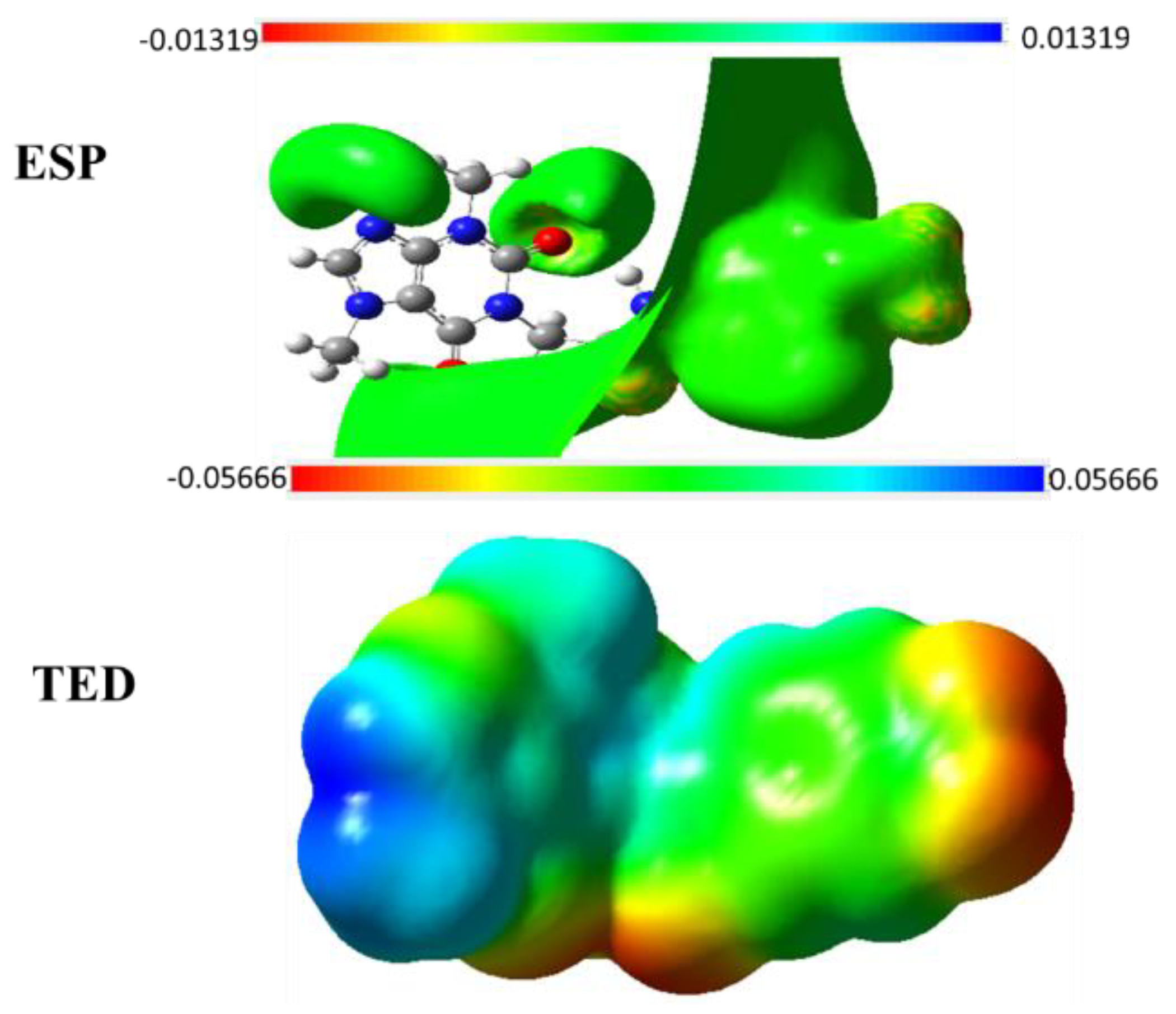
2.5.4. TED and ESP Surface Potential Maps
2.6. ADMET Profiling Study
2.7. In Silico Toxicity Studies
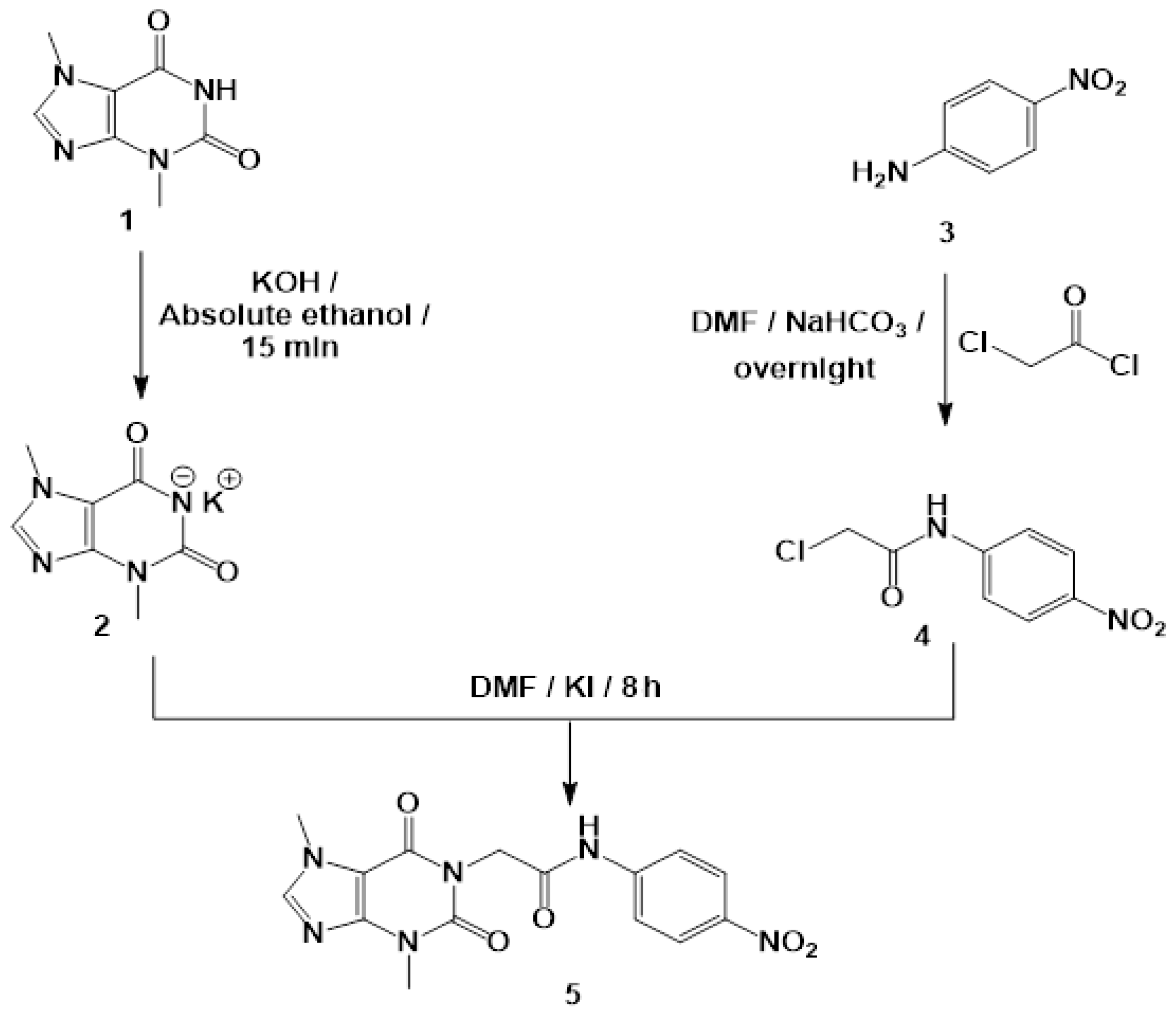
2.8. Chemistry
2.9. Biological Evaluation
2.9.1. In Vitro EGFR Inhibition
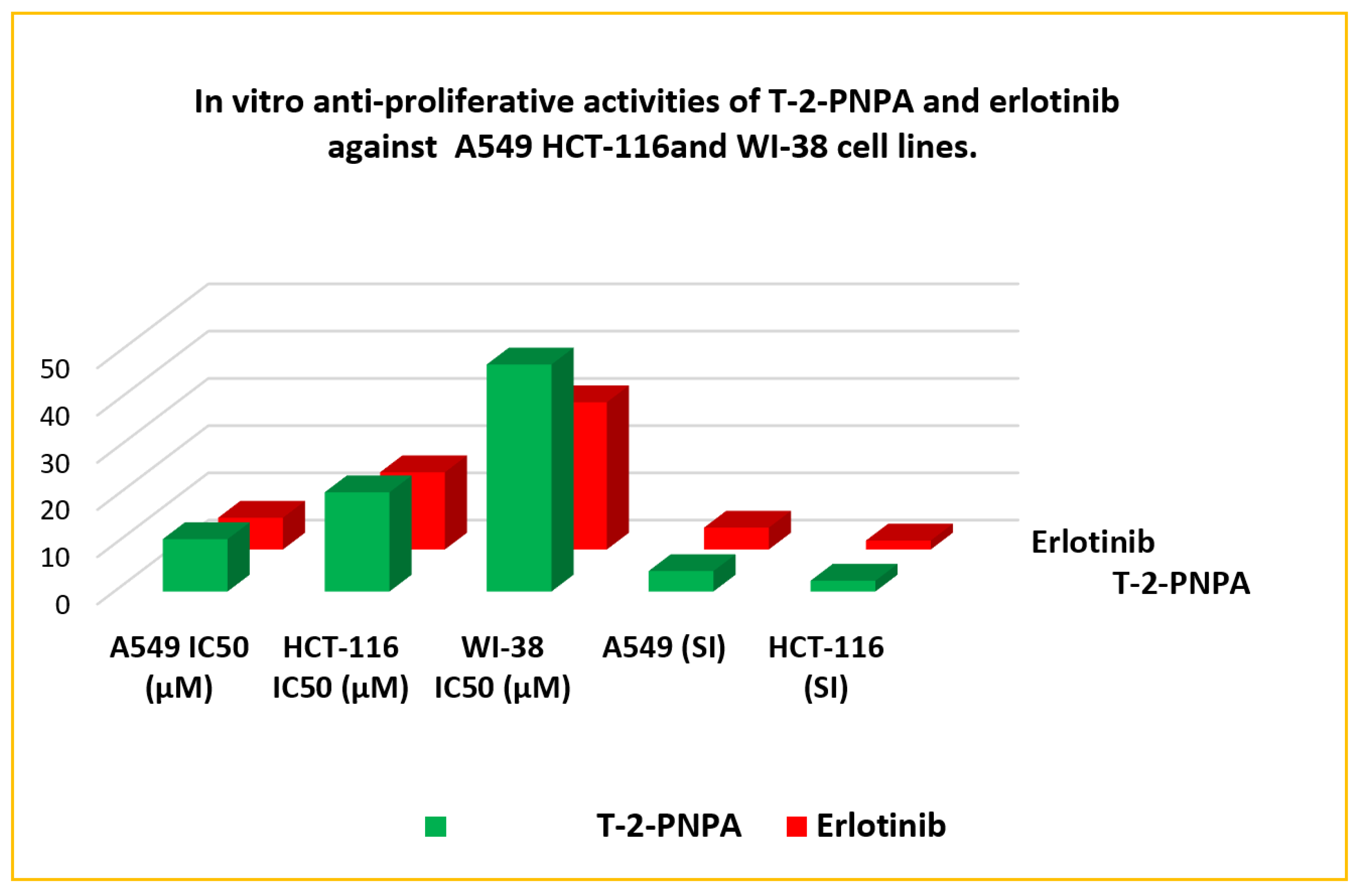
2.9.2. Cytotoxicity
3. Experimental Section
3.1. In Silico Studies
3.1.1. Docking Studies
3.1.2. MD Simulations
3.1.3. MM-GBSA
3.1.4. DFT
3.1.5. ADMET Studies
3.1.6. Toxicity Studies
3.2. Synthesis of T-2-PNPA
3.3. Biological Studies
3.3.1. In Vitro EGFR Inhibition
3.3.2. In Vitro Antiproliferative Activity
3.3.3. Safety Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- WHO Cancer, Fact Sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 13 June 2022).
- Chaudhari, P.; Bari, S.; Surana, S.; Shirkhedkar, A.; Wakode, S.; Shelar, S.; Racharla, S.; Ugale, V.; Ghodke, M. Logical synthetic strategies and structure-activity relationship of indolin-2-one hybrids as small molecule anticancer agents: An Overview. J. Mol. Struct. 2021, 1247, 131280. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Dash, Y.; Elzayat, E.; Abdou, A.M.; Hassan, R.A. Novel thienopyrimidine-aminothiazole hybrids: Design, synthesis, antimicrobial screening, anticancer activity, effects on cell cycle profile, caspase-3 mediated apoptosis and VEGFR-2 inhibition. Bioorganic Chem. 2021, 114, 105137. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, R.I.; Gee, J.M.W.; Harper, M.E. EGFR and cancer prognosis. Eur. J. Cancer 2001, 37, 9–15. [Google Scholar] [CrossRef]
- Hashmi, A.A.; Naz, S.; Hashmi, S.K.; Irfan, M.; Hussain, Z.F.; Khan, E.Y.; Asif, H.; Faridi, N. Epidermal growth factor receptor (EGFR) overexpression in triple-negative breast cancer: Association with clinicopathologic features and prognostic parameters. Surg. Exp. Pathol. 2019, 2, 6. [Google Scholar] [CrossRef]
- Spano, J.-P.; Lagorce, C.; Atlan, D.; Milano, G.; Domont, J.; Benamouzig, R.; Attar, A.; Benichou, J.; Martin, A.; Morere, J.-F. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann. Oncol. 2005, 16, 102–108. [Google Scholar] [CrossRef]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef]
