Gut Microbiota Differences in Infants with Cow-Milk-Induced Allergic Proctocolitis: A Comparative Cross-Sectional Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Eligibility Criteria
- Exclusively breastfed, <6 months old;
- Born at term (≥37 weeks’ gestation), vaginally delivered;
- No history of diarrhoea or gastrointestinal infection;
- No autoimmune, atopic, or metabolic disease in infant or mother;
- No exposure to antibiotics, probiotics, immunosuppressants, or chemotherapeutic agents (infant or mother during pregnancy).
2.3. Faecal Sample Collection
2.4. DNA Extraction and 16S rRNA Amplicon Sequencing
2.5. Bioinformatics and Data Quality Control
2.6. Ethics Approval
3. Statistical Analysis
3.1. Baseline Characteristics
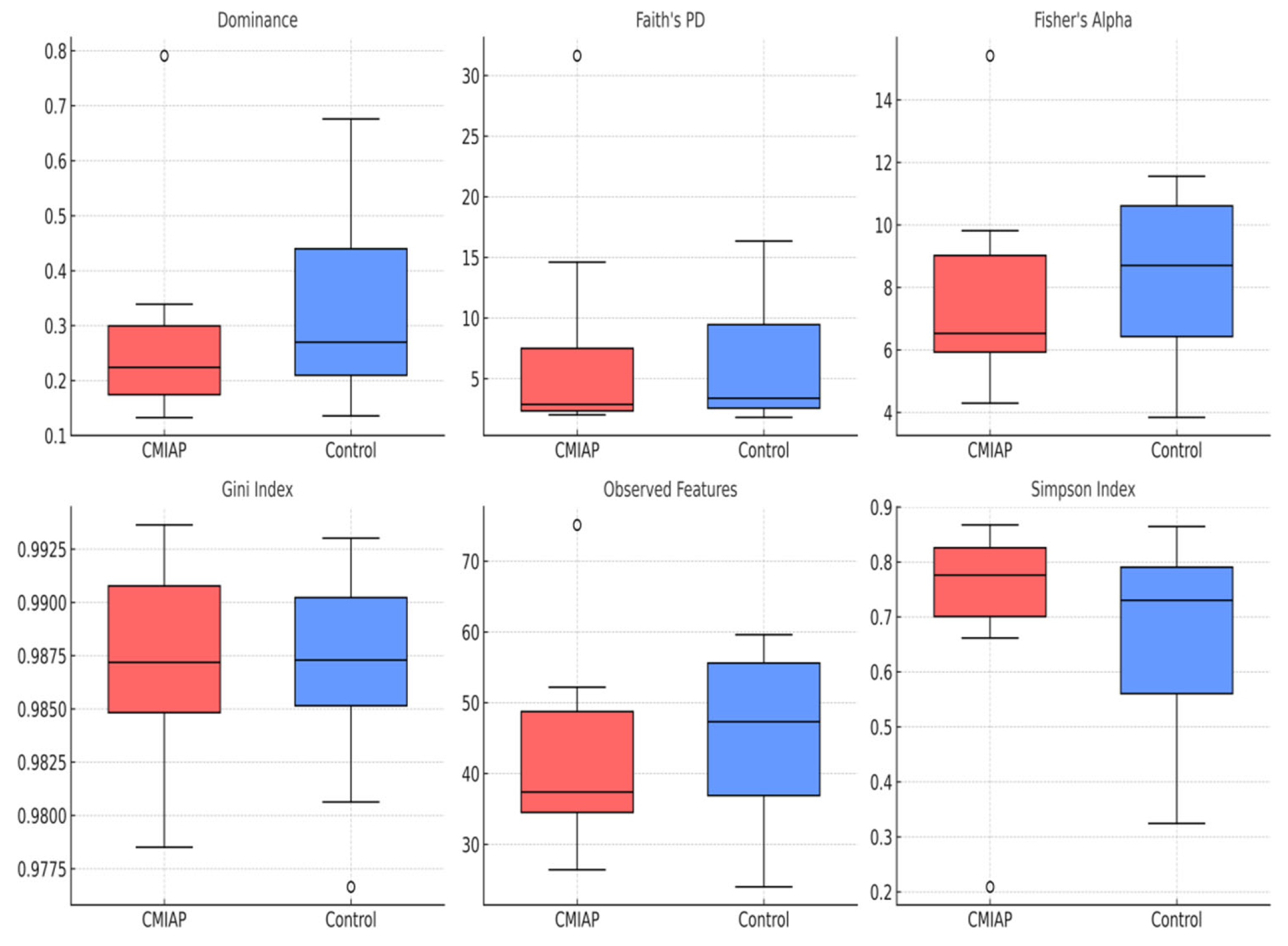
3.2. Alpha-Diversity
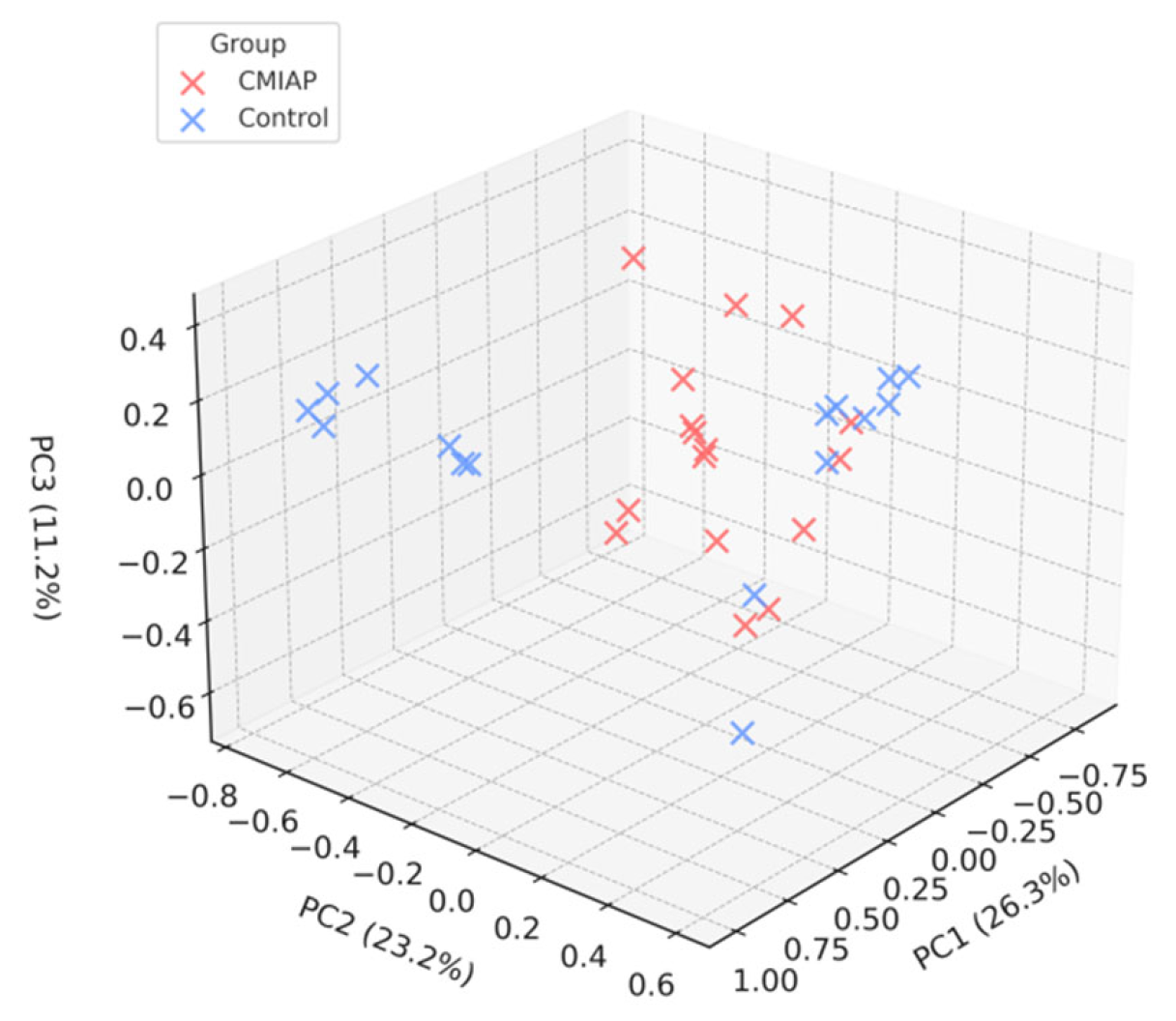
3.3. Beta-Diversity
3.4. Differential Abundance
4. Results
| Characteristic | CMIAP (n = 16) | Control (n = 16) | p-Value |
|---|---|---|---|
| Female sex, n (%) | 8 (50%) | 8 (50%) | 1.00 |
| Gestational age (weeks), mean ± SD | 40 ± 1.2 | 39 ± 1.3 | 0.08 |
| Vaginal delivery, n (%) | 16 (100%) | 16 (100%) | — |
| Exclusively breastfed, n (%) | 16 (100%) | 16 (100%) | — |
| Age at sampling (months), mean ± SD (range) | 2.0 ± 0.44 (1.5–3.0) | 2.4 ± 0.66 (1.5–3.5) | 0.15 |
| Total dairy intake of mothers (mL/day) | 250 ± 80 | 240 ± 75 | 0.72 |
| Yoghurt consumption of mothers (days/week) | 2.3 ± 1.0 | 2.5 ± 1.2 | 0.72 |
4.1. Comparison of Alpha-Diversity Indices
4.2. Beta-Diversity Analysis
4.3. Differential Abundance of Microbial Taxa
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AP | Allergic proctocolitis |
| CMIAP | Cow-milk-induced allergic proctocolitis |
| IgE | Immunoglobulin E |
| SCFA | Shortchain fatty acid |
| Treg | Regulatory T cell |
| IL-10 | Interleukin-10 |
| IL-25 | Interleukin-25 |
| IL-33 | Interleukin-33 |
| TSLP | Thymic stromal lymphopoietin |
| Th2 | T-helper type 2 |
| Th17 | T-helper type 17 |
| ASV | Amplicon sequence variant |
| QIIME 2 | Quantitative insights into microbial ecology version 2 |
| ANCOM–BC | Analysis of composition of microbiomes with bias correction |
