A Pilot Study Using Natural Language Processing to Explore Textual Electronic Mental Healthcare Data
Abstract
1. Introduction
2. Natural Language Processing Pipeline
- Data preprocessing
- De-identification and anonymization to remove personally identifiable information while preserving clinical relevance.
- Text normalization, including correction of spelling errors, handling of acronyms, and standardization of abbreviations.
- Tokenization, where free-text clinical notes are segmented into individual words or phrases to facilitate analysis.
- 2.
- Annotation and labeling
- Clinical experts annotate a subset of the dataset to identify medical concepts such as diagnoses, symptoms, medications, and treatment responses.
- Inter-annotator agreement measures to ensure consistency and reliability in the labeling.
- 3.
- Model selection and training
- Named-entity recognition (NER) to identify key medical concepts from unstructured text.
- Sentiment analysis to assess emotional and psychological indicators within the notes.
- Topic modeling to uncover hidden themes and patterns in mental health records.
- Text summarization to generate concise representations of extensive patient histories.
- 4.
- Model validation and evaluation
- Precision: The proportion of correctly identified concepts out of all identified instances.
- Recall: The proportion of correctly identified concepts out of all actual instances in the dataset.
- F1 score: The harmonic mean of precision and recall, ensuring a balance between both.
- 5.
- Interpretation and integration into clinical research
- Efficient data retrieval for mental health research.
- Integration with existing structured EHR fields to enhance clinical decision-making.
- Identification of previously under-documented conditions and symptoms in mental health records.
3. Overview of Key Methods and Architectures
- Named-Entity Recognition (NER)
- Sentiment Analysis
- Text Summarization
- Keyword extraction
- Aspect-Based Opinion Mining
- Topic modeling
- NLP architectures for exploring EHR data
4. Southern Health NHS Foundation Trust’s NLP Approach for UK-CRIS
5. Recent Advances in NLP Methods in Healthcare
6. Large-Scale Language Models in Healthcare
7. Challenges and Limitations
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | artificial intelligence |
| ADEs | adverse drug events |
| AOBM | aspect-based opinion mining |
| BTS | biomedical text summarization |
| IE | information extraction |
| IR | information retrieval |
| EHR | electronic health records |
| GATE | general architecture for text engineering |
| LDA | latent Dirichlet allocation |
| NER | named-entity recognition |
| NHS | National Health Service |
| NLP | natural language processing |
| SHFT | Southern Health NHS Foundation Trust |
| UK | United Kingdom |
| UK-CRIS | UK Clinical Record Interactive Search |
References
- Vigo, D.; Thornicroft, G.; Atun, R. Estimating the true global burden of mental illness. Lancet Psychiatry 2016, 3, 171–178. [Google Scholar] [CrossRef] [PubMed]
- NHS Digital. Mental Health Bulletin 2020–2021 Annual Report. 2020. Available online: https://digital.nhs.uk/data-and-information/publications/statistical/mental-health-bulletin/2020-21-annual-report (accessed on 14 October 2024).
- Dorning, H.; Davies, A.; Blunt, I. Focus on: People with Mental Ill Health and Hospital Use. Exploring Disparities in Hospital Use for Physical Healthcare; The Nuffield Trust: London, UK, 2015. [Google Scholar]
- Lin, J.; Jiao, T.; Biskupiak, J.E.; McAdam-Marx, C. Application of electronic medical record data for health outcomes research: A review of recent literature. Expert Rev. Pharmacoecon. Outcomes Res. 2013, 13, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.; Soguero-Ruiz, C.; Oyvind Mikalsen, K.; Lindsetmo, R.O.; Kouskoumvekaki, I.; Girolami, M.; Olav Skrovseth, S.; Augestad, K.M. Analysis of free text in electronic health records for identification of cancer patient trajectories. Sci. Rep. 2017, 7, 46226. [Google Scholar] [CrossRef] [PubMed]
