Visual Analytics for Electronic Health Records: A Review
Abstract
1. Introduction
2. Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
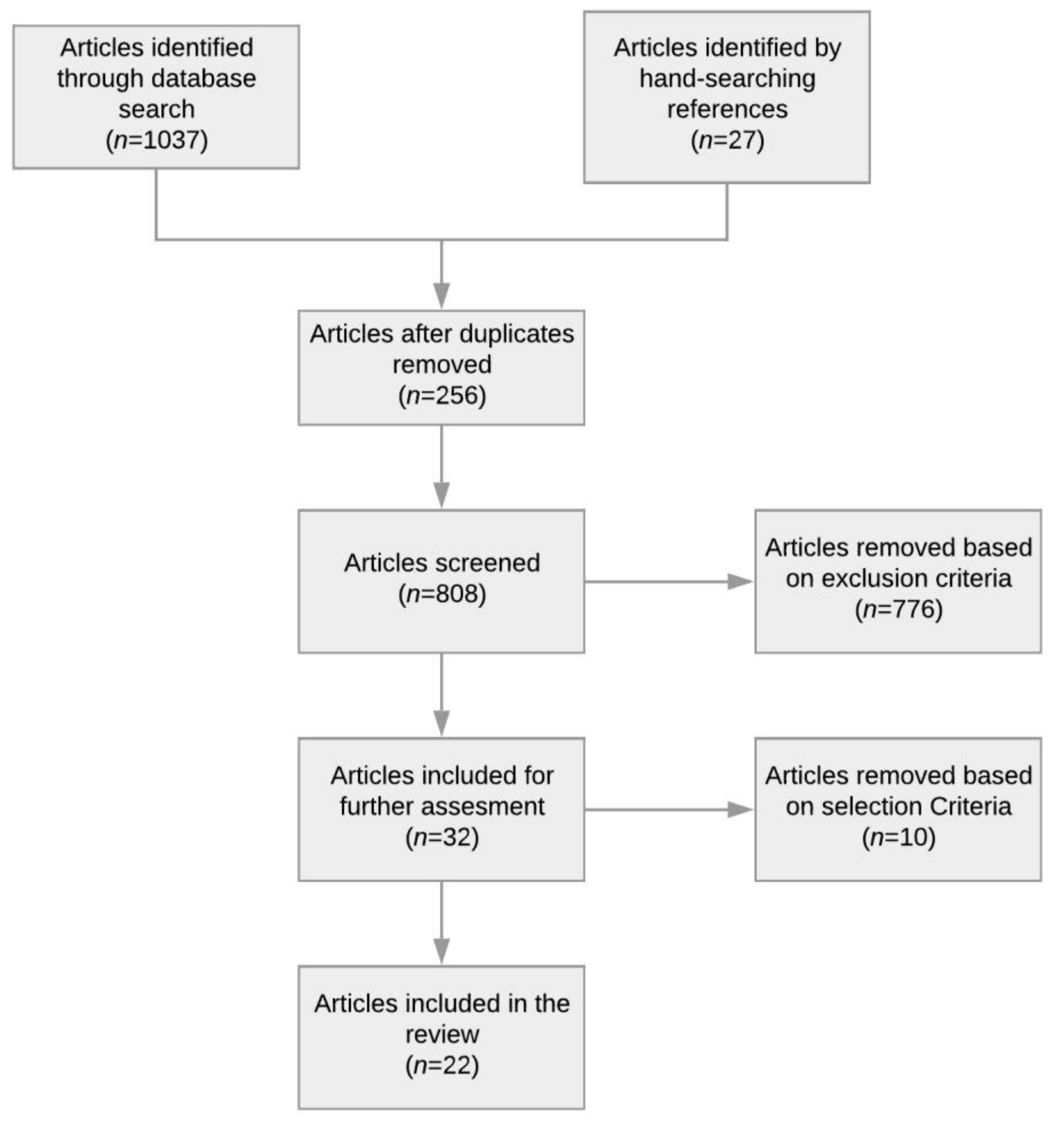
2.3. Article Selection and Analysis
2.4. Results
3. EHR-Based Visual Analytics Systems
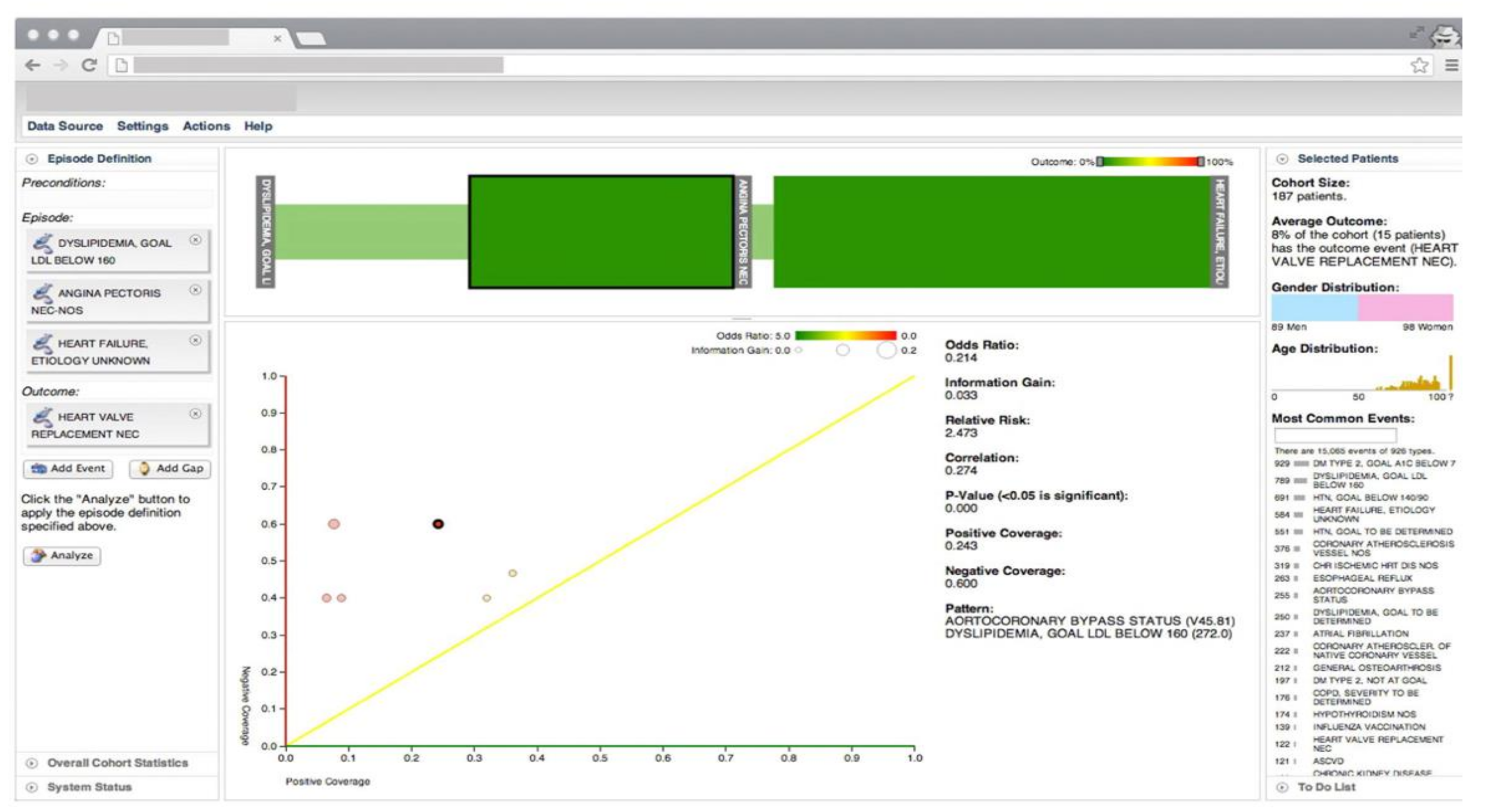
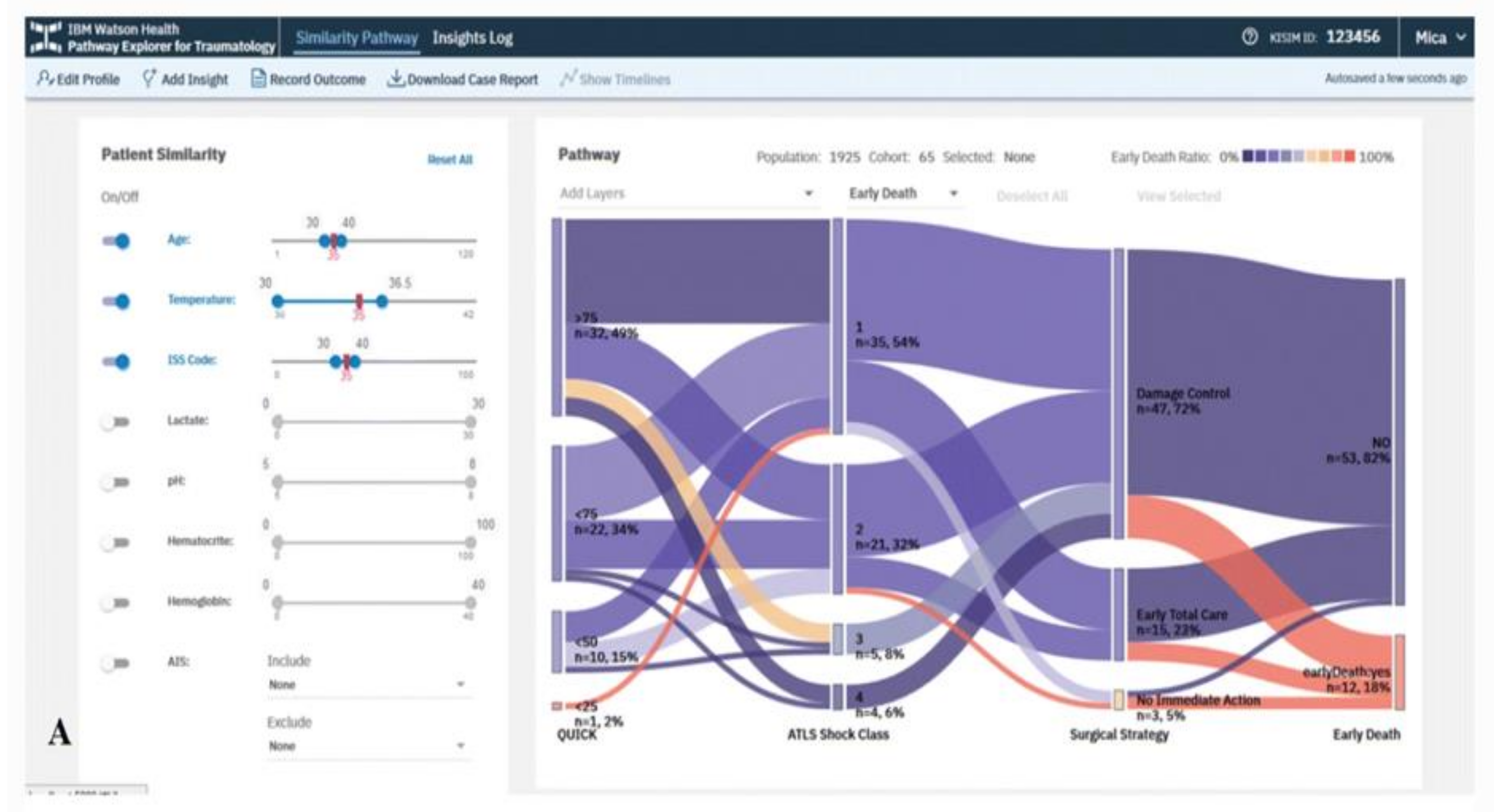
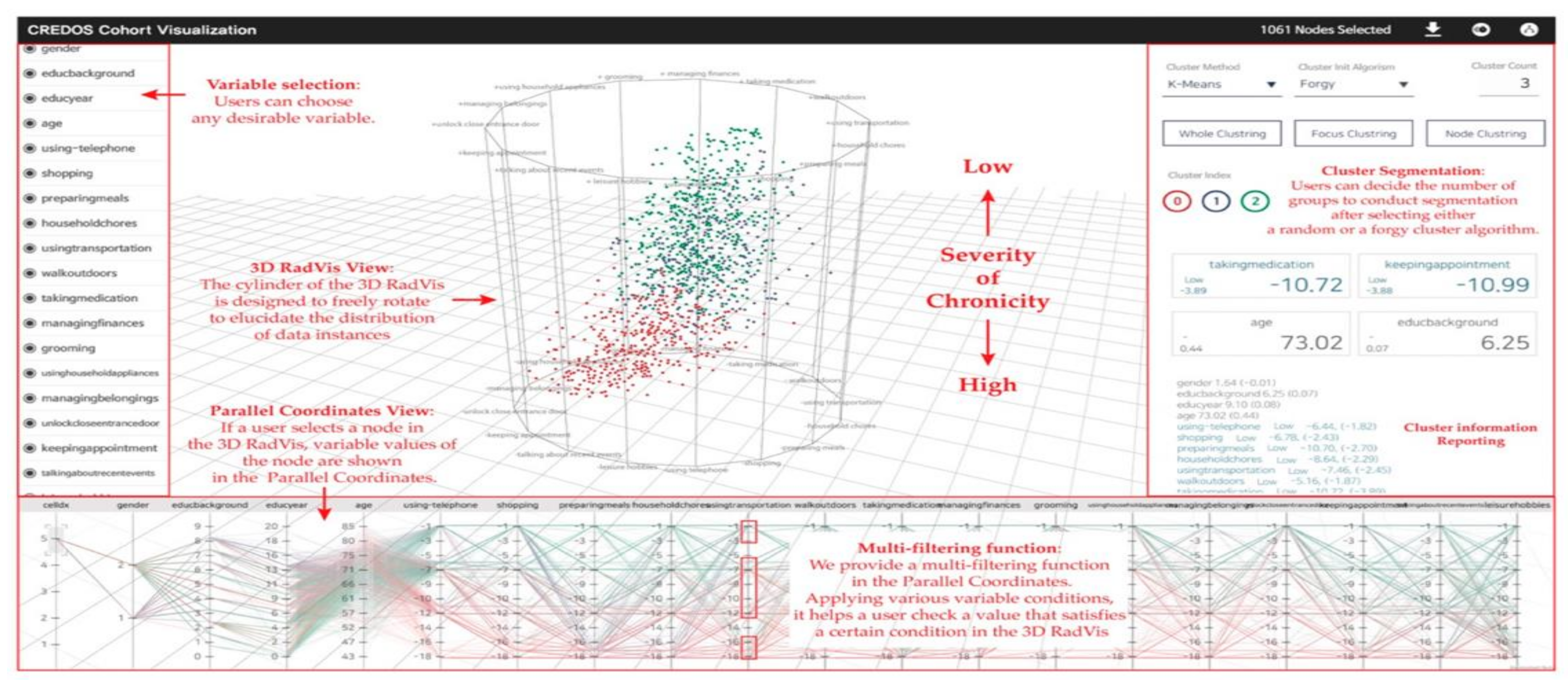
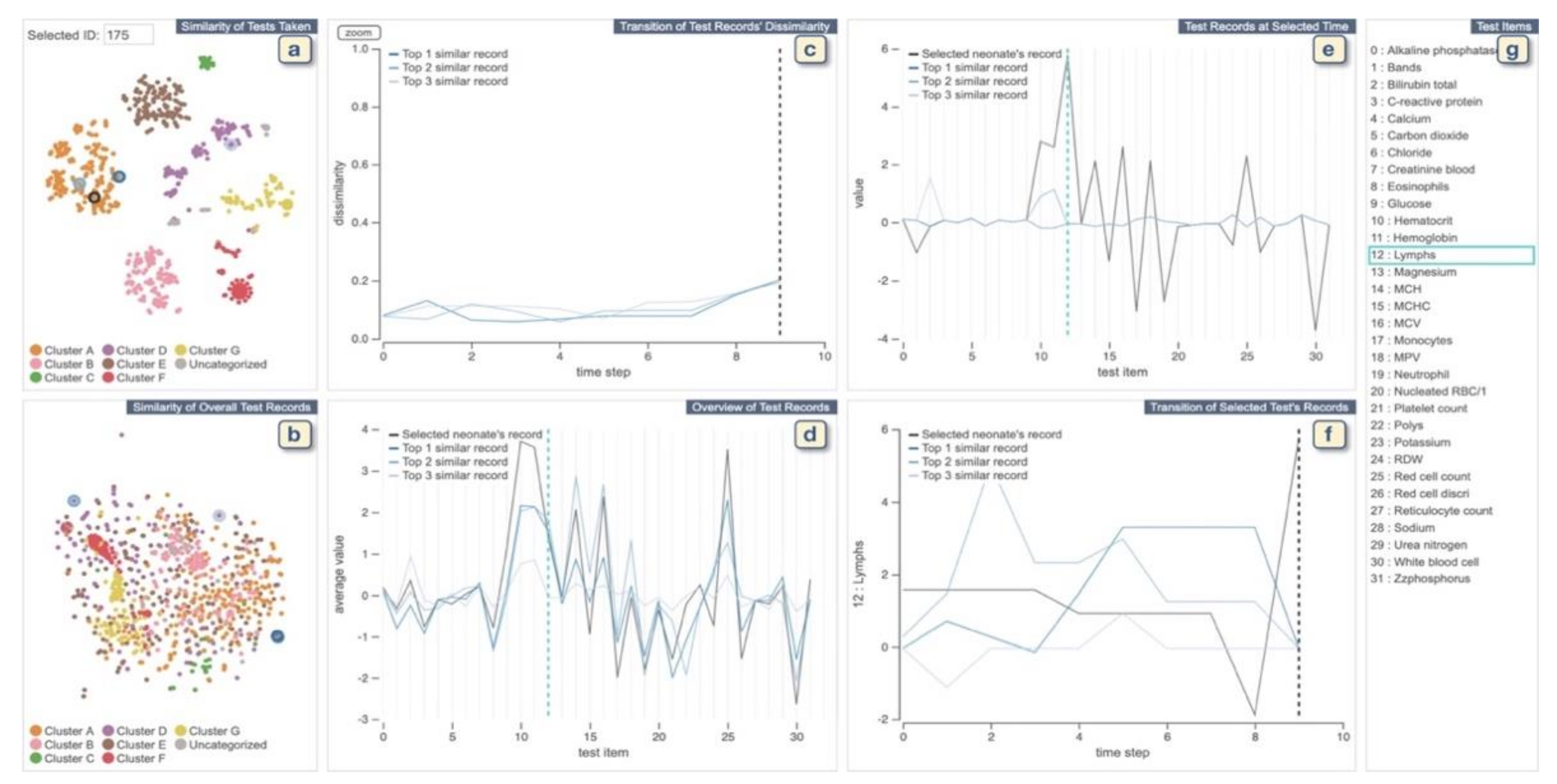
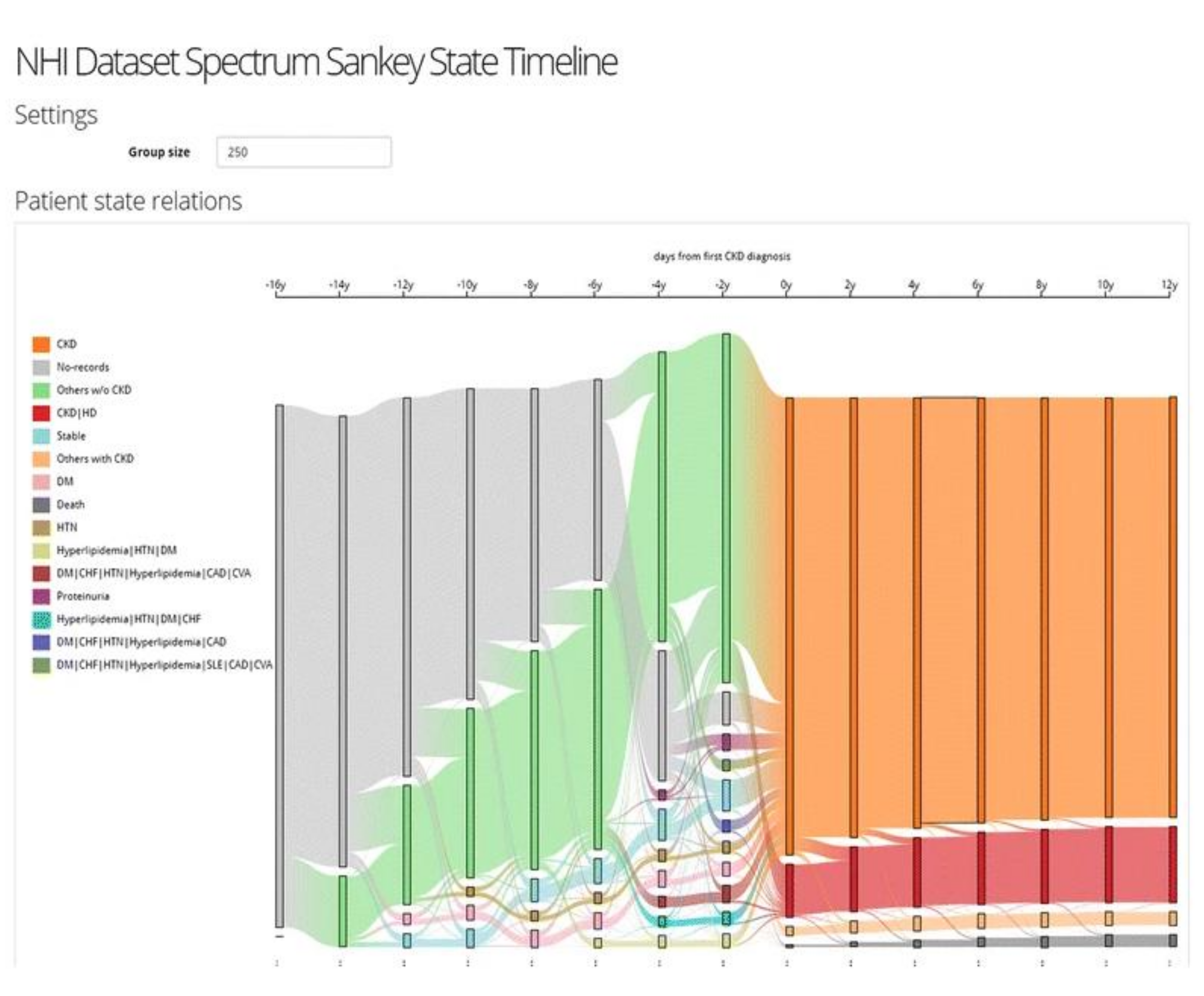
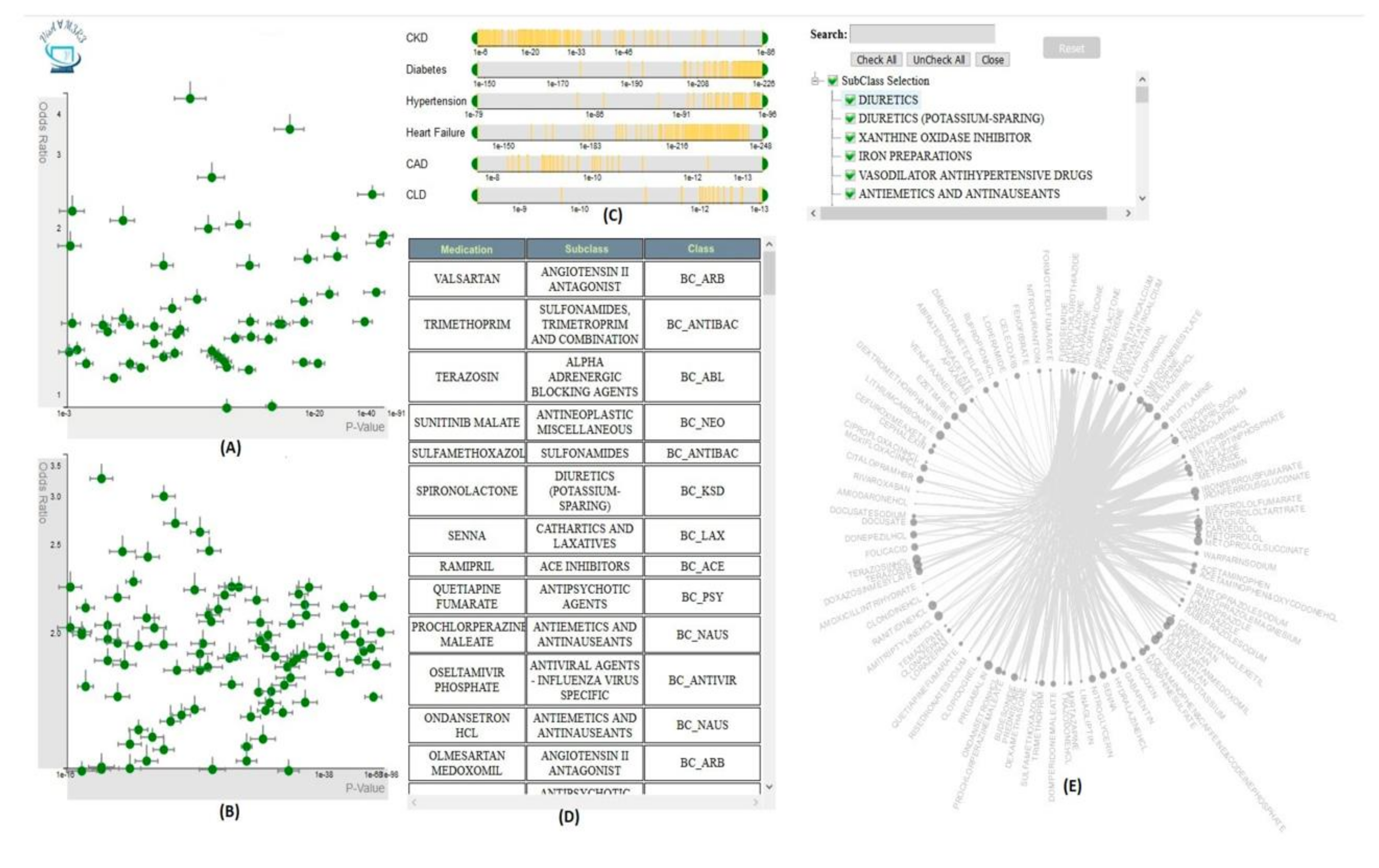
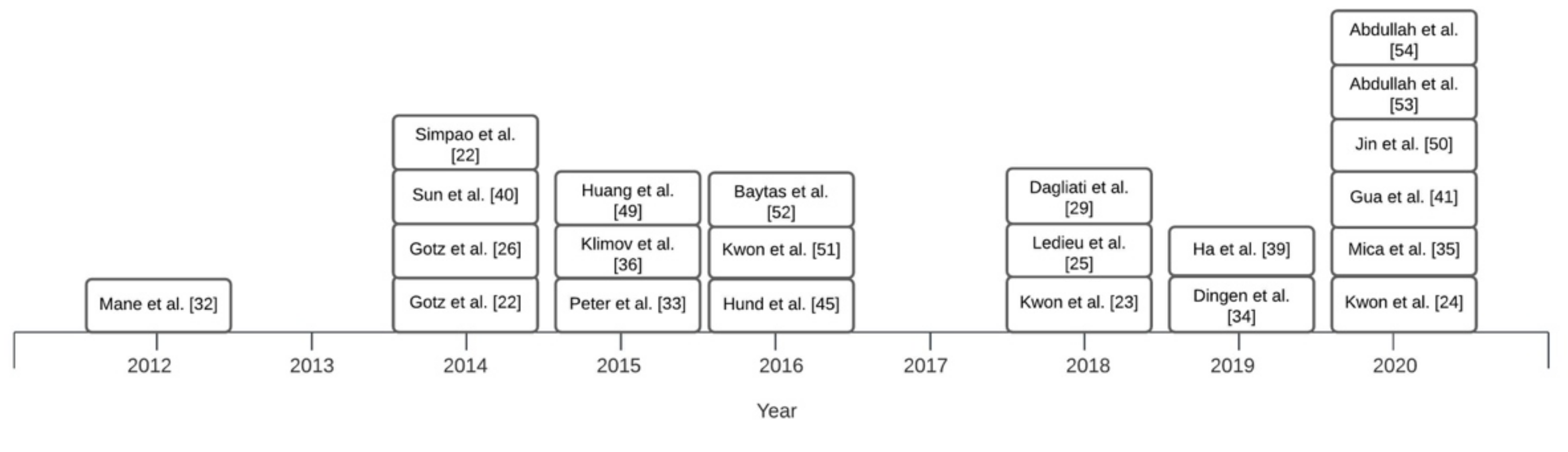
Overview of Systems
4. Design Space
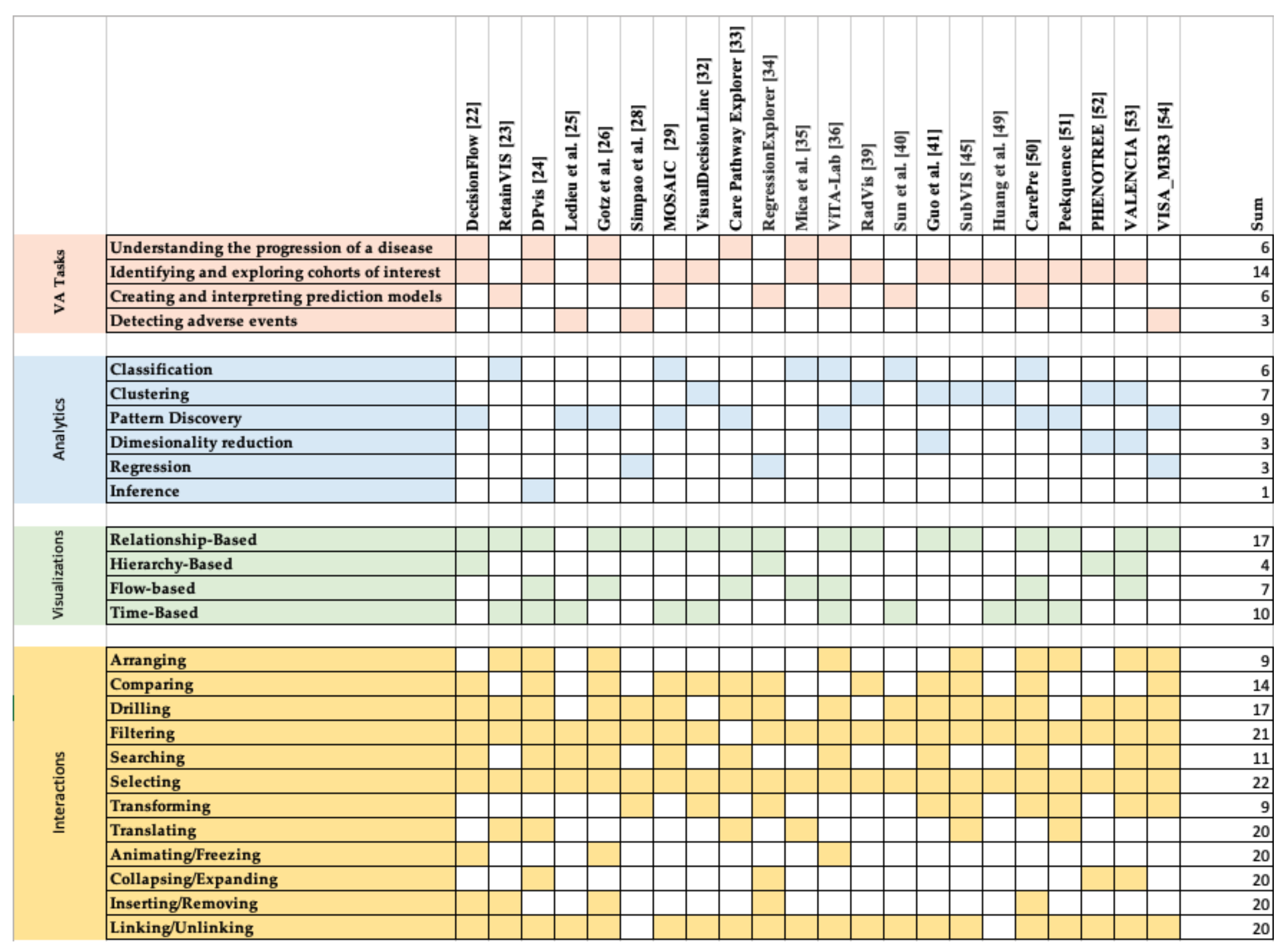
4.1. VA Tasks
4.2. Analytics
4.3. Visualizations
4.4. Interactions
5. Discussion and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Murdoch, T.B.; Detsky, A.S. The Inevitable Application of Big Data to Health Care. JAMA J. Am. Med. Assoc. 2013, 309, 1351–1352. [Google Scholar] [CrossRef]
- Doupi, P. Using EHR Data for Monitoring and Promoting Patient Safety: Reviewing the Evidence on Trigger Tools. Stud. Health Technol. Inf. 2012, 180, 786–790. [Google Scholar]
- Agrawal, A. Medication Errors: Prevention Using Information Technology Systems. Br. J. Clin. Pharmacol. 2009, 67, 681–686. [Google Scholar] [CrossRef]
- Dey, S.; Luo, H.; Fokoue, A.; Hu, J.; Zhang, P. Predicting Adverse Drug Reactions through Interpretable Deep Learning Framework. BMC Bioinform. 2018, 19, 476. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.S.; Rostamzadeh, N.; Sedig, K.; Lizotte, D.J.; Garg, A.X.; McArthur, E. Machine Learning for Identifying Medication-Associated Acute Kidney Injury. Informatics 2020, 7, 18. [Google Scholar] [CrossRef]
- Tang, P.C.; McDonald, C.J. Electronic health record systems. In Biomedical Informatics: Computer Applications in Health Care and Biomedicine; Shortliffe, E.H., Cimino, J.J., Eds.; Health Informatics; Springer: New York, NY, USA, 2006; pp. 447–475. ISBN 978-0-387-36278-6. [Google Scholar]
- Christensen, T.; Grimsmo, A. Instant Availability of Patient Records, but Diminished Availability of Patient Information: A Multi-Method Study of GP’s Use of Electronic Patient Records. BMC Med. Inform. Decis. Mak. 2008, 8, 12. [Google Scholar] [CrossRef]
- Rostamzadeh, N.; Abdullah, S.S.; Sedig, K. Data-Driven Activities Involving Electronic Health Records: An Activity and Task Analysis Framework for Interactive Visualization Tools. Multimodal Technol. Interact. 2020, 4, 7. [Google Scholar] [CrossRef]
- Heisey-Grove, D.; Danehy, L.N.; Consolazio, M.; Lynch, K.; Mostashari, F. A National Study of Challenges to Electronic Health Record Adoption and Meaningful Use. Med. Care 2014, 52, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Lau, F.; Price, M.; Boyd, J.; Partridge, C.; Bell, H.; Raworth, R. Impact of Electronic Medical Record on Physician Practice in Office Settings: A Systematic Review. BMC Med. Inform. Decis. Mak. 2012, 12, 10. [Google Scholar] [CrossRef]
