Genotype–Phenotype Analysis of Children with Epilepsy Referred for Whole-Exome Sequencing at a Tertiary Care University Hospital
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection Tool
2.2. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fisher, R.S.; Acevedo, C.; Arzimanoglou, A.; Bogacz, A.; Cross, J.H.; Elger, C.E.; Engel, J., Jr.; Forsgren, L.; French, J.A.; Glynn, M.; et al. ILAE official report: A practical clinical definition of epilepsy. Epilepsia 2014, 55, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Camfield, P.; Camfield, C. Incidence, prevalence and aetiology of seizures and epilepsy in children. Epileptic Disord. 2015, 17, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Cowan, L.D. The epidemiology of the epilepsies in children. Ment. Retard. Dev. Disabil. Res. Rev. 2002, 8, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Al Rajeh, S.; Awada, A.; Bademosi, O.; Ogunniyi, A. The prevalence of epilepsy and other seizure disorders in an Arab population: A community-based study. Seizure 2001, 10, 410–414. [Google Scholar] [CrossRef]
- Strzelczyk, A.; Reese, J.P.; Dodel, R.; Hamer, H.M. Cost of epilepsy: A systematic review. Pharmacoeconomics 2008, 26, 463–476. [Google Scholar] [CrossRef]
- Begley, C.E.; Durgin, T.L. The direct cost of epilepsy in the United States: A systematic review of estimates. Epilepsia 2015, 56, 1376–1387. [Google Scholar]
- Selassie, A.W.; Wilson, D.A.; Wagner, J.L.; Smith, G.; Wannamaker, B.B. Population-based comparative analysis of risk of death in children and adolescents with epilepsy and migraine. Epilepsia 2015, 56, 1957–1965. [Google Scholar]
- Camfield, C.S.; Camfield, P.R.; Veugelers, P.J. Death in children with epilepsy: A population-based study. Lancet 2002, 359, 1891–1895. [Google Scholar] [CrossRef]
- Wirrell, E.C.; Grossardt, B.R.; Wong-Kisiel, L.C.; Nickels, K.C. Incidence and classification of new-onset epilepsy and epilepsy syndromes in children in Olmsted County, Minnesota from 1980 to 2004: A population-based study. Epilepsy Res. 2011, 95, 110–118. [Google Scholar] [CrossRef]
- Brodie, M.J.; Zuberi, S.M.; Scheffer, I.E.; Fisher, R.S. The 2017 ILAE classification of seizure types and the epilepsies: What do people with epilepsy and their caregivers need to know? Epileptic Disord. 2018, 20, 77–87. [Google Scholar] [CrossRef]