- Cohen, K.B.; Hunter, L. Natural language processing and systems biology. In Artificial Intelligence Methods and Tools for Systems Biology; Springer: Dordrecht, The Netherlands, 2004; pp. 147–173. [Google Scholar]
- Locke, W.N.; Booth, A.D. (Eds.) Machine Translation of Languages; Fourteen essays; John Wiley: New York, NY, USA, 1955; ISBN 9780262120029. [Google Scholar]
- Tenney, I.; Xia, P.; Chen, B.; Wang, A.; Poliak, A.; McCoy, R.T.; Kim, N.; Van Durme, B.; Bowman, S.R.; Das, D.; et al. What do you learn from context? Probing for sentence structure in contextualized word representations. arXiv 2019, arXiv:1905.06316. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar] [CrossRef]
- Carlini, N.; Tramer, F.; Wallace, E.; Jagielski, M.; Herbert-Voss, A.; Lee, K.; Roberts, A.; Brown, T.; Song, D.; Erlingsson, U.; et al. Extracting training data from large language models. In Proceedings of the 30th USENIX Security Symposium, Vancouver, BC, Canada, 11–13 August 2021; pp. 2633–2650. [Google Scholar] [CrossRef]
- Lawson, N.; Eustice, K.; Perkowitz, M.; Yetisgen-Yildiz, M. Annotating large email datasets for named entity recognition with mechanical turk. In Proceedings of the NAACL HLT 2010 Workshop on Creating Speech and Language Data with Amazon’s Mechanical Turk, Los Angeles, CA, USA, 6 June 2010; pp. 71–79. [Google Scholar]
- Lu, W.; Guttentag, A.; Elbel, B.; Kiszko, K.; Abrams, C.; Kirchner, T.R. Crowdsourcing for food purchase receipt annotation via amazon mechanical Turk: A feasibility study. J. Med. Int. Res. 2019, 21, e12047. [Google Scholar] [CrossRef] [PubMed]
- Entzeridou, E.; Markopoulou, E.; Mollaki, V. Public and physician’s expectations and ethical concerns about electronic health record: Benefits outweigh risks except for information security. Int. J. Med. Inform. 2018, 110, 98–107. [Google Scholar] [CrossRef]
- Kormilitzin, A.; Vaci, N.; Liu, Q.; Nevado-Holgado, A. Med7: A transferable clinical natural language processing model for electronic health records. Artif. Intell. Med. 2021, 118, 102086. [Google Scholar] [CrossRef]
- Carson, L.; Jewell, A.; Downs, J.; Stewart, R. Multisite data linkage projects in mental health research. Lancet Psychiatry 2020, 7, e61. [Google Scholar] [CrossRef]
- Botelle, R.; Bhavsar, V.; Kadra-Scalzo, G.; Mascio, A.; Williams, M.V.; Roberts, A.; Velupillai, S.; Stewart, R. Can natural language processing models extract and classify instances of interpersonal violence in mental healthcare electronic records: An applied evaluative study. BMJ Open 2022, 12, e052911. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rizzo, A.A.; Lange, B.; Buckwalter, J.G.; Forbell, E.; Kim, J.; Sagae, K.; Williams, J.; Rothbaum, B.O.; Difede, J.; Reger, G.; et al. An intelligent virtual human system for providing healthcare information and support. Stud. Health Technol. Inform. 2011, 163, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Bhakta, R.; Savin-Baden, M.; Tombs, G. Sharing Secrets with Robots? 2014. Available online: https://www.learntechlib.org/primary/p/147797/ (accessed on 14 October 2024).
- Meeker, D.; Cerully, J.L.; Johnson, M.; Iyer, N.; Kurz, J.; Scharf, D.M. SimCoach evaluation: A virtual human intervention to encourage service-member help-seeking for posttraumatic stress disorder and depression. Rand Health Q. 2016, 5, 13. [Google Scholar]
- McCoy, T.H.; Castro, V.M.; Cagan, A.; Roberson, A.M.; Kohane, I.S.; Perlis, R.H. Sentiment measured in hospital discharge notes is associated with readmission and mortality risk: An electronic health record study. PLoS ONE 2015, 10, e0136341. [Google Scholar] [CrossRef]