- Ola, O.; Sedig, K. The Challenge of Big Data in Public Health: An Opportunity for Visual Analytics. Online J. Public Health Inf. 2014, 5, 223. [Google Scholar] [CrossRef][Green Version]
- Keim, D.A.; Mansmann, F.; Thomas, J. Visual Analytics: How Much Visualization and How Much Analytics? ACM SIGKDD Explor. Newsl. 2010, 11, 5. [Google Scholar] [CrossRef]
- Sedig, K.; Parsons, P.; Babanski, A. Towards a Characterization of Interactivity in Visual Analytics. J. Multimed. Process. Technol. 2012, 3, 12–28. [Google Scholar]
- Ribarsky, W.; Fisher, B.; Pottenger, W.M. Science of Analytical Reasoning. Inf. Vis. 2009. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of Machine Learning in Drug Discovery and Development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Cortez, P.; Embrechts, M.J. Using Sensitivity Analysis and Visualization Techniques to Open Black Box Data Mining Models. Inf. Sci. 2013, 225, 1–17. [Google Scholar] [CrossRef]
- Keim, D.A.; Munzner, T.; Rossi, F.; Verleysen, M. Bridging Information Visualization with Machine Learning (Dagstuhl Seminar 15101). Dagstuhl Rep. 2015, 5, 1–27. [Google Scholar] [CrossRef]
- Rajwan, Y.G.; Barclay, P.W.; Lee, T.; Sun, I.-F.; Passaretti, C.; Lehmann, H. Visualizing Central Line –Associated Blood Stream Infection (CLABSI) Outcome Data for Decision Making by Health Care Consumers and Practitioners—An Evaluation Study. Online J. Public Health Inf. 2013, 5, 218. [Google Scholar] [CrossRef]
- Goldsmith, M.-R.; Transue, T.R.; Chang, D.T.; Tornero-Velez, R.; Breen, M.S.; Dary, C.C. PAVA: Physiological and Anatomical Visual Analytics for Mapping of Tissue-Specific Concentration and Time-Course Data. J. Pharm. Pharm. 2010, 37, 277–287. [Google Scholar] [CrossRef]
- Perer, A.; Sun, J. MatrixFlow: Temporal Network Visual Analytics to Track Symptom Evolution during Disease Progression. AMIA Annu. Symp. Proc. 2012, 2012, 716–725. [Google Scholar] [PubMed]
- Lo, Y.-S.; Lee, W.-S.; Liu, C.-T. Utilization of Electronic Medical Records to Build a Detection Model for Surveillance of Healthcare-Associated Urinary Tract Infections. J. Med. Syst. 2013, 37, 9923. [Google Scholar] [CrossRef]
- Gotz, D.; Stavropoulos, H. Decisionflow: Visual Analytics for High-Dimensional Temporal Event Sequence Data. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1783–1792. [Google Scholar] [CrossRef]