- Pal, D.K.; Pong, A.W.; Chung, W.K. Genetic evaluation and counseling for epilepsy. Nat. Rev. Neurol. 2010, 6, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, I.E.; Berkovic, S.; Capovilla, G.; Connolly, M.B.; French, J.; Guilhoto, L.; Hirsch, E.; Jain, S.; Mathern, G.W.; Moshé, S.L.; et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017, 58, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, Z.-J.; Liu, L.; Xu, H.-Q.; Shi, Y.-W.; Yi, Y.-H.; He, N.; Liao, W.-P. Epilepsy-associated genes. Seizure 2017, 44, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Brunklaus, A.; Schorge, S.; Smith, A.D.; Ghanty, I.; Stewart, K.; Gardiner, S.; Du, J.; Pérez-Palma, E.; Symonds, J.D.; Collier, A.C.; et al. SCN1A variants from bench to bedside-improved clinical prediction from functional characterization. Hum. Mutat. 2019, 41, 363–374. [Google Scholar] [CrossRef]
- Nakamura, K.; Kato, M.; Osaka, H.; Yamashita, S.; Nakagawa, E.; Haginoya, K.; Tohyama, J.; Okuda, M.; Wada, T.; Shimakawa, S.; et al. Clinical spectrum of SCN2A mutations expanding to Ohtahara syndrome. Neurology 2013, 81, 992–998. [Google Scholar] [CrossRef]
- Ream, M.A.; Patel, A.D. Obtaining genetic testing in pediatric epilepsy. Epilepsia 2015, 56, 1505–1514. [Google Scholar] [CrossRef]
- Kuersten, M.; Tacke, M.; Gerstl, L.; Hoelz, H.; Stülpnagel, C.; Borggraefe, I. Antiepileptic therapy approaches in KCNQ2 related epilepsy: A systematic review. Eur. J. Med. Genet. 2020, 63, 103628. [Google Scholar] [CrossRef]
- Bindu, P.S.; Sonam, K.; Govindaraj, P.; Govindaraju, C.; Chiplunkar, S.; Nagappa, M.; Kumar, R.; Vekhande, C.C.; Arvinda, H.R.; Gayathri, N.; et al. Outcome of epilepsy in patients with mitochondrial disorders: Phenotype genotype and magnetic resonance imaging correlations. Clin. Neurol. Neurosurg. 2018, 164, 182–189. [Google Scholar] [CrossRef]
- Wu, C.-C.; Tsai, M.-H.; Chu, Y.-J.; Weng, W.-C.; Fan, P.-C.; Lee, W.-T. The role of targeted gene panel in pediatric drug-resistant epilepsy. Epilepsy Behav. 2020, 106, 107003. [Google Scholar] [CrossRef]