- Denecke, K.; Deng, Y. Sentiment analysis in medical settings: New opportunities and challenges. Artif. Intell. Med. 2015, 64, 17–27. [Google Scholar] [CrossRef]
- Martin, S.A.; Sinsky, C.A. The map is not the territory: Medical records and 21st century practice. Lancet 2016, 388, 2053–2056. [Google Scholar] [CrossRef]
- Liu, Y.H.; Song, X.; Chen, S.F. Long story short: Finding health advice with informative summaries on health social media. Aslib J. Inf. Manag. 2019, 71, 821–840. [Google Scholar] [CrossRef]
- Yin, Y.; Zhang, Y.; Liu, X.; Zhang, Y.; Xing, C.; Chen, H. HealthQA: A Chinese QA summary system for smart health. In Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2014; pp. 51–62. [Google Scholar] [CrossRef]
- Wang, M.; Wang, M.; Yu, F.; Yang, Y.; Walker, J.; Mostafa, J. A systematic review of automatic text summarization for biomedical literature and EHRs. J. Am. Med. Inform. Assoc. 2021, 28, 2287–2297. [Google Scholar] [CrossRef]
- Wu, P.H.; Yu, A.; Tsai, C.W.; Koh, J.L.; Kuo, C.C.; Chen, A.L. Keyword extraction and structuralization of medical reports. Health Inf. Sci. Syst. 2020, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Gandhi, P.; Kabir, M.A.; Zou, C.; Blakey, J.; Luo, X. Progress notes classification and keyword extraction using attention-based deep learning models with BERT. arXiv 2019, arXiv:1910.05786. [Google Scholar] [CrossRef]
- Zeng, Z.; Deng, Y.; Li, X.; Naumann, T.; Luo, Y. Natural Language Processing for EHR-Based Computational Phenotyping. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ding, Y.; Tang, J.; Dong, X.; He, B.; Qiu, J.; Wild, D.J. Finding complex biological relationships in recent PubMed articles using Bio-LDA. PLoS ONE 2011, 6, e17243. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Leroy, G. Large-scale analysis of free-text data for mental health surveillance with topic modelling. In 26th Americas Conference on Information Systems, AMCIS 2020; Association for Information Systems: Atlanta, GA, USA, 2020; ISBN 9781733632546. [Google Scholar]
- Fernandes, A.C.; Cloete, D.; Broadbent, M.; Hayes, R.D.; Chang, C.K.; Jackson, R.G.; Roberts, A.; Tsang, J.; Soncul, M.; Liebscher, J.; et al. Development and evaluation of a de-identification procedure for a case register sourced from mental health electronic records. BMC Med. Inform. Decis. Mak. 2013, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Goodday, S.M.; Kormilitzin, A.; Vaci, N.; Liu, Q.; Cipriani, A.; Smith, T.; Nevado-Holgado, A. Maximizing the use of social and behavioural information from secondary care mental health electronic health records. J. Biomed. Inform. 2020, 107, 103429. [Google Scholar] [CrossRef]
- Callard, F.; Broadbent, M.; Denis, M.; Hotopf, M.; Soncul, M.; Wykes, T.; Lovestone, S.; Stewart, R. Developing a new model for patient recruitment in mental health services: A cohort study using Electronic Health Records. BMJ Open 2014, 4, e005654. [Google Scholar] [CrossRef]
- Walker, S.; Potts, J.; Martos, L.; Barrera, A.; Hancock, M.; Bell, S.; Geddes, J.; Cipriani, A.; Henshall, C. Consent to discuss participation in research: A pilot study. Evid.-Based Ment. Health 2020, 23, 77–82. [Google Scholar] [CrossRef]
- Vaci, N.; Koychev, I.; Kim, C.H.; Kormilitzin, A.; Liu, Q.; Lucas, C.; Dehghan, A.; Nenadic, G.; Nevado-Holgado, A. Real-world effectiveness, its predictors and onset of action of cholinesterase inhibitors and memantine in dementia: Retrospective health record study. Br. J. Psychiatry 2021, 218, 261–267. [Google Scholar] [CrossRef]
- Jackson, R.G.; Ball, M.; Patel, R.; Hayes, R.D.; Dobson, R.J.; Stewart, R. TextHunter—A User Friendly Tool for Extracting Generic Concepts from Free Text in Clinical Research. AMIA Annu. Symp. Proc. 2014, 2014, 729–738. [Google Scholar] [PubMed] [PubMed Central]