| DESeq2 | Differential expression analysis for sequence count data |
| PERMDISP | Permutational analysis of multivariate dispersion |
| PERMANOVA | Permutational multivariate analysis of variance |
| PCoA | Principal coordinate analysis |
| DNA | Deoxyribonucleic acid |
| PCR | Polymerase chain reaction |
| IRB | Institutional review board |
| log2FC | Log2 fold change |
| LPS | Lipopolysaccharide |
| TLR4 | Toll-like receptor 4 |
| FMT | Faecal microbiota transplantation |
| GOS | Galacto-oligosaccharides |
| FOS | Fructo-oligosaccharides |
| SD | Standard deviation |
| padj | Adjusted p-value |
| lfcSE | Log2 fold change standard error |
| Wald stat | Wald test statistic |
| diff_abn | Differentially abundant |
| FC | Fold Change |
References
- Tam, J.S. Food Protein-Induced Proctocolitis and Enteropathy. J. Food Allergy 2020, 2, 55–58. [Google Scholar] [CrossRef]
- Barni, S.; Mori, F.; Giovannini, M.; Liotti, L.; Mastrorilli, C.; Pecoraro, L.; Saretta, F.; Castagnoli, R.; Arasi, S.; Caminiti, L.; et al. Allergic Proctocolitis: Literature Review and Proposal of a Diagnostic–Therapeutic Algorithm. Life 2023, 13, 1824. [Google Scholar] [CrossRef] [PubMed]
- Bahçeci, S.; Töz, P.; Çelik, F.; Can, D. A Different Starting Line for Allergic March: Food Protein-Induced Allergic Proctocolitis. Allergol. Immunopathol. 2023, 51, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Nardo, D. Allergic Proctocolitis Is a Risk Factor for Functional Gastrointestinal Disorders in Children. Pediatrics 2018, 142, S245–S246. [Google Scholar] [CrossRef]
- Carucci, L.; Nocerino, R.; Coppola, S.; Bedogni, G.; Capasso, P.; Giglio, V.; Canani, R. Factors Influencing the Natural History of Non-IgE-Mediated Gastrointestinal Food Allergies in Paediatric Age: A Prospective Multicentre Cohort Study. BMJ Paediatr. Open 2025, 9, e003203. [Google Scholar] [CrossRef] [PubMed]
- Matangkasombut, P.; Padungpak, S.; Thaloengsok, S.; Kamchaisatian, W.; Sasisakulporn, C.; Jotikasthira, W.; Benjaponpitak, S.; Manuyakorn, W. Detection of β-Lactoglobulin in Human Breast Milk 7 Days after Cow Milk Ingestion. Paediatr. Int. Child Health 2017, 37, 199–203. [Google Scholar] [CrossRef]