- Alonazi, N.A.; Alnemri, A.; El Melegy, E.; Mohamed, N.; Talaat, I.; Hosny, A.; Alonazi, A.; Mohamed, S. Clinical characteristics and aetiology of early childhood epilepsy: A single centre experience in Saudi Arabia. Sudan. J. Paediatr. 2018, 18, 57–62. [Google Scholar]
- Babtain, F.A. Impact of a family history of epilepsy on the diagnosis of epilepsy in Southern Saudi Arabia. Seizure 2013, 22, 542–547. [Google Scholar] [CrossRef][Green Version]
- Nashabat, M.; Al Qahtani, X.S.; Almakdob, S.; Altwaijri, W.; Ba-Armah, D.M.; Hundallah, K.; Al Hashem, A.; Al Tala, S.; Maddirevula, S.; Alkuraya, F.S.; et al. The landscape of early infantile epileptic encephalopathy in a consanguineous population. Seizure 2019, 69, 154–172. [Google Scholar] [CrossRef]
- Subki, A.H.; Alasmari, A.S.; Jan, F.M.; Moria, F.A.; Jan, M.M. Reflex Seizures Triggered by Diaper Change in Dravet Syndrome. Can. J. Neurol. Sci. 2016, 43, 585–587. [Google Scholar] [CrossRef][Green Version]
- AlSaif, S.; Umair, M.; Alfadhel, M. Biallelic SCN2A Gene Mutation Causing Early Infantile Epileptic Encephalopathy: Case Report and Review. J. Cent. Nerv. Syst. Dis. 2019, 11, 1179573519849938. [Google Scholar]
- Mir, A.; Chaudhary, M.; Alkhaldi, H.; Alhazmi, R.; Albaradie, R.; Housawi, Y. Epilepsy in patients with EAST syndrome caused by mutation in the KCNJ10. Brain Dev. 2019, 41, 706–715. [Google Scholar] [CrossRef]
- Faheem, M.; INaseer, M.; GChaudhary, A.; AKumosani, T.; Rasool, M.; AAlgahtani, H.; Bibi, F.; AKamal, M.; Al-Qahtani, M. Array-comparative genomic hybridization analysis of a cohort of Saudi patients with epilepsy. CNS Neurol. Disord. Drug Targets 2015, 14, 468–475. [Google Scholar] [CrossRef]
- Naseer, M.I.; Alwasiyah, M.K.; Abdulkareem, A.A.; Bajammal, R.A.; Trujillo, C.; Abu-Elmagd, M.; Jafri, M.A.; Chaudhary, A.G.; Al-Qahtani, M.H. A novel homozygous mutation in SZT2 gene in Saudi family with developmental delay, macrocephaly and epilepsy. Genes Genom. 2018, 40, 1149–1155. [Google Scholar]
- Almobarak, S.; Almuhaizea, M.; Abukhaled, M.; Alyamani, S.; Dabbagh, O.; Chedrawi, A.; Khan, S.; Aldhalaan, H. Tuberous Sclerosis Complex: Clinical Spectrum and Epilepsy: A Retrospective Chart Review Study. Transl. Neurosci. 2018, 9, 154–160. [Google Scholar] [CrossRef]