- Iqbal, E.; Mallah, R.; Jackson, R.G.; Ball, M.; Ibrahim, Z.M.; Broadbent, M.; Dzahini, O.; Stewart, R.; Johnston, C.; Dobson, R.J. Identification of Adverse Drug Events from Free Text Electronic Patient Records and Information in a Large Mental Health Case Register. PLoS ONE 2015, 10, e0134208. [Google Scholar] [CrossRef] [PubMed]
- Kadra, G.; Stewart, R.; Shetty, H.; Jackson, R.G.; Greenwood, M.A.; Roberts, A.; Chang, C.K.; MacCabe, J.H.; Hayes, R.D. Extracting antipsychotic polypharmacy data from electronic health records: Developing and evaluating a novel process. BMC Psychiatry 2015, 15, 166. [Google Scholar] [CrossRef]
- Patel, R.; Jayatilleke, N.; Broadbent, M.; Chang, C.K.; Foskett, N.; Gorrell, G.; Hayes, R.D.; Jackson, R.; Johnston, C.; Shetty, H.; et al. Negative symptoms in schizophrenia: A study in a large clinical sample of patients using a novel automated method. BMJ Open 2015, 5, e007619. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Wilson, R.; Jackson, R.; Ball, M.; Shetty, H.; Broadbent, M.; Stewart, R.; McGuire, P.; Bhattacharyya, S. Cannabis use and treatment resistance in first episode psychosis: A natural language processing study. Lancet 2015, 385, S79. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.L.; Stewart, R.; Ogden, J.; Broadbent, M.; Pasupathy, D.; Howard, L.M. The characteristics and health needs of pregnant women with schizophrenia compared with bipolar disorder and affective psychoses. BMC Psychiatry 2015, 15, 88. [Google Scholar] [CrossRef] [PubMed]
- Downs, J.; Velupillai, S.; George, G.; Holden, R.; Kikoler, M.; Dean, H.; Fernandes, A.; Dutta, R. Detection of Suicidality in Adolescents with Autism Spectrum Disorders: Developing a Natural Language Processing Approach for Use in Electronic Health Records. AMIA Annu. Symp. Proc. 2018, 2017, 641–649. [Google Scholar] [PubMed]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Devlin, J. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Gururangan, S.; Marasović, A.; Swayamdipta, S.; Lo, K.; Beltagy, I.; Downey, D.; Smith, N.A. Don’t stop pretraining: Adapt language models to domains and tasks. arXiv 2020, arXiv:2004.10964. [Google Scholar]
- Wang, X.; Zhang, Y.; Ren, X.; Zhang, Y.; Jiang, J.; Han, J. An empirical study of pre-trained transformer models for biomedical text mining. ACM Trans. Comput. Biol. Bioinform. 2021, 18, 1432–1445. [Google Scholar]
- Doshi-Velez, F.; Kim, B. Towards a rigorous science of interpretable machine learning. arXiv 2017, arXiv:1702.08608. [Google Scholar]
- Bender, E.M.; Gebru, T.; McMillan-Major, A.; Shmitchell, S. On the dangers of stochastic parrots: Can language models be too big? In Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency (FAccT’21), Athens, Greece, 3–10 March 2021. [Google Scholar]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.S.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2021, 25, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.B.; Hodas, N.O.; Vishnu, A. Deep learning for healthcare: Review, opportunities, and challenges. Brief. Bioinform. 2021, 22, 858–875. [Google Scholar] [CrossRef]
- He, J.; Liu, Z.; Xia, Y.; Wang, J.; Zhang, X.; Liu, Y. Analyzing the potential of ChatGPT-like models in healthcare: Opportunities and challenges. J. Med. Internet Res. 2019, 21, e16279. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Jiang, X.; Wang, J.; Liu, Y. Advances in Natural Language Processing for Clinical Text: Applications and Challenges. J. Biomed. Inform. 2021, 118, 103799. [Google Scholar] [CrossRef]
- Qin, C.; Zhang, Z.; Chen, J.; Yasunaga, M.; Yang, D. Is chatgpt a general-purpose natural language processing task solver? arXiv 2023, arXiv:2302.06476. [Google Scholar]
- Franciscu, S. ChatGPT: A Natural Language Generation Model for Chatbots. 2023. Available online: https://doi.org/10.13140/RG.2.2.24777.83044 (accessed on 14 October 2024).