- Kwon, B.C.; Choi, M.-J.; Kim, J.T.; Choi, E.; Kim, Y.B.; Kwon, S.; Sun, J.; Choo, J. Retainvis: Visual Analytics with Interpretable and Interactive Recurrent Neural Networks on Electronic Medical Records. IEEE Trans. Vis. Comput. Graph. 2018, 25, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Kwon, B.C.; Anand, V.; Severson, K.A.; Ghosh, S.; Sun, Z.; Frohnert, B.I.; Lundgren, M.; Ng, K. DPVis: Visual Analytics with Hidden Markov Models for Disease Progression Pathways. IEEE Trans. Vis. Comput. Graph. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ledieu, T.; Bouzille, G.; Plaisant, C.; Thiessard, F.; Polard, E.; Cuggia, M. Mining Clinical Big Data for Drug Safety: Detecting Inadequate Treatment with a DNA Sequence Alignment Algorithm. AMIA Annu. Symp. Proc. 2018, 2018, 1368–1376. [Google Scholar] [PubMed]
- Gotz, D.; Wang, F.; Perer, A. A Methodology for Interactive Mining and Visual Analysis of Clinical Event Patterns Using Electronic Health Record Data. J. Biomed. Inform. 2014, 48, 148–159. [Google Scholar] [CrossRef]
- Ayres, J.; Flannick, J.; Gehrke, J.; Yiu, T. Sequential Pattern Mining Using a Bitmap Representation. In Proceedings of the Eighth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Edmonton, AB, Canada, 23 July 2002; pp. 429–435. [Google Scholar]
- Simpao, A.F.; Ahumada, L.M.; Desai, B.R.; Bonafide, C.P.; Galvez, J.A.; Rehman, M.A.; Jawad, A.F.; Palma, K.L.; Shelov, E.D. Optimization of Drug-Drug Interaction Alert Rules in a Pediatric Hospital’s Electronic Health Record System Using a Visual Analytics Dashboard. J. Am. Med. Inform. Assoc. 2014, 22, 361–369. [Google Scholar] [CrossRef]
- Dagliati, A.; Sacchi, L.; Tibollo, V.; Cogni, G.; Teliti, M.; Martinez-Millana, A.; Traver, V.; Segagni, D.; Posada, J.; Ottaviano, M.; et al. A Dashboard-Based System for Supporting Diabetes Care. J. Am. Med. Inf. Assoc. 2018, 25, 538–547. [Google Scholar] [CrossRef]
- Sacchi, L.; Capozzi, D.; Bellazzi, R.; Larizza, C. JTSA: An Open Source Framework for Time Series Abstractions. Comput. Methods Programs Biomed. 2015, 121, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Dagliati, A.; Sacchi, L.; Zambelli, A.; Tibollo, V.; Pavesi, L.; Holmes, J.H.; Bellazzi, R. Temporal Electronic Phenotyping by Mining Careflows of Breast Cancer Patients. J. Biomed. Inf. 2017, 66, 136–147. [Google Scholar] [CrossRef]
- Mane, K.K.; Bizon, C.; Schmitt, C.; Owen, P.; Burchett, B.; Pietrobon, R.; Gersing, K. VisualDecisionLinc: A Visual Analytics Approach for Comparative Effectiveness-Based Clinical Decision Support in Psychiatry. J. Biomed. Inform. 2012, 45, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Perer, A.; Wang, F.; Hu, J. Mining and Exploring Care Pathways from Electronic Medical Records with Visual Analytics. J. Biomed. Inform. 