- Anazi, S.; Maddirevula, S.; Faqeih, E.; AlSedairy, H.; Alzahrani, F.; Shamseldin, H.E.; Patel, N.; Hashem, M.; Ibrahim, N.; Abdulwahab, F.; et al. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry 2017, 22, 615–624. [Google Scholar] [CrossRef]
- Pastorino, G.M.G.; Operto, F.F.; Padovano, C.; Vivenzio, V.; Scuoppo, C.; Pastorino, N.; Roccella, M.; Vetri, L.; Carotenuto, M.; Coppola, G. Social Cognition in Neurodevelopmental Disorders and Epilepsy. Front. Neurol. 2021, 14, 658823. [Google Scholar] [CrossRef]
- Bashiri, F.A.; Hudairi, A.; Al Ghamdi, M.; Mahmoud, A.A. A Case of Neonatal Epileptic Encephalopathy due to SCN2A Mutation Responsive to a Ketogenic Diet. J. Pediatr. Epilepsy 2018, 7, 148–151. [Google Scholar] [CrossRef]
- Han, C.; Alkhater, R.; Froukh, T.; Minassian, A.G.; Galati, M.; Liu, R.H.; Fotouhi, M.; Sommerfeld, J.; Alfrook, A.J.; Marshall, C.; et al. Epileptic Encephalopathy Caused by Mutations in the Guanine Nucleotide Exchange Factor DENND5A. Am. J. Hum. Genet. 2016, 99, 1359–1367. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ferri, L.; Funghini, S.; Fioravanti, A.; Biondi, E.; la Marca, G.; Guerrini, R.; Donati, M.; Morrone, A. Aminoacylase I deficiency due to ACY1 mRNA exon skipping. Clin. Genet. 2014, 86, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Samanta, D. PCDH19-Related Epilepsy Syndrome: A Comprehensive Clinical Review. Pediatr. Neurol. 2020, 105, 3–9. [Google Scholar] [CrossRef]
- Hesse, A.N.; Bevilacqua, J.; Shankar, K.; Reddi, H.V. Retrospective genotype-phenotype analysis in a 305 patient cohort referred for testing of a targeted epilepsy panel. Epilepsy Res. 2018, 144, 53–61. [Google Scholar] [CrossRef]
- Jain, P.; Andrade, D.; Donner, E.; Dyment, D.; Prasad, A.N.; Goobie, S.; Boycott, K.; Lines, M.; Snead, O.C. Development of Criteria for Epilepsy Genetic Testing in Ontario, Canada. Can. J. Neurol. Sci. 2019, 46, 7–13. [Google Scholar] [CrossRef]
- Alsharif, M.M.; El-Fetoh, N.M.A.; Ali, G.Y.; Alanazi, K.F.; Alanazi, A.N.; FalahAlanazi, O.; Alshalan, M.H.; Alfuhigi, Z.D.; Alruwaili, A.E.; Alhazmi, R.S.; et al. Epilepsy as a health problem among school children in Turaif, Northern Saudi Arabia, 2017. Electron. Physician 2017, 9, 5036–5042. [Google Scholar] [CrossRef][Green Version]