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.-W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Uzuner, O.; South, B.R.; Shen, S.; DuVall, S.L. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Inform. Assoc. 2011, 18, 552–556. [Google Scholar] [CrossRef]
- Stewart, R.; Soremekun, M.; Perera, G.; Broadbent, M.; Callard, F.; Denis, M.; Hotopf, M.; Lovestone, S. The South London and Maudsley NHS Foundation Trust Biomedical Research Centre (SLAM BRC) case register: Development and descriptive data. BMC Psychiatry 2009, 9, 51. [Google Scholar] [CrossRef]
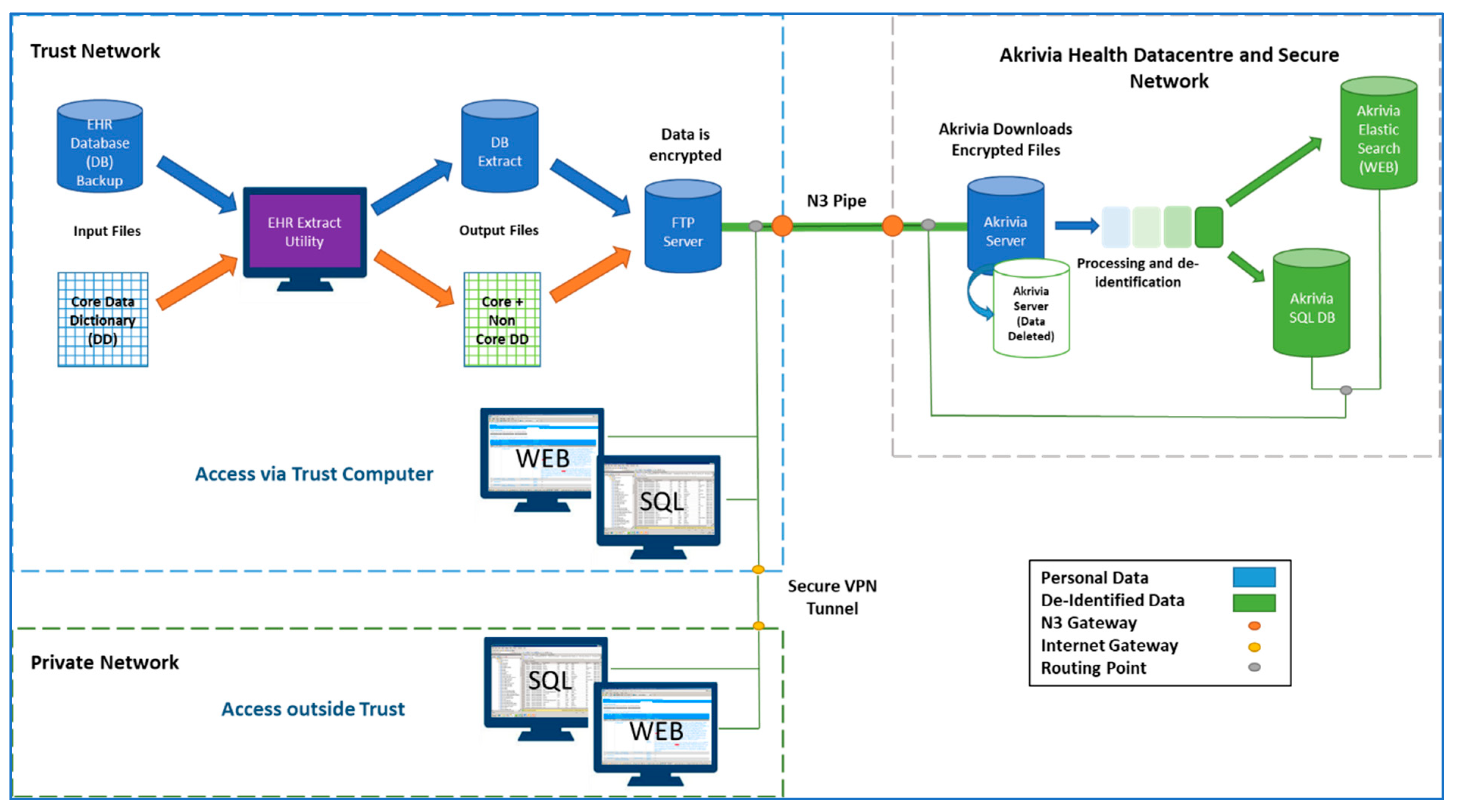
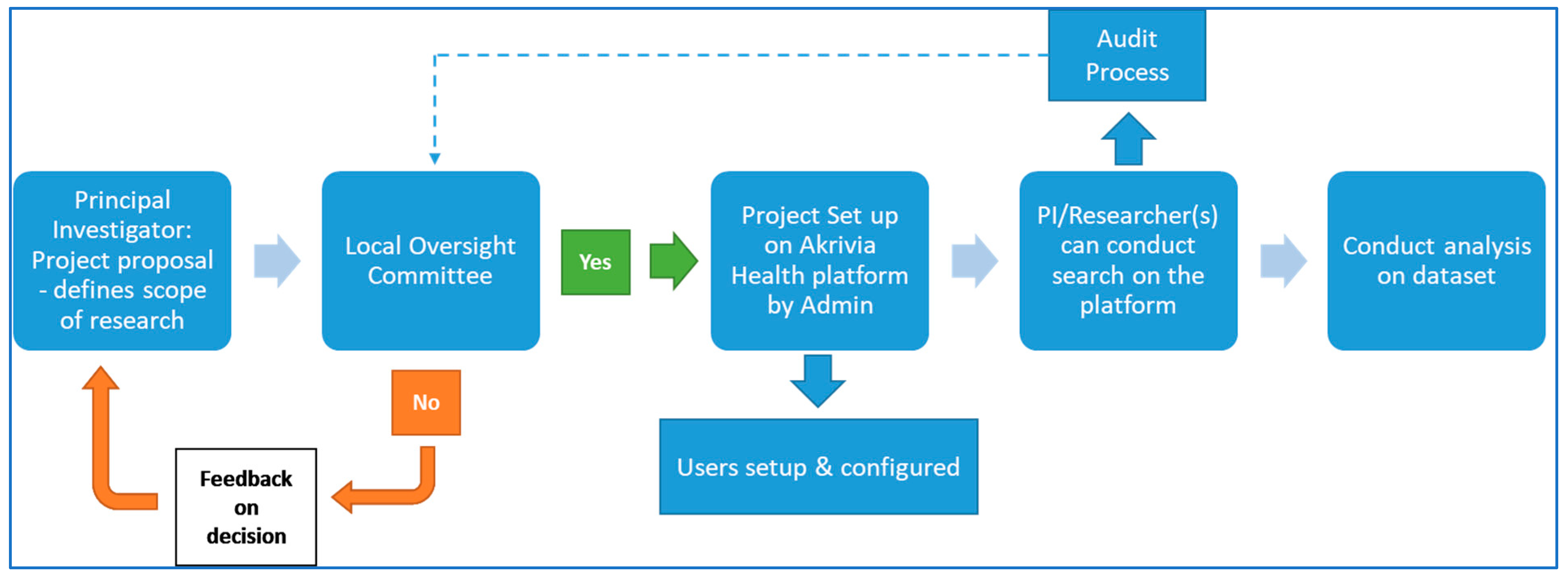
| Advantages | Disadvantages/Limitations |
|---|---|
| Ability to extract relevant information from unstructured text [5,14]. | Challenges processing free text include spelling and grammar errors, non-standard abbreviations and punctuation, acronyms, hedge phrases, and the variability in information recorded from practitioner to practitioner [5,14]. |
| Ability to deal with large volume of data [11,15]. | Need for large volume of training data to initially build models that are robust and can provide clinically meaningful outcomes [11,15]. |
| Can save time, as NLP tools can help provide clinicians with easy and rapid access to relevant patient data [4,5]. | NLP tools can miss out on some context-sensitive meanings and temporal relationships across sentences in a long clinical text. There is still a need for quality control and validation techniques to ensure practical usability of NLP outputs [4,9]. |
| Can identify pertinent clinicopathological parameters of the diseases and key indicators of follow-up outcomes [8]. | Issues with incomplete data, missing information, inconsistencies in describing disease status/staging within or between hospital sites. Differences in practice and followed guidance, and scoring systems are real examples [4,5,11]. |
| Rule-Based NLP Uses manually defined rules, regular expressions, and medical lexicons such as SNOMED CT and UMLS Pros: High precision for structured tasks Cons: Poor scalability, struggles with complex language patterns |
| Traditional Machine Learning (ML) Employs statistical models (support vector machine, naïve Bayes, random forest) Pros: Works well with small datasets Cons: Requires domain-specific feature extraction |
| Deep Learning Utilizes neutral networks (LSTM, GRU, CNN) for text processing Pros: Captures complex patterns and sequential dependencies Cons: Needs large labeled datasets and high computational power |