- Xia, Y.; Cao, H.; Zheng, J.; Chen, L. Claudin-1 Mediated Tight Junction Dysfunction as a Contributor to Atopic March. Front. Immunol. 2022, 13, 927465. [Google Scholar] [CrossRef]
- Nowak-Węgrzyn, A.; Katz, Y.; Mehr, S.S.; Koletzko, S. Non–IgE-Mediated Gastrointestinal Food Allergies. J. Allergy Clin. Immunol. Pract. 2015, 3, 721–728. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, X.; Chen, F.; Rolnik, B.M.; Chleilat, F.; Ling, Z.; Snyder, M.P.; Zhou, X. The Roles and Mechanisms of Gut Microbiota in Food Allergy. Adv. Gut Microbiome Res. 2023, 2023, 9575410. [Google Scholar] [CrossRef]
- Martin, V.M.; Virkud, Y.V.; Dahan, E.; Seay, H.L.; Itzkovits, D.; Vlamakis, H.; Xavier, R.; Shreffler, W.G.; Yuan, Q.; Yassour, M. Longitudinal Disease-Associated Gut Microbiome Differences in Infants with Food Protein-Induced Allergic Proctocolitis. Microbiome 2022, 10, 154. [Google Scholar] [CrossRef]
- Huang, Y.J.; Marsland, B.J.; Bunyavanich, S.; O’Mahony, L.; Leung, D.Y.; Muraro, A.; Fleisher, T.A. The Microbiome in Allergic Disease: Current Understanding and Future Opportunities. J. Allergy Clin. Immunol. 2017, 139, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Gilbert, J.A.; Nagler, C.R. The Role of the Commensal Microbiota in the Regulation of Tolerance to Dietary Allergens. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Nevoral, J.; Rada, V.; Vlková, E.; Bláhová, K.; Bronský, J.; Bubáková, D.; Killer, J. Intestinal Microbiota in Exclusively Breast-Fed Infants with Blood-Streaked Stools. Folia Microbiol. 2009, 54, 167–171. [Google Scholar] [CrossRef]
- Kumagai, H.; Maisawa, S.; Tanaka, M.; Takahashi, M.; Takasago, Y.; Nishijima, A.; Watanabe, S. Intestinal Microbiota and Secretory Immunoglobulin A in Feces of Exclusively Breast-Fed Infants with Blood-Streaked Stools. Microbiol. Immunol. 2012, 56, 657–663. [Google Scholar] [CrossRef]