- Margari, L.; Legrottaglie, A.R.; Vincenti, A.; Coppola, G.; Operto, F.F.; Buttiglione, M.; Cassano, A.; Bartolomeo, N.; Mariggiò, M.A. Association between SCN1A gene polymorphisms and drug resistant epilepsy in pediatric patients. Seizure 2018, 55, 30–35. [Google Scholar] [CrossRef] [PubMed]
| Mutation (N = 45) | No Mutation (N = 8) | Total (N = 53) | p-Value | |
|---|---|---|---|---|
| Age (years) | ||||
| Mean ± standard deviation | 9.4 ± 7.1 | 9.3 ± 7.3 | 9.3 ± 7.2 | 0.952 |
| Median and inter-quartile range | 7 (4–15) | 9 (3.5–12.5) | 7 (4–15) | 0.893 |
| Gender | ||||
| Male | 22 (48.9%) | 4 (50.0%) | 26 (49.1%) | >0.99 |
| Female | 23 (51.1%) | 4 (50.0%) | 27 (50.9%) | |
| Seizure type at initial diagnosis | ||||
| Generalized, tonic–clonic | 9 (20.5%) | 1 (12.5%) | 10 (19.2%) | 0.120 |
| Generalized, absence | 3 (6.8%) | 0 (0.0%) | 3 (5.8%) | |
| Generalized, non-specified | 1 (2.3%) | 0 (0.0%) | 1 (1.9%) | |
| Focal | 14 (31.8%) | 1 (12.5%) | 15 (28.8%) | |
| Infantile spasm | 7 (15.9%) | 1 (12.5%) | 8 (15.4%) | |
| Drop attack | 2 (4.5%) | 0 (0.0%) | 2 (3.8%) | |
| Nocturnal seizure | 0 (0.0%) | 2 (25.0%) | 2 (3.8%) | |
| Others | 8 (18.2%) | 3 (37.5%) | 11 (21.2%) | |
| Other seizure type | ||||
| Generalized, tonic–clonic | 12 (48.0%) | 0 (0.0%) | 12 (44.4%) | 0.140 |
| Generalized, absence | 1 (4.0%) | 0 (0.0%) | 1 (3.7%) | |
| Generalized, non-specified | 1 (4.0%) | 0 (0.0%) | 1 (3.7%) | |
| Focal | 6 (24.0%) | 1 (50.0%) | 7 (25.9%) | |
| Infantile spasm | 0 (0.0%) | 1 (50.0%) | 1 (3.7%) | |
| Drop attack | 2 (8.0%) | 0 (0.0%) | 2 (7.4%) | |
| Nocturnal seizure | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| Others | 3 (12.0%) | 0 (0.0%) | 3 (11.1%) | |
| Age at onset of epilepsy (months) | ||||
| Median and inter-quartile range | 4 (1.4–13.5) | 6 (6–17.2) | 5 (2.0–13.5) | 0.286 |
| Development before seizure onset | ||||
| Normal | 12 (30.0%) | 3 (37.5%) | 15 (31.3%) | 0.692 |
| Delayed | 28 (70.0%) | 5 (62.5%) | 33 (68.8%) | |
| History | ||||
| Parents consanguinity | 14 (31.8%) | 1 (12.5%) | 15 (28.8%) | 0.412 |
| Family history of epilepsy | 15 (33.3%) | 2 (25.0%) | 17 (32.1%) | >0.99 |
| Mutation (N = 45) | No Mutation (N = 8) | Total (N = 53) | p-Value | |
|---|---|---|---|---|
| Neurological features | ||||
| None | 11 (24.4%) | 3 (37.5%) | 14 (26.4%) | 0.882 |
| 1–2 features | 22 (48.9%) | 3 (37.5%) | 25 (47.2%) | |