2015, 56, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Dingen, D.; van’t Veer, M.; Houthuizen, P.; Mestrom, E.H.J.; Korsten, E.H.H.M.; Bouwman, A.R.A.; van Wijk, J. RegressionExplorer: Interactive Exploration of Logistic Regression Models with Subgroup Analysis. IEEE Trans. Vis. Comput. Graph. 2019, 25, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Mica, L.; Niggli, C.; Bak, P.; Yaeli, A.; McClain, M.; Lawrie, C.M.; Pape, H.-C. Development of a Visual Analytics Tool for Polytrauma Patients: Proof of Concept for a New Assessment Tool Using a Multiple Layer Sankey Diagram in a Single-Center Database. World J. Surg. 2020, 44, 764–772. [Google Scholar] [CrossRef]
- Klimov, D.; Shknevsky, A.; Shahar, Y. Exploration of Patterns Predicting Renal Damage in Patients with Diabetes Type II Using a Visual Temporal Analysis Laboratory. J. Am. Med. Inform. Assoc. 2015, 22, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Moskovitch, R.; Shahar, Y. Classification of Multivariate Time Series via Temporal Abstraction and Time Intervals Mining. Knowl. Inf. Syst. 2015, 45, 35–74. [Google Scholar] [CrossRef]
- Moskovitch, R.; Shahar, Y. Fast Time Intervals Mining Using the Transitivity of Temporal Relations. Knowl. Inf. Syst. 2015, 42, 21–48. [Google Scholar] [CrossRef]
- Ha, H.; Lee, J.; Han, H.; Bae, S.; Son, S.; Hong, C.; Shin, H.; Lee, K. Dementia Patient Segmentation Using EMR Data Visualization: A Design Study. Int. J. Environ. Res. Public Health 2019, 16, 3438. [Google Scholar] [CrossRef]
- Sun, J.; McNaughton, C.D.; Zhang, P.; Perer, A.; Gkoulalas-Divanis, A.; Denny, J.C.; Kirby, J.; Lasko, T.; Saip, A.; Malin, B.A. Predicting Changes in Hypertension Control Using Electronic Health Records from a Chronic Disease Management Program. J. Am. Med. Inf. Assoc. 2014, 21, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Fujiwara, T.; Li, Y.; Lima, K.M.; Sen, S.; Tran, N.K.; Ma, K.-L. Comparative Visual Analytics for Assessing Medical Records with Sequence Embedding. Vis. Inform. 2020, 4, 72–85. [Google Scholar] [CrossRef]
- Gower, J.C.; Warrens, M.J. Similarity, Dissimilarity, and Distance, Measures Of. Wiley StatsRef Stat. Ref. Online 2014, 1–11. [Google Scholar] [CrossRef]
- Kramer, M.A. Nonlinear Principal Component Analysis Using Autoassociative Neural Networks. AICHE J. 1991, 37, 233–243. [Google Scholar] [CrossRef]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to Sequence Learning with Neural Networks. Adv. Neural Inf. Process. Syst. 2014, 27, 3104–3112. [Google Scholar]
- Hund, M.; Böhm, D.; Sturm, W.; Sedlmair, M.; Schreck, T.; Ullrich, T.; Keim, D.A.; Majnaric, L.; Holzinger, A. Visual Analytics for Concept Exploration in Subspaces of Patient Groups. Brain Inf. 2016, 3, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Müller, E.; Günnemann, S.; Assent, I.; Seidl, T. Evaluating Clustering in Subspace Projections of High Dimensional Data. Proc. VLDB Endow. 2009, 2, 1270–1281. [Google Scholar] [CrossRef]