- Ayati, A.; Moghimi, S.; Salarinejad, S.; Safavi, M.; Pouramiri, B.; Foroumadi, A. A review on progression of epidermal growth factor receptor (EGFR) inhibitors as an efficient approach in cancer targeted therapy. Bioorg. Chem. 2020, 99, 103811. [Google Scholar] [CrossRef]
- Metwaly, A.M.; Ghoneim, M.M.; Eissa, I.H.; Elsehemy, I.A.; Mostafa, A.E.; Hegazy, M.M.; Afifi, W.M.; Dou, D. Traditional ancient Egyptian medicine: A review. Saudi J. Biol. Sci. 2021, 28, 5823–5832. [Google Scholar] [CrossRef]
- Han, X.; Yang, Y.; Metwaly, A.M.; Xue, Y.; Shi, Y.; Dou, D. The Chinese herbal formulae (Yitangkang) exerts an antidiabetic effect through the regulation of substance metabolism and energy metabolism in type 2 diabetic rats. J. Ethnopharmacol. 2019, 239, 111942. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barcz, E.; Sommer, E.; Janik, P.; Marianowski, L.; Skopinska-Rozewska, E. Adenosine receptor antagonism causes inhibition of angiogenic activity of human ovarian cancer cells. Oncol. Rep. 2000, 7, 1285–1376. [Google Scholar] [CrossRef] [PubMed]
- Kakuyamanee Iwazaki, A.; Sadzuka, Y. Effect of methylxanthine derivatives on doxorubicin transport and antitumor activity. Curr. Drug Metab. 2001, 2, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Sultani, H.N.; Ghazal, R.A.; Hayallah, A.M.; Abdulrahman, L.K.; Abu-Hammour, K.; AbuHammad, S.; Taha, M.O.; Zihlif, M.A. Inhibitory effects of new mercapto xanthine derivatives in human mcf7 and k562 cancer cell lines. J. Heterocycl. Chem. 2017, 54, 450–456. [Google Scholar] [CrossRef]
- Woskresensky, A. Ueber das Theobromin. Justus Liebigs Ann. Der Chem. 1842, 41, 125–127. [Google Scholar] [CrossRef] [Green Version]
- Smit, H.J. Theobromine and the pharmacology of cocoa. Methylxanthines 2011, 201–234. [Google Scholar]
- Sugimoto, N.; Miwa, S.; Hitomi, Y.; Nakamura, H.; Tsuchiya, H.; Yachie, A. Theobromine, the primary methylxanthine found in Theobroma cacao, prevents malignant glioblastoma proliferation by negatively regulating phosphodiesterase-4, extracellular signal-regulated kinase, Akt/mammalian target of rapamycin kinase, and nuclear factor-kappa B. Nutr. Cancer 2014, 66, 419–423. [Google Scholar]
- Gil, M.; Skopińska-Rózewska, E.; Radomska, D.; Demkow, U.; Skurzak, H.; Rochowska, M.; Beuth, J.; Roszkowski, K. Effect of purinergic receptor antagonists suramin and theobromine on tumor-induced angiogenesis in BALB/c mice. Folia Biol. 1993, 39, 63–68. [Google Scholar]