- Dong, P.; Feng, J.J.; Yan, D.Y.; Lyu, Y.J.; Xu, X. Early-Life Gut Microbiome and Cow’s Milk Allergy: A Prospective Case-Control 6-Month Follow-Up Study. Saudi J. Biol. Sci. 2018, 25, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Li, Z.; Liu, X.; Cheng, Y.; Luo, Y.; Tong, X.; Yuan, L.; Wang, Y.; Sun, J.; Li, L.; et al. Altered Fecal Microbiota Composition Associated with Food Allergy in Infants. Appl. Environ. Microbiol. 2014, 80, 2546–2554. [Google Scholar] [CrossRef]
- Mennini, M.; Fiocchi, A.; Cafarotti, A.; Montesano, M.; Mauro, A.; Villa, M.; Di Nardo, G. Food Protein-Induced Allergic Proctocolitis in Infants: Literature Review and Proposal of a Management Protocol. World Allergy Organ. J. 2020, 13, 100471. [Google Scholar] [CrossRef]
- Venter, C.; Brown, T.; Meyer, R.; Walsh, J.; Shah, N.; Nowak-Węgrzyn, A.; Chen, T.; Fleischer, D.; Heine, R.; Levin, M.; et al. Better Recognition, Diagnosis, and Management of Non–IgE-Mediated Cow’s Milk Allergy in Infancy: iMAP—An International Interpretation of the MAP (Milk Allergy in Primary Care) Guideline. Clin. Transl. Allergy 2017, 7, 26. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An Ultra-Fast All-in-One FASTQ Pre-Processor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 1 May 2024).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.; Wagner, H. Vegan: Community Ecology Package, R Package Version 2.6-4. Available online: https://CRAN.R-project.org/package=vegan (accessed on 1 May 2024).
- Moriki, D.; Francino, M.P.; Koumpagioti, D.; Boutopoulou, B.; Rufián-Henares, J.Á.; Priftis, K.N.; Douros, K. The Role of the Gut Microbiome in Cow’s Milk Allergy: A Clinical Approach. Nutrients 2022, 14, 4537. [Google Scholar] [CrossRef] [PubMed]
- Gavzy, S.J.; Kensiski, A.; Lee, Z.L.; Mongodin, E.F.; Ma, B.; Bromberg, J.S. Bifidobacterium mechanisms of immune modulation and tolerance. Gut Microbes 2023, 15, 2291164. [Google Scholar] [CrossRef]
- Chua, H.H.; Chou, H.C.; Tung, Y.L.; Chiang, B.L.; Liao, C.C.; Liu, H.H.; Ni, Y.H. Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology 2018, 154, 154–167. [Google Scholar] [CrossRef]
- Meadows, V.; Antonio, J.M.; Ferraris, R.P.; Gao, N. Ruminococcus gnavus in the gut: Driver, contributor, or innocent bystander in steatotic liver disease? FEBS J. 2025, 292, 1252–1264. [Google Scholar] [CrossRef]