- Cox, M.A.A.; Cox, T.F. Multidimensional Scaling. In Handbook of Data Visualization; Chen, C., Härdle, W., Unwin, A., Eds.; Springer Handbooks Comp. Statistics; Springer: Berlin/Heidelberg, Germany, 2008; pp. 315–347. ISBN 978-3-540-33037-0. [Google Scholar]
- Rao, R.; Card, S.K. The Table Lens: Merging Graphical and Symbolic Representations in an Interactive Focus+ Context Visualization for Tabular Information. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, Boston, MA, USA, 24–28 April 1994; pp. 318–322. [Google Scholar]
- Huang, C.-W.; Lu, R.; Iqbal, U.; Lin, S.-H.; Nguyen, P.A.A.; Yang, H.-C.; Wang, C.-F.; Li, J.; Ma, K.-L.; Li, Y.-C.J.; et al. A Richly Interactive Exploratory Data Analysis and Visualization Tool Using Electronic Medical Records. BMC Med. Inform. Decis. Mak. 2015, 15, 92. [Google Scholar] [CrossRef]
- Jin, Z.; Cui, S.; Guo, S.; Gotz, D.; Sun, J.; Cao, N. CarePre: An Intelligent Clinical Decision Assistance System. ACM Trans. Comput. Healthc. 2020, 1, 1–20. [Google Scholar] [CrossRef]
- Kwon, B.C.; Verma, J.; Perer, A. Peekquence: Visual Analytics for Event Sequence Data. In Proceedings of the ACM SIGKDD 2016 Workshop on Interactive Data Exploration and Analytics, San Francisco, CA, USA, 14 August 2016; Volume 1. [Google Scholar]
- Baytas, I.M.; Lin, K.; Wang, F.; Jain, A.K.; Zhou, J. PhenoTree: Interactive Visual Analytics for Hierarchical Phenotyping from Large-Scale Electronic Health Records. IEEE Trans. Multimed. 2016, 18, 2257–2270. [Google Scholar] [CrossRef]
- Abdullah, S.S.; Rostamzadeh, N.; Sedig, K.; Garg, A.X.; McArthur, E. Visual Analytics for Dimension Reduction and Cluster Analysis of High Dimensional Electronic Health Records. Informatics 2020, 7, 17. [Google Scholar] [CrossRef]
- Abdullah, S.S.; Rostamzadeh, N.; Sedig, K.; Garg, A.X.; McArthur, E. Multiple Regression Analysis and Frequent Itemset Mining of Electronic Medical Records: A Visual Analytics Approach Using VISA_M3R3. Data 2020, 5, 33. [Google Scholar] [CrossRef]
- Sedig, K.; Parsons, P. Design of Visualizations for Human-Information Interaction: A Pattern-Based Framework. Synth. Lect. Vis. 2016, 4, 1–185. [Google Scholar] [CrossRef][Green Version]
- Yadav, P.; Pruinelli, L.; Hangsleben, A.; Dey, S.; Hauwiller, K.; Westra, B.L.; Delaney, C.W.; Kumar, V.; Steinbach, M.S.; Simon, G.J. Modelling Trajectories for Diabetes Complications. In Proceedings of the 4th Workshop on Data Mining for Medicine and Healthcare. 2015 SIAM International Conference on Data Mining, Vancouver, BC, Canada, 30 April–2 May 2015. [Google Scholar]