- Barcz, E.; Sommer, E.; Sokolnicka, I.; Gawrychowski, K.; Roszkowska-Purska, K.; Janik, P.; Skopinska-Rózewska, E. The influence of theobromine on angiogenic activity and proangiogenic cytokines production of human ovarian cancer cells. Oncol. Rep. 1998, 5, 517–537. [Google Scholar] [CrossRef]
- da Rosa, R.; Schenkel, E.P.; Bernardes, L.S.C. Semisynthetic and newly designed derivatives based on natural chemical scaffolds: Moving beyond natural products to fight Trypanosoma cruzi. Phytochem. Rev. 2020, 19, 105–122. [Google Scholar] [CrossRef]
- Mahmoud, A.H.; Masters, M.R.; Yang, Y.; Lill, M.A. Elucidating the multiple roles of hydration for accurate protein-ligand binding prediction via deep learning. Commun. Chem. 2020, 3, 19. [Google Scholar] [CrossRef] [Green Version]
- Elton, D.C.; Boukouvalas, Z.; Fuge, M.D.; Chung, P.W. Engineering, Deep learning for molecular design—A review of the state of the art. Mol. Syst. Des. Eng. 2019, 4, 828–849. [Google Scholar] [CrossRef] [Green Version]
- Keith, J.A.; Vassilev-Galindo, V.; Cheng, B.; Chmiela, S.; Gastegger, M.; Müller, K.R.; Tkatchenko, A. Combining machine learning and computational chemistry for predictive insights into chemical systems. Chem. Rev. 2021, 121, 9816–9872. [Google Scholar] [CrossRef]
- Cáceres, E.L.; Tudor, M.; Cheng, A.C. Deep learning approaches in predicting ADMET properties. Future Med. Chem. 2020, 12, 1995–1999. [Google Scholar] [CrossRef] [PubMed]
- Ciallella, H.L.; Zhu, H. Advancing computational toxicology in the big data era by artificial intelligence: Data-driven and mechanism-driven modeling for chemical toxicity. Chem. Res. Toxicol. 2019, 32, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Tielens, F.; Gierada, M.; Handzlik, J.; Calatayud, M. Characterization of amorphous silica based catalysts using DFT computational methods. Catal. Today 2020, 354, 3–18. [Google Scholar] [CrossRef]
- Pracht, P.; Bohle, F.; Grimme, S. Automated exploration of the low-energy chemical space with fast quantum chemical methods. Phys. Chem. Chem. Phys. 2020, 22, 7169–7192. [Google Scholar] [CrossRef]
- Seidel, T.; Schuetz, D.A.; Garon, A.; Langer, T. The pharmacophore concept and its applications in computer-aided drug design. Prog. Chem. Org. Nat. Prod. 2019, 110, 99–141. [Google Scholar]