| 3 or more features | 12 (26.7%) | 2 (25.0%) | 14 (26.4%) | |
| Neurological features | ||||
| Developmental delay | 20 (44.4%) | 2 (25.0%) | 22 (41.5%) | 0.445 |
| Microcephaly | 9 (20.0%) | 1 (12.5%) | 10 (18.9%) | >0.99 |
| Hypotonia | 7 (15.6%) | 1 (12.5%) | 8 (15.1%) | >0.99 |
| Dysmorphic features | 7 (15.6%) | 0 (0.0%) | 7 (13.2%) | 0.577 |
| Movement disorders | 5 (11.1%) | 1 (12.5%) | 6 (11.3%) | >0.99 |
| Vision problems | 5 (11.1%) | 0 (0.0%) | 5 (9.4%) | >0.99 |
| Choreoathetosis | 2 (4.4%) | 2 (25.0%) | 4 (7.5%) | 0.104 |
| Nystagmus | 3 (6.7%) | 1 (12.5%) | 4 (7.5%) | 0.491 |
| Speech problems | 3 (6.7%) | 1 (12.5%) | 4 (7.5%) | 0.491 |
| Ataxia | 0 (0.0%) | 2 (25.0%) | 2 (3.8%) | 0.02 |
| Spastic quadriplegia | 2 (4.4%) | 0 (0.0%) | 2 (3.8%) | >0.99 |
| Others | 13 (28.9%) | 1 (12.5%) | 14 (26.4%) | 0.665 |
| Cognitive/language delay | ||||
| Cognitive | 11 (24.4%) | 2 (25.0%) | 13 (24.5%) | 0.953 |
| Language | 10 (22.2%) | 2 (25.0%) | 12 (22.6%) | |
| Cognitive/language | 8 (17.8%) | 2 (25.0%) | 10 (18.9%) | |
| Neither | 16 (35.6%) | 2 (25.0%) | 18 (34.0%) | |
| Abnormal behavioral features | ||||
| Any feature | 13 (28.9%) | 1 (12.5%) | 14 (26.4%) | 0.665 |
| Attention deficit hyperactivity disorder (ADHD) | 4 (8.9%) | 0 (0.0%) | 4 (7.5%) | >0.99 |
| Autism spectrum disorder (ASD) | 2 (4.4%) | 0 (0.0%) | 2 (3.8%) | >0.99 |
| Hyperactivity disorder | 2 (4.4%) | 0 (0.0%) | 2 (3.8%) | >0.99 |
| Aggressive behavior | 1 (2.2%) | 0 (0.0%) | 1 (1.9%) | >0.99 |
| Anxiety disorder | 1 (2.2%) | 0 (0.0%) | 1 (1.9%) | >0.99 |
| Phobia | 1 (2.2%) | 0 (0.0%) | 1 (1.9%) | >0.99 |
| Other abnormal behavior | 3 (6.7%) | 1 (12.5%) | 4 (7.5%) | 0.491 |
| Mutation (N = 45) | No Mutation (N = 8) | Total (N = 53) | p-Value | |
|---|---|---|---|---|
| EEG at initial diagnosis | ||||
| Normal | 10 (22.2%) | 1 (12.5%) | 11 (20.8%) | 0.490 |
| Abnormal | 31 (68.9%) | 5 (62.5%) | 36 (67.9%) | |
| Not available | 4 (8.9%) | 2 (25.0%) | 6 (11.3%) | |
| EEG at initial diagnosis, positive findings | ||||
| Abnormal background activity | 5 (16.1%) | 1 (20.0%) | 6 (16.7%) | 0.450 |
| Epileptiform discharges | 11 (35.5%) | 3 (60.0%) | 14 (38.9%) | |
| Both | 15 (48.4%) | 1 (20.0%) | 16 (44.4%) | |
| Follow-up EEG | ||||