- Oh, W.; Kim, E.; Castro, M.R.; Caraballo, P.J.; Kumar, V.; Steinbach, M.S.; Simon, G.J. Type 2 Diabetes Mellitus Trajectories and Associated Risks. Big Data 2016, 4, 25–30. [Google Scholar] [CrossRef]
- Mathias, J.S.; Gossett, D.; Baker, D.W. Use of Electronic Health Record Data to Evaluate Overuse of Cervical Cancer Screening. J. Am. Med. Inf. Assoc. 2012, 19, e96–e101. [Google Scholar] [CrossRef]
- Strom, B.L.; Schinnar, R.; Jones, J.; Bilker, W.B.; Weiner, M.G.; Hennessy, S.; Leonard, C.E.; Cronholm, P.F.; Pifer, E. Detecting Pregnancy Use of Non-Hormonal Category X Medications in Electronic Medical Records. J. Am. Med. Inf. Assoc. 2011, 18, i81–i86. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galea, M.H.; Blamey, R.W.; Elston, C.E.; Ellis, I.O. The Nottingham Prognostic Index in Primary Breast Cancer. Breast Cancer Res Treat. 1992, 22, 207–219. [Google Scholar] [CrossRef]
- Knaus, W.A.; Wagner, D.P.; Draper, E.A.; Zimmerman, J.E.; Bergner, M.; Bastos, P.G.; Sirio, C.A.; Murphy, D.J.; Lotring, T.; Damiano, A.; et al. The APACHE III Prognostic System: Risk Prediction of Hospital Mortality for Critically III Hospitalized Adults. Chest 1991, 100, 1619–1636. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, D.; Testa, A.C.; Bourne, T.; Ferrazzi, E.; Ameye, L.; Konstantinovic, M.L.; Van Calster, B.; Collins, W.P.; Vergote, I.; Van Huffel, S.; et al. Logistic Regression Model to Distinguish Between the Benign and Malignant Adnexal Mass Before Surgery: A Multicenter Study by the International Ovarian Tumor Analysis Group. JCO 2005, 23, 8794–8801. [Google Scholar] [CrossRef] [PubMed]
- Nashef, S.A.; Roques, F.; Michel, P.; Gauducheau, E.; Lemeshow, S.; Salamon, R.; EuroSCORE Study Group. European System for Cardiac Operative Risk Evaluation (EuroSCORE). Eur. J. Cardiothorac. Surg. 1999, 16, 9–13. [Google Scholar] [CrossRef]
- Chalmers, J.; Pullan, M.; Fabri, B.; McShane, J.; Shaw, M.; Mediratta, N.; Poullis, M. Validation of EuroSCORE II in a Modern Cohort of Patients Undergoing Cardiac Surgery. Eur. J Cardiothorac. Surg. 2013, 43, 688–694. [Google Scholar] [CrossRef]
- Gaziano, T.A.; Bitton, A.; Anand, S.; Abrahams-Gessel, S.; Murphy, A. Growing Epidemic of Coronary Heart Disease in Low- and Middle-Income Countries. Curr. Probl. Cardiol. 2010, 35, 72–115. [Google Scholar] [CrossRef]
- Munzner, T. Visualization Analysis and Design; CRC Press: Boca Raton, FL, USA, 2014; ISBN 978-1-4987-5971-7. [Google Scholar]
- Treisman, A. Preattentive Processing in Vision. Comput. Vis. Graph. Image Process. 1985, 31, 156–177. [Google Scholar] [CrossRef]
- Ware, C. Information Visualization: Perception for Design; Morgan Kaufmann: Burlington, MA, USA, 2019; ISBN 978-0-12-812876-3. [Google Scholar]