- Bonomi, P. Erlotinib: A new therapeutic approach for non-small cell lung cancer. Expert Opin. Investig. Drugs 2003, 12, 1395–1401. [Google Scholar] [CrossRef]
- Ou, S.-H.I. Second-generation irreversible epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs): A better mousetrap? A review of the clinical evidence. Crit. Rev. Oncol. Hematol. 2012, 83, 407–421. [Google Scholar] [CrossRef]
- Jänne, P.A.; Yang, J.C.-H.; Kim, D.-W.; Planchard, D.; Ohe, Y.; Ramalingam, S.S.; Ahn, M.-J.; Kim, S.-W.; Su, W.-C.; Horn, L. AZD9291 in EGFR inhibitor–resistant non–small-cell lung cancer. N. Engl. J. Med. 2015, 372, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Xu, X. Parallel phase 1 clinical trials in the US and in China: Accelerating the test of avitinib in lung cancer as a novel inhibitor selectively targeting mutated EGFR and overcoming T790M—Induced resistance. Cancer Commun. 2015, 34, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traxler, P.; Bold, G.; Frei, J.; Lang, M.; Lydon, N.; Mett, H.; Buchdunger, E.; Meyer, T.; Mueller, M.; Furet, P. Use of a pharmacophore model for the design of EGF-R tyrosine kinase inhibitors: 4-(phenylamino) pyrazolo [3, 4-d] pyrimidines. J. Med. Chem. 1997, 40, 3601–3616. [Google Scholar] [CrossRef] [PubMed]
- Ducray, R.; Ballard, P.; Barlaam, B.C.; Hickinson, M.D.; Kettle, J.G.; Ogilvie, D.J.; Trigwell, C.B. Novel 3-alkoxy-1H-pyrazolo [3, 4-d] pyrimidines as EGFR and erbB2 receptor tyrosine kinase inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 959–962. [Google Scholar] [CrossRef]
- Gaber, A.A.; Bayoumi, A.H.; El-Morsy, A.M.; Sherbiny, F.F.; Mehany, A.B.; Eissa, I.H. Design, synthesis and anticancer evaluation of 1H-pyrazolo [3, 4-d] pyrimidine derivatives as potent EGFRWT and EGFRT790M inhibitors and apoptosis inducers. Bioorg. Chem. 2018, 80, 375–395. [Google Scholar] [CrossRef]
- Elmetwally, S.A.; Saied, K.F.; Eissa, I.H.; Elkaeed, E.B. Design, synthesis and anticancer evaluation of thieno [2, 3-d] pyrimidine derivatives as dual EGFR/HER2 inhibitors and apoptosis inducers. Bioorg. Chem. 2019, 88, 102944. [Google Scholar] [CrossRef]