| Transformers Advanced architectures like BERT, BioBERT, ClinicalBERT, and GPT Pros: Context-aware, state-of-the-art performance Cons: Computationally expensive, requires fine-tuning for medical applications |
| Hybrid Approaches Combine rule-based NLP with deep learning for optimal accuracy Pros: Leverages domain knowledge and data-driven insights Cons: Integration complexity and computational overhead |
| Concept | F1 Score |
|---|---|
| Symptoms and signs | 0.80 |
| Medication | 0.94 |
| Dosage | 0.95 |
| Mental health diagnosis | 0.83 |
| Sleep quality | 0.85 |
| Study Title | Authors | NLP Method Used | Outcome/Validation |
|---|---|---|---|
| Development and evaluation of a de-identification procedure for a case register sourced from mental health electronic records | Fernandes et al. [31] | Pattern matching | De-identified psychiatric database sourced from HER, which protects patient anonymity and increases data availability for research. |
| TextHunter—A User Friendly Tool for Extracting Generic Concepts from Free Text in Clinical Research | Jackson et al. [36] | Concept extraction model | A tool for the creation of training data to construct concept extraction models. Confidence thresholds on accuracy measures like precision and recall were used for validation. |
| Identification of Adverse Drug Events from Free Text Electronic Patient Records and Information in a Large Mental Health Case Register | Iqbal et al. [37] | Text mining | Mined instances of adverse drug events (ADEs) related to antipsychotic therapy from free text content. The tool identified extrapyramidal side effects with >0.85 precision and >0.86 recall during testing. |
| Extracting antipsychotic polypharmacy data from electronic health records: developing and evaluating a novel process | Kadra et al. [38] | Named-entity extraction | Individual instances of antipsychotic prescribing and co-prescriptions were extracted from both structured and free text fields in EHR. Validity was assessed against a manually coded gold standard to establish precision and recall. |
| Negative symptoms in schizophrenia: a study in a large clinical sample of patients using a novel automated method | Patel et al. [39] | Text mining, Aspect based opinion mining | 10 different negative symptoms were ascertained from the clinical records of patients with schizophrenia. Further, associations between demographic aspects (like age, gender, marital status) and hospitalization aspects (like likelihood of admission, readmission, length of admission) were determined. |