- Zhan, Z.; Liu, W.; Pan, L.; Bao, Y.; Yan, Z.; Hong, L. Overabundance of Veillonella parvula promotes intestinal inflammation by activating macrophages via the LPS–TLR4 pathway. Cell Death Discov. 2022, 8, 251. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Sarwar, P.F.; Potter, C.; Comstock, S.S.; Klepac-Ceraj, V. Role of human-milk-oligosaccharide-metabolising bacteria in the development of atopic dermatitis/eczema. Front. Pediatr. 2023, 11, 1090048. [Google Scholar] [CrossRef]
- Hendrickx, D.M.; An, R.; Boeren, S.; Mutte, S.K.; Lambert, J.M.; Belzer, C. Assessment of infant outgrowth of cow’s milk allergy in relation to the faecal microbiome and metaproteome. Sci. Rep. 2023, 13, 12029. [Google Scholar] [CrossRef]
- Wurm, P.; Stampfer, L.; Greimel, T.; Leitner, E.; Zechner, E.L.; Bauchinger, S.; Hauer, A.C.P.; Gorkiewicz, G.; Högenauer, C.; Hoffmann, K.M. Gut microbiota dysbiosis in suspected food protein-induced proctocolitis—A prospective comparative cohort trial. J. Pediatr. Gastroenterol. Nutr. 2023, 77, 31–38. [Google Scholar] [CrossRef]
- d’Hennezel, E.; Abubucker, S.; Murphy, L.O.; Cullen, T.W. Total lipopolysaccharide from the human gut microbiome silences Toll-like receptor signalling. mSystems 2017, 2, e00046-17. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Tsolis, R.M.; Bäumler, A.J. Dysbiotic Proteobacteria Expansion: A Microbial Signature of Epithelial Dysfunction. Curr. Opin. Microbiol. 2017, 39, 1–6. [Google Scholar] [CrossRef] [PubMed]
| Purpose | Product | Supplier (City, Country) | Catalogue No. |
|---|---|---|---|
| DNA extraction | QIAamp Fast DNA Stool Mini Kit | Qiagen (Hilden, Germany) | 51604 |
| DNA quantification | Qubit 3 Fluorometer + dsDNA HS Assay Kit | Thermo Fisher Scientific (Waltham, MA, USA) | Q33216/Q32851 |
| PCR amplification | KAPA HiFi HotStart ReadyMix (2×) | Roche (KAPA Biosystems) (Switzerland) | KK2602/KK2601 |
| 16S V3–V4 primers | Custom primers | Oligomer (Custom Synthesis) | — |
| Amplicon cleanup | AMPure XP magnetic beads | Beckman Coulter (Brea, CA, USA) | A63880 |
| Indexing | Nextera XT Index Kit v2 Set A/B | Illumina (San Diego, CA, USA) | FC-131-2001/2002 |