- Elzahabi, H.S.; Nossier, E.S.; Alasfoury, R.A.; El-Manawaty, M.; Sayed, S.M.; Elkaeed, E.B.; Metwaly, A.M.; Hagras, M.; Eissa, I.H. Design, synthesis, and anti-cancer evaluation of new pyrido [2, 3-d] pyrimidin-4 (3H)-one derivatives as potential EGFRWT and EGFRT790M inhibitors and apoptosis inducers. J. Enzym. Inhib. Med. Chem. 2022, 37, 1053–1076. [Google Scholar] [CrossRef]
- Gandin, V.; Ferrarese, A.; Dalla Via, M.; Marzano, C.; Chilin, A.; Marzaro, G. Targeting kinases with anilinopyrimidines: Discovery of N-phenyl-N’-[4-(pyrimidin-4-ylamino) phenyl] urea derivatives as selective inhibitors of class III receptor tyrosine kinase subfamily. Sci. Rep. 2015, 5, 16750. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Gray, N.S. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006, 2, 358–364. [Google Scholar] [CrossRef]
- Furet, P.; Caravatti, G.; Lydon, N.; Priestle, J.P.; Sowadski, J.M.; Trinks, U.; Traxler, P. Modelling study of protein kinase inhibitors: Binding mode of staurosporine and origin of the selectivity of CGP 52411. J. Comput. Aided Mol. Des. 1995, 9, 465–472. [Google Scholar] [CrossRef]
- Eldehna, W.M.; El Hassab, M.A.; Elsayed, Z.M.; Al-Warhi, T.; Elkady, H.; Abo-Ashour, M.F.; Abourehab, M.A.; Eissa, I.H.; Abdel-Aziz, H.A. Design, synthesis, in vitro biological assessment and molecular modeling insights for novel 3-(naphthalen-1-yl)-4, 5-dihydropyrazoles as anticancer agents with potential EGFR inhibitory activity. Sci. Rep. 2022, 12, 12821. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.A.; Santos, L.S.; Prata, D.M.; Peixoto, F.C.; da Cunha, E.F.; Ramalho, T.C. Optimal wavelet signal compression as an efficient alternative to investigate molecular dynamics simulations: Application to thermal and solvent effects of MRI probes. Theor. Chem. Acc. 2017, 136, 15. [Google Scholar] [CrossRef]
- Husein, D.Z.; Hassanien, R.; Khamis, M. Cadmium oxide nanoparticles/graphene composite: Synthesis, theoretical insights into reactivity and adsorption study. RSC Adv. 2021, 11, 27027–27041. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Husein, D.Z. Novel synthesis of multicomponent porous nano-hybrid composite, theoretical investigation using DFT and dye adsorption applications: Disposing of waste with waste. Environ. Sci. Pollut. Res. 2022, 1–28. [Google Scholar] [CrossRef] [PubMed]