| Cannabis use and treatment resistance in first episode psychosis: a natural language processing study | Patel et al. [40] | Keyword extraction | Cannabis use as documented in free-text clinical records was identified and extraction to determine its association with hospital admissions. |
| The characteristics and health needs of pregnant women with schizophrenia compared with bipolar disorder and affective psychoses | Taylor et al. [41] | General Architecture for Text Engineering (GATE) software | Information on medication was extracted using structured indicators describing medication from free text for 3 months before and the first trimester of pregnancy. Two raters cross-checked 5 cases each week until satisfactory reliability was obtained and then a consecutive 22 cases (26 pregnancies) were independently rated for reliability analyses. |
| Detection of Suicidality in Adolescents with Autism Spectrum Disorders: Developing a Natural Language Processing Approach for Use in Electronic Health Records | Downs et al. [42] | Topic modeling | An NLP tool was developed to capture suicidality within clinical texts. Evaluation against human annotators using precision, recall, and F1 score was shown to be above 0.8. |
| Med7: a transferable clinical natural language processing model for electronic health records | Kormilitzin et al. [14] | Named-entity recognition | A model was trained to recognize attributes related to medication. Through transfer learning and fine tuning, the model was shown to achieve an F1 score of 0.944. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delanerolle, G.; Bouchareb, Y.; Shetty, S.; Cavalini, H.; Phiri, P. A Pilot Study Using Natural Language Processing to Explore Textual Electronic Mental Healthcare Data. Informatics 2025, 12, 28. https://doi.org/10.3390/informatics12010028
Delanerolle G, Bouchareb Y, Shetty S, Cavalini H, Phiri P. A Pilot Study Using Natural Language Processing to Explore Textual Electronic Mental Healthcare Data. Informatics. 2025; 12(1):28. https://doi.org/10.3390/informatics12010028
Chicago/Turabian StyleDelanerolle, Gayathri, Yassine Bouchareb, Suchith Shetty, Heitor Cavalini, and Peter Phiri. 2025. "A Pilot Study Using Natural Language Processing to Explore Textual Electronic Mental Healthcare Data" Informatics 12, no. 1: 28. https://doi.org/10.3390/informatics12010028
APA StyleDelanerolle, G., Bouchareb, Y., Shetty, S., Cavalini, H., & Phiri, P. (2025). A Pilot Study Using Natural Language Processing to Explore Textual Electronic Mental Healthcare Data. Informatics, 12(1), 28. https://doi.org/10.3390/informatics12010028


_Bryant.png)