| Normal | 13 (31.7%) | 1 (12.5%) | 14 (28.6%) | 0.473 |
| Abnormal | 19 (46.3%) | 4 (50.0%) | 23 (46.9%) | |
| No follow-up | 9 (22.0%) | 3 (37.5%) | 12 (24.5%) | |
| Follow-up EEG, positive findings | ||||
| Abnormal background activity | 6 (31.6%) | 0 (0.0%) | 6 (26.1%) | 0.419 |
| Epileptiform discharges | 6 (31.6%) | 1 (25.0%) | 7 (30.4%) | |
| Both | 7 (36.8%) | 3 (75.0%) | 10 (43.5%) | |
| Brain MRI | ||||
| Normal | 22 (48.9%) | 2 (25.0%) | 24 (45.3%) | 0.373 |
| Abnormal | 22 (48.9%) | 6 (75.0%) | 28 (52.8%) | |
| Not performed | 1 (2.2%) | 0 (0.0%) | 1 (1.9%) | |
| Positive MRI findings | ||||
| Generalized brain atrophy | 10 (45.5%) | 0 (0.0%) | 10 (35.7%) | 0.062 |
| Ventriculomegaly | 5 (22.7%) | 1 (16.7%) | 6 (21.4%) | >0.99 |
| Abnormal signal intensity | 6 (27.3%) | 0 (0.0%) | 6 (21.4%) | 0.289 |
| Thinning/dysgenesis of corpus callosum | 6 (27.3%) | 0 (0.0%) | 6 (21.4%) | 0.289 |
| Cerebellar atrophy | 2 (9.1%) | 1 (16.7%) | 3 (10.7%) | 0.530 |
| Lack of/delayed myelination | 1 (4.5%) | 1 (16.7%) | 2 (7.1%) | 0.389 |
| Hematoma/hemorrhage | 2 (9.1%) | 0 (0.0%) | 2 (7.1%) | >0.99 |
| Others | 8 (36.4%) | 4 (66.7%) | 12 (42.9%) | 0.354 |
| Calcification | 1 (4.5%) | 0 (0.0%) | 1 (3.6%) | -- |
| Cerebral and cerebellar edema | 1 (4.5%) | 0 (0.0%) | 1 (3.6%) | -- |
| Chiari II malformation | 0 (0.0%) | 1 (16.7%) | 1 (3.6%) | -- |
| FLAIR vascular hyperintensity | 0 (0.0%) | 1 (16.7%) | 1 (3.6%) | -- |
| Gliosis | 0 (0.0%) | 1 (16.7%) | 1 (3.6%) | -- |
| Gray matter heterotropia | 0 (0.0%) | 1 (16.7%) | 1 (3.6%) | -- |
| Hemosiderosis | 1 (4.5%) | 0 (0.0%) | 1 (3.6%) | -- |
| Hydrocephalus | 1 (4.5%) | 0 (0.0%) | 1 (3.6%) | -- |
| Ischemia | 1 (4.5%) | 0 (0.0%) | 1 (3.6%) | -- |
| Mesial temporal sclerosis | 1 (4.5%) | 0 (0.0%) | 1 (3.6%) | -- |
| Simplified gyral pattern | 0 (0.0%) | 1 (16.7%) | 1 (3.6%) | -- |
| Subdural hygroma | 1 (4.5%) | 0 (0.0%) | 1 (3.6%) | -- |
| White matter atrophy | 1 (4.5%) | 0 (0.0%) | 1 (3.6%) | -- |
| Mutation (N = 45) | No Mutation (N = 8) | Total (N = 53) | p-Value | |
|---|---|---|---|---|
| Current ASMs | ||||
| Levetiracetam | 22 (48.9%) | 5 (62.5%) | 27 (50.9%) | 0.704 |
| Topiramate | 13 (28.9%) | 0 (0.0%) | 13 (24.5%) | 0.176 |
| Valproic acid | 9 (20.0%) | 2 (25.0%) | 11 (20.8%) | 0.665 |