- La Porta, F.A.; Ramalho, T.C.; Santiago, R.T.; Rocha, M.V.; da Cunha, E.F. Orbital signatures as a descriptor of regioselectivity and chemical reactivity: The role of the frontier orbitals on 1, 3-dipolar cycloadditions. J. Phys. Chem. A 2011, 115, 824–833. [Google Scholar] [CrossRef]
- Coelho, J.V.; Freitas, M.P.; Ramalho, T.C.; Martins, C.R.; Bitencourt, M.; Cormanich, R.A.; Tormena, C.F.; Rittner, R. The case of infrared carbonyl stretching intensities of 2-bromocyclohexanone: Conformational and intermolecular interaction insights. Chem. Phys. Lett. 2010, 494, 26–30. [Google Scholar] [CrossRef]
- Ferreira, L.L.; Andricopulo, A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today 2019, 24, 1157–1165. [Google Scholar] [CrossRef]
- Norinder, U.; Bergström, C.A. Prediction of ADMET properties. ChemMedChem Chem. Enabling Drug Discov. 2006, 1, 920–937. [Google Scholar]
- Dearden, J.C. In silico prediction of drug toxicity. J. Comput.-Aided Mol. Des. 2003, 17, 119–127. [Google Scholar] [CrossRef]
- Idakwo, G.; Luttrell, J.; Chen, M.; Hong, H.; Zhou, Z.; Gong, P.; Zhang, C. A review on machine learning methods for in silico toxicity prediction. J. Environ. Sci. Health Part C 2018, 36, 169–191. [Google Scholar] [CrossRef]
- Kruhlak, N.; Benz, R.; Zhou, H.; Colatsky, T. (Q) SAR modeling and safety assessment in regulatory review. Clin. Pharmacol. Ther. 2012, 91, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Elkaeed, E.B.; Yousef, R.G.; Elkady, H.; Alsfouk, A.A.; Husein, D.Z.; Ibrahim, I.M.; Metwaly, A.M.; Eissa, I.H. New Anticancer Theobromine Derivative Targeting EGFRWT and EGFRT790M: Design, Semi-Synthesis, In Silico, and In Vitro Anticancer Studies. Molecules 2022, 27, 5859. [Google Scholar] [CrossRef] [PubMed]
- Eissa, I.H.; Alesawy, M.S.; Saleh, A.M.; Elkaeed, E.B.; Alsfouk, B.A.; El-Attar, A.-A.M.; Metwaly, A.M. Ligand and structure-based in silico determination of the most promising SARS-CoV-2 nsp16-nsp10 2′-o-Methyltransferase complex inhibitors among 3009 FDA approved drugs. Molecules 2022, 27, 2287. [Google Scholar] [CrossRef] [PubMed]
- Suleimen, Y.M.; Jose, R.A.; Suleimen, R.N.; Arenz, C.; Ishmuratova, M.; Toppet, S.; Dehaen, W.; Alsfouk, A.A.; Elkaeed, E.B.; Eissa, I.H. Isolation and In Silico Anti-SARS-CoV-2 Papain-Like Protease Potentialities of Two Rare 2-Phenoxychromone Derivatives from Artemisia spp. Molecules 2022, 27, 1216. [Google Scholar] [CrossRef]