- Institute of Medicine (US) Committee on Quality of Health Care in America; Kohn, L.T.; Corrigan, J.M.; Donaldso, M.S. To Err Is Human: Building a Safer Health System; National Academies Press: Washington, DC, USA, 2000; ISBN 978-0-309-26174-6. [Google Scholar]
- Brennan, T.A.; Leape, L.L.; Laird, N.M.; Hebert, L.; Localio, A.R.; Lawthers, A.G.; Newhouse, J.P.; Weiler, P.C.; Hiatt, H.H. Incidence of Adverse Events and Negligence in Hospitalized Patients. N. Engl. J. Med. 1991, 324, 370–376. [Google Scholar] [CrossRef]
- Leape, L.L.; Brennan, T.A.; Laird, N.; Lawthers, A.G.; Localio, A.R.; Barnes, B.A.; Hebert, L.; Newhouse, J.P.; Weiler, P.C.; Hiatt, H. The Nature of Adverse Events in Hospitalized Patients. N. Engl. J. Med. 1991, 324, 377–384. [Google Scholar] [CrossRef]
- Thomas, E.J.; Studdert, D.M.; Burstin, H.R.; Orav, E.J.; Zeena, T.; Williams, E.J.; Howard, K.M.; Weiler, P.C.; Brennan, T.A. Incidence and Types of Adverse Events and Negligent Care in Utah and Colorado. Med. Care 2000, 38, 261–271. [Google Scholar] [CrossRef]
- Wilson, R.M.; Runciman, W.B.; Gibberd, R.W.; Harrison, B.T.; Newby, L.; Hamilton, J.D. The Quality in Australian Health Care Study. Med. J. Aust. 1995, 163, 458–471. [Google Scholar] [CrossRef]
- Thomas, E.J.; Studdert, D.M.; Newhouse, J.P.; Zbar, B.I.W.; Howard, K.M.; Williams, E.J.; Brennan, T.A. Costs of Medical Injuries in Utah and Colorado. Inquiry 1999, 36, 255–264. [Google Scholar] [PubMed]
- Torio, C.M.; Elixhauser, A.; Andrews, R.M. Trends in Potentially Preventable Hospital Admissions among Adults and Children, 2005–2010: Statistical Brief #151. In Healthcare Cost and Utilization Project (HCUP) Statistical Briefs; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, 2006. [Google Scholar]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000; ISBN 978-0-521-78019-3. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar]
- Lewis, D.D. Naive (Bayes) at Forty: The Independence Assumption in Information Retrieval. In Proceedings of the European Conference on Machine Learning; Springer: Berlin/Heidelberg, Germany, 1998; pp. 4–15. [Google Scholar]
- Daniel, G.G. Artificial Neural Network. In Encyclopedia of Sciences and Religions; Runehov, A.L.C., Oviedo, L., Eds.; Springer: Dordrecht, The Netherlands, 2013; p. 143. ISBN 978-1-4020-8265-8. [Google Scholar]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-Means Clustering Algorithm. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Jain, A.K. Data Clustering: 50 Years beyond K-Means. Pattern Recognit. Lett. 2010, 31, 651–666. [Google Scholar] [CrossRef]
- Nielsen, F. Hierarchical Clustering. In Introduction to HPC with MPI for Data Science; Nielsen, F., Ed.; Undergraduate Topics in Computer Science; Springer International Publishing: Cham, Switzerland, 2016; pp. 195–211. ISBN 978-3-319-21903-5. [Google Scholar]