| Phenobarbitone | 8 (17.8%) | 1 (12.5%) | 9 (17.0%) | >0.99 |
| Carbamazepine | 7 (15.6%) | 1 (12.5%) | 8 (15.1%) | >0.99 |
| Lamotrigine | 7 (15.6%) | 0 (0.0%) | 7 (13.2%) | 0.577 |
| Clonazepam | 6 (13.3%) | 0 (0.0%) | 6 (11.3%) | 0.574 |
| Lorazepam/clobazam | 3 (6.7%) | 0 (0.0%) | 3 (5.7%) | >0.99 |
| Vigabatrin | 2 (4.4%) | 0 (0.0%) | 2 (3.8%) | >0.99 |
| Phenytoin | 1 (2.2%) | 0 (0.0%) | 1 (1.9%) | >0.99 |
| Oxcarbazepine | 1 (2.2%) | 0 (0.0%) | 1 (1.9%) | >0.99 |
| Others | 8 (17.8%) | 0 (0.0%) | 8 (15.1%) | 0.333 |
| Tried ASMs | ||||
| Phenobarbitone | 9 (20.0%) | 1 (12.5%) | 10 (18.9%) | >0.99 |
| Carbamazepine | 6 (13.3%) | 1 (12.5%) | 7 (13.2%) | >0.99 |
| Levetiracetam | 5 (11.1%) | 1 (12.5%) | 6 (11.3%) | >0.99 |
| Topiramate | 5 (11.1%) | 1 (12.5%) | 6 (11.3%) | >0.99 |
| Valproic acid | 5 (11.1%) | 0 (0.0%) | 5 (9.4%) | >0.99 |
| Clonazepam | 3 (6.7%) | 0 (0.0%) | 3 (5.7%) | >0.99 |
| Vigabatrin | 3 (6.7%) | 0 (0.0%) | 3 (5.7%) | >0.99 |
| ACTH | 3 (6.7%) | 0 (0.0%) | 3 (5.7%) | >0.99 |
| Lamotrigine | 2 (4.4%) | 0 (0.0%) | 2 (3.8%) | >0.99 |
| Phenytoin | 1 (2.2%) | 0 (0.0%) | 1 (1.9%) | >0.99 |
| Others | 2 (4.4%) | 1 (12.5%) | 3 (5.7%) | 0.394 |
| Seizure outcome | ||||
| Controlled with ASMs | 5 (11.9%) | 2 (25.0%) | 7 (14.0%) | 0.717 |
| Controlled without ASMs | 20 (47.6%) | 5 (62.5%) | 25 (50.0%) | |
| Partially controlled | 7 (16.7%) | 0 (0.0%) | 7 (14.0%) | |
| Not controlled | 7 (16.7%) | 1 (12.5%) | 8 (16.0%) | |
| Death | 3 (7.1%) | 0 (0.0%) | 3 (6.0%) |
| Mutation | N (%) | Age at Onset | Presentation at Initial Diagnosis | DD | ID | S/LD | PC | FH | Control with ASMs |
|---|---|---|---|---|---|---|---|---|---|
| SCN1A | 4 (8.9%) | 2 to 12 months | Dravet syndrome (1/4) Febrile then non-febrile GTC (1/4) GTC (1/4) Focal seizures (1/4) | 1/4 | 2/4 | 3/4 | 2/4 | 2/4 | 2/4 |
| DENND5A | 3 (6.7%) | 2 to 4 months | Focal seizures (2/3) Apnea and starring (1/3) | 3/3 | 2/3 | 3/3 | 2/3 | 2/3 | 2/3 |
| KCNQ2 | 2 (4.4%) | 2 days to 4 months | Benign familial neonatal convulsions (1/2) Focal seizures (2/2) | 0/2 | 0/2 | 0/2 | 0/2 | 2/2 | 2/2 |
| ACY1 | 2 (4.4%) | 4 months | Infantile spasms (2/2) | 1/2 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 |
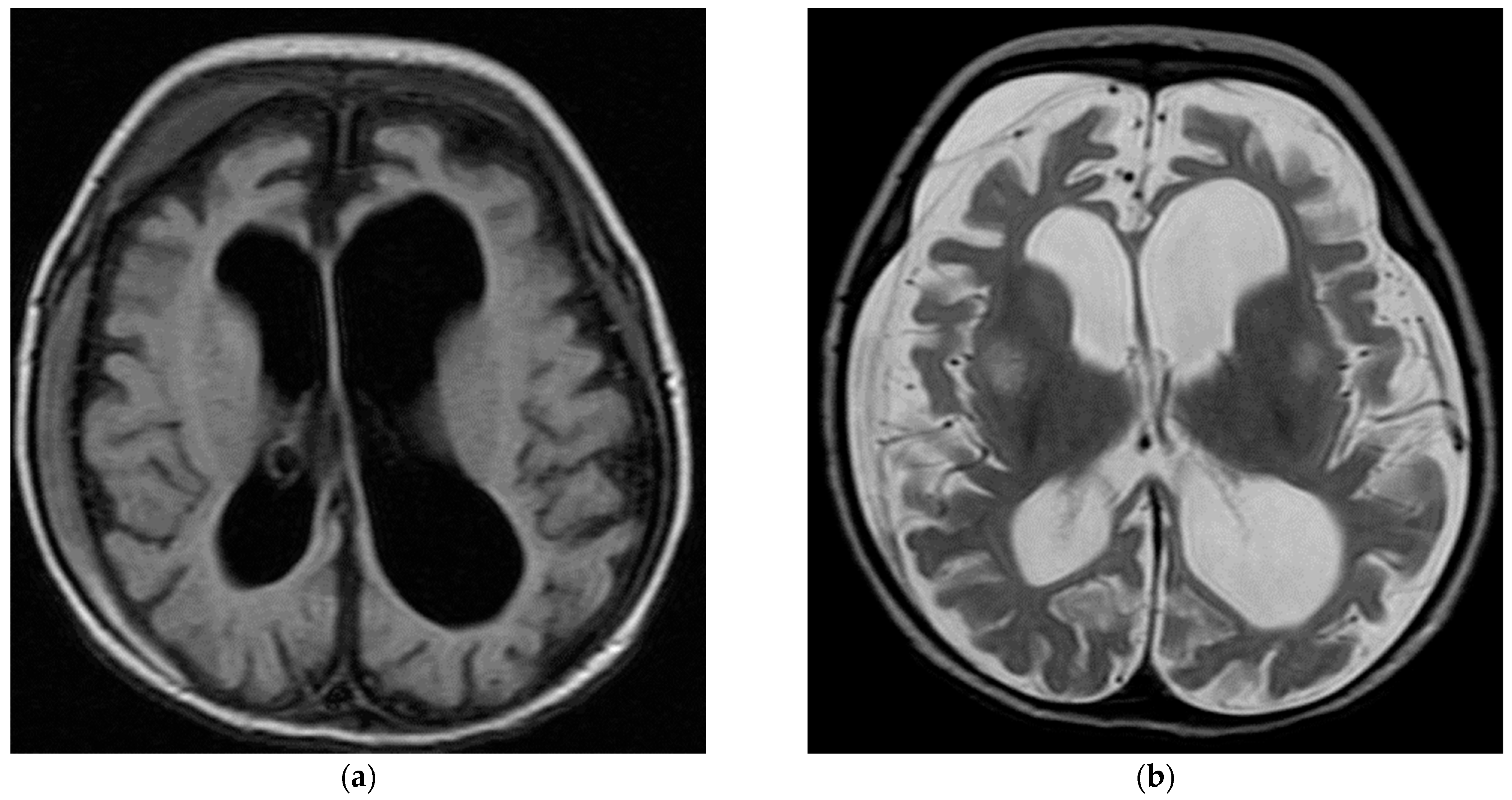