| Library quantification | KAPA Library Quantification Kit for Illumina | Roche (Basel, Switzerland) | KK4824 |
| Size qualification | High-Sensitivity D5000 ScreenTape | Agilent (Santa Clara, CA, USA) | 5067-5592 |
| Sequencing | NovaSeq 6000 System | Illumina (San Diego, CA, USA) | SY-401-1001 |
| Genus (Phylum) | baseMean | log2 Fold Change (CMIAP vs. Control) | p-Value | adj. p-Value |
|---|---|---|---|---|
| Veillonella (Firmicutes) | 8485.01 | +4.73 | 0.0016 | 0.0221 |
| Hungatella (Firmicutes) | 57.24 | +24.99 | <0.0001 | <0.0001 |
| Citrobacter (Proteobacteria) | 303.72 | +10.44 | 0.0007 | 0.0124 |
| Collinsella (Actinobacteriota) | 202.36 | –29.35 | <0.0001 | <0.0001 |
| Limosilactobacillus (Firmicutes) | 40.75 | –8.01 | 0.0025 | 0.0285 |
| Genus (Phylum) | log2 Fold Change (CMIAP vs. Control) | p-Value | q-Value |
|---|---|---|---|
| BifidobacteriumActinobacteriota) | –2.27 | 0.001 | 0.022 |
| (Firmicutes) | +2.69 | <0.0001 | <0.0001 |
| Collinsella (Actinobacteriota) | –2.65 | <0.0001 | 0.097 |
| Limosilactobacillus (Firmicutes) | –1.40 | <0.0001 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haytoglu, Z.; Ozcan, D.; Altintas, D.U. Gut Microbiota Differences in Infants with Cow-Milk-Induced Allergic Proctocolitis: A Comparative Cross-Sectional Study. Children 2025, 12, 734. https://doi.org/10.3390/children12060734
Haytoglu Z, Ozcan D, Altintas DU. Gut Microbiota Differences in Infants with Cow-Milk-Induced Allergic Proctocolitis: A Comparative Cross-Sectional Study. Children. 2025; 12(6):734. https://doi.org/10.3390/children12060734
Chicago/Turabian StyleHaytoglu, Zeliha, Dilek Ozcan, and Derya Ufuk Altintas. 2025. "Gut Microbiota Differences in Infants with Cow-Milk-Induced Allergic Proctocolitis: A Comparative Cross-Sectional Study" Children 12, no. 6: 734. https://doi.org/10.3390/children12060734
APA StyleHaytoglu, Z., Ozcan, D., & Altintas, D. U. (2025). Gut Microbiota Differences in Infants with Cow-Milk-Induced Allergic Proctocolitis: A Comparative Cross-Sectional Study. Children, 12(6), 734. https://doi.org/10.3390/children12060734