- Fraley, C.; Raftery, A.E. Model-Based Clustering, Discriminant Analysis, and Density Estimation. J. Am. Stat. Assoc. 2002, 97, 611–631. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. Kdd 1996, 96, 226–231. [Google Scholar]
- Agrawal, R.; Imielinski, T.; Swami, A. Mining Association Rules between Sets of Items in Large Databases. In Proceedings of the 1993 ACM SIGMOD International Conference on Management of Data, Washington, DC, USA, 26–28 May 1993; pp. 207–216. [Google Scholar]
- Ismail, B.; Anil, M. Regression Methods for Analyzing the Risk Factors for a Life Style Disease among the Young Population of India. Indian Heart J. 2014, 66, 587–592. [Google Scholar] [CrossRef]
- Stopar, L.; Skraba, P.; Grobelnik, M.; Mladenic, D. StreamStory: Exploring Multivariate Time Series on Multiple Scales. IEEE Trans. Vis. Comput. Graph. 2019, 25, 1788–1802. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjya, D.; Shanmugam, K.; Gao, T.; Mattei, N.; Varshney, K.; Subramanian, D. Event-Driven Continuous Time Bayesian Networks. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 3 April 2020; Volume 34, pp. 3259–3266. [Google Scholar]
- Siwek, K.; Osowski, S.; Markiewicz, T.; Korytkowski, J. Analysis of Medical Data Using Dimensionality Reduction Techniques. Przegląd Elektrotechniczny 2013, 89, 279–281. [Google Scholar]
- Sedig, K.; Parsons, P. Interaction Design for Complex Cognitive Activities with Visual Representations: A Pattern-Based Approach. AIS Trans. Hum. Comput. Interact. 2013, 5, 84–133. [Google Scholar] [CrossRef]


| KEYWORDS: (K1) AND (K2) AND (K3) | |
|---|---|
| within each group, the keywords are combined by the “OR” operator | |
| K1 (Visualization) | Visualization or visual |
| K2 (Analytics) | Analytics or analysis or data mining or machine learning |
| K3 (EHR 1) | EHR or electronic health record or electronic medical record or EMR 2 or healthcare record or patient record or clinical data |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rostamzadeh, N.; Abdullah, S.S.; Sedig, K. Visual Analytics for Electronic Health Records: A Review. Informatics 2021, 8, 12. https://doi.org/10.3390/informatics8010012
Rostamzadeh N, Abdullah SS, Sedig K. Visual Analytics for Electronic Health Records: A Review. Informatics. 2021; 8(1):12. https://doi.org/10.3390/informatics8010012
Chicago/Turabian StyleRostamzadeh, Neda, Sheikh S. Abdullah, and Kamran Sedig. 2021. "Visual Analytics for Electronic Health Records: A Review" Informatics 8, no. 1: 12. https://doi.org/10.3390/informatics8010012
APA StyleRostamzadeh, N., Abdullah, S. S., & Sedig, K. (2021). Visual Analytics for Electronic Health Records: A Review. Informatics, 8(1), 12. https://doi.org/10.3390/informatics8010012