- Suleimen, Y.M.; Jose, R.A.; Suleimen, R.N.; Arenz, C.; Ishmuratova, M.Y.; Toppet, S.; Dehaen, W.; Alsfouk, B.A.; Elkaeed, E.B.; Eissa, I.H. Jusanin, a New Flavonoid from Artemisia commutata with an In Silico Inhibitory Potential against the SARS-CoV-2 Main Protease. Molecules 2022, 27, 1636. [Google Scholar] [CrossRef]
- Mohammed, S.O.; El Ashry, E.S.H.; Khalid, A.; Amer, M.R.; Metwaly, A.M.; Eissa, I.H.; Elkaeed, E.B.; Elshobaky, A.; Hafez, E.E. Expression, Purification, and Comparative Inhibition of Helicobacter pylori Urease by Regio-Selectively Alkylated Benzimidazole 2-Thione Derivatives. Molecules 2022, 27, 865. [Google Scholar] [CrossRef]
- Alanazi, M.M.; Elkady, H.; Alsaif, N.A.; Obaidullah, A.J.; Alkahtani, H.M.; Alanazi, M.M.; Alharbi, M.A.; Eissa, I.H.; Dahab, M.A. New quinoxaline-based VEGFR-2 inhibitors: Design, synthesis, and antiproliferative evaluation with in silico docking, ADMET, toxicity, and DFT studies. RSC Adv. 2021, 11, 30315–30328. [Google Scholar] [CrossRef]
- Alanazi, M.M.; Elkady, H.; Alsaif, N.A.; Obaidullah, A.J.; Alanazi, W.A.; Al-Hossaini, A.M.; Alharbi, M.A.; Eissa, I.H.; Dahab, M.A. Discovery of new quinoxaline-based derivatives as anticancer agents and potent VEGFR-2 inhibitors: Design, synthesis, and in silico study. J. Mol. Struct. 2021, 1253, 132220. [Google Scholar] [CrossRef]





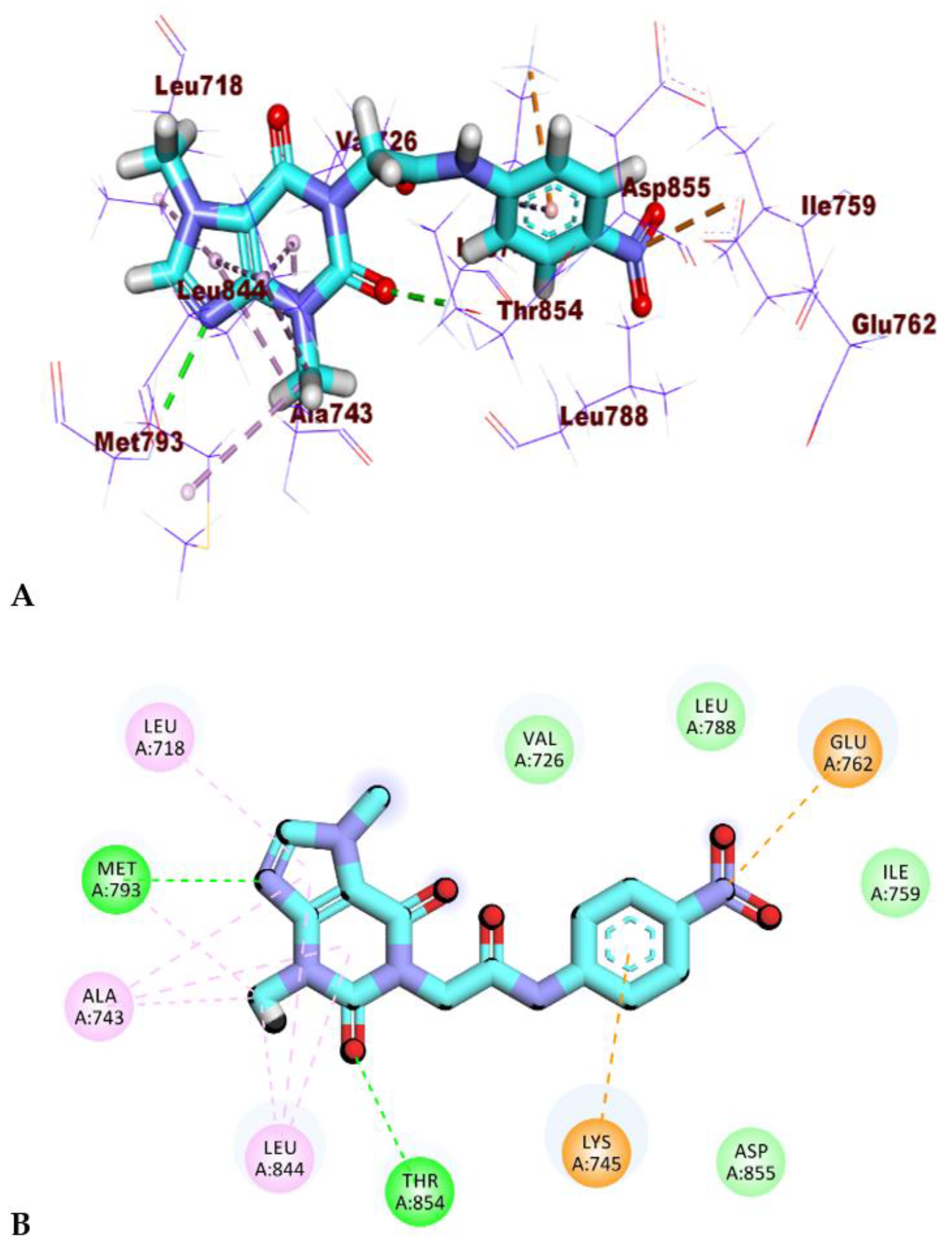




| Cluster Number | Number of HIs | Amino Acids in Receptor | Number of HBs | Amino Acids in Receptor |
|---|---|---|---|---|
| C1 | 2 | L694-L820 | 0 | None |
| C2 | 3 | A719-K721-T766 | 0 | None |
| C3 | 3 | V702-K721-T830 | 1 | S696 |
| C4 | 3 | V702 (2)-T830 | 2 | R817-N818 |
| IP | EA | μ (eV) | χ (eV) | η (eV) | σ (eV) | ω (eV) | Dm (Debye) | TE (eV) | ∆Nmax | ∆E (eV) |
|---|---|---|---|---|---|---|---|---|---|---|
| −2.616 | −6.876 | −4.746 | 4.746 | −2.130 | −0.469 | −23.989 | 11.321 | −34966.4 | −2.228 | 23.989 |
| Comp. | BBB Level | Solubility Level | Absorption Level | Hepatotoxic Prediction | CYP2D6 Prediction | Plasma Protein Binding |
|---|---|---|---|---|---|---|
| T-2-PNPA | Very low | Good | Medium | Nontoxic | Noninhibitor | <90% |
| Erlotinib | High | Low | Good | Toxic | Noninhibitor | 90% |
| Comp. | FDA Rodent Carcinogenicity (Mouse—Female) | Carcinogenic Potency TD50 (Mouse) * | Ames Mutagenicity | Rat Maximum Tolerated Dose (Feed) ** | Rat Oral LD50 ** | Rat Chronic LOAEL ** | Skin Irritancy | Ocular Irritancy |
|---|---|---|---|---|---|---|---|---|
| T-2-PNPA | Non-Carcinogen | 36.246 | Non-Mutagen | 0.017 | 1.730 | 0.018 | Mild | Mild |
| Erlotinib | 39.771 | 0.083 | 0.662 | 0.036 | Non-Irritant |
| Comp. | In Vitro Cytotoxicity IC50 (µM) a | A549 (SI) | HCT-116 (SI) | EGFRWT IC50 (nM) | EGFRT790M IC50 (nM) | ||
|---|---|---|---|---|---|---|---|
| A549 | HCT-116 | WI-38 | |||||
| T-2-PNPA | 11.09 | 21.01 | 48.06 | 4.3 | 2.3 | 7.05 | 126.20 |
| Erlotinib | 6.73 | 16.35 | 31.17 | 4.6 | 1.9 | 5.91 | 202.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elkaeed, E.B.; Yousef, R.G.; Elkady, H.; Alsfouk, A.A.; Husein, D.Z.; Ibrahim, I.M.; Alswah, M.; Elzahabi, H.S.A.; Metwaly, A.M.; Eissa, I.H. A New Theobromine-Based EGFRWT and EGFRT790M Inhibitor and Apoptosis Inducer: Design, Semi-Synthesis, Docking, DFT, MD Simulations, and In Vitro Studies. Processes 2022, 10, 2290. https://doi.org/10.3390/pr10112290
Elkaeed EB, Yousef RG, Elkady H, Alsfouk AA, Husein DZ, Ibrahim IM, Alswah M, Elzahabi HSA, Metwaly AM, Eissa IH. A New Theobromine-Based EGFRWT and EGFRT790M Inhibitor and Apoptosis Inducer: Design, Semi-Synthesis, Docking, DFT, MD Simulations, and In Vitro Studies. Processes. 2022; 10(11):2290. https://doi.org/10.3390/pr10112290
Chicago/Turabian StyleElkaeed, Eslam B., Reda G. Yousef, Hazem Elkady, Aisha A. Alsfouk, Dalal Z. Husein, Ibrahim M. Ibrahim, Mohamed Alswah, Heba S. A. Elzahabi, Ahmed M. Metwaly, and Ibrahim H. Eissa. 2022. "A New Theobromine-Based EGFRWT and EGFRT790M Inhibitor and Apoptosis Inducer: Design, Semi-Synthesis, Docking, DFT, MD Simulations, and In Vitro Studies" Processes 10, no. 11: 2290. https://doi.org/10.3390/pr10112290
APA StyleElkaeed, E. B., Yousef, R. G., Elkady, H., Alsfouk, A. A., Husein, D. Z., Ibrahim, I. M., Alswah, M., Elzahabi, H. S. A., Metwaly, A. M., & Eissa, I. H. (2022). A New Theobromine-Based EGFRWT and EGFRT790M Inhibitor and Apoptosis Inducer: Design, Semi-Synthesis, Docking, DFT, MD Simulations, and In Vitro Studies. Processes, 10(11), 2290. https://doi.org/10.3390/pr10112290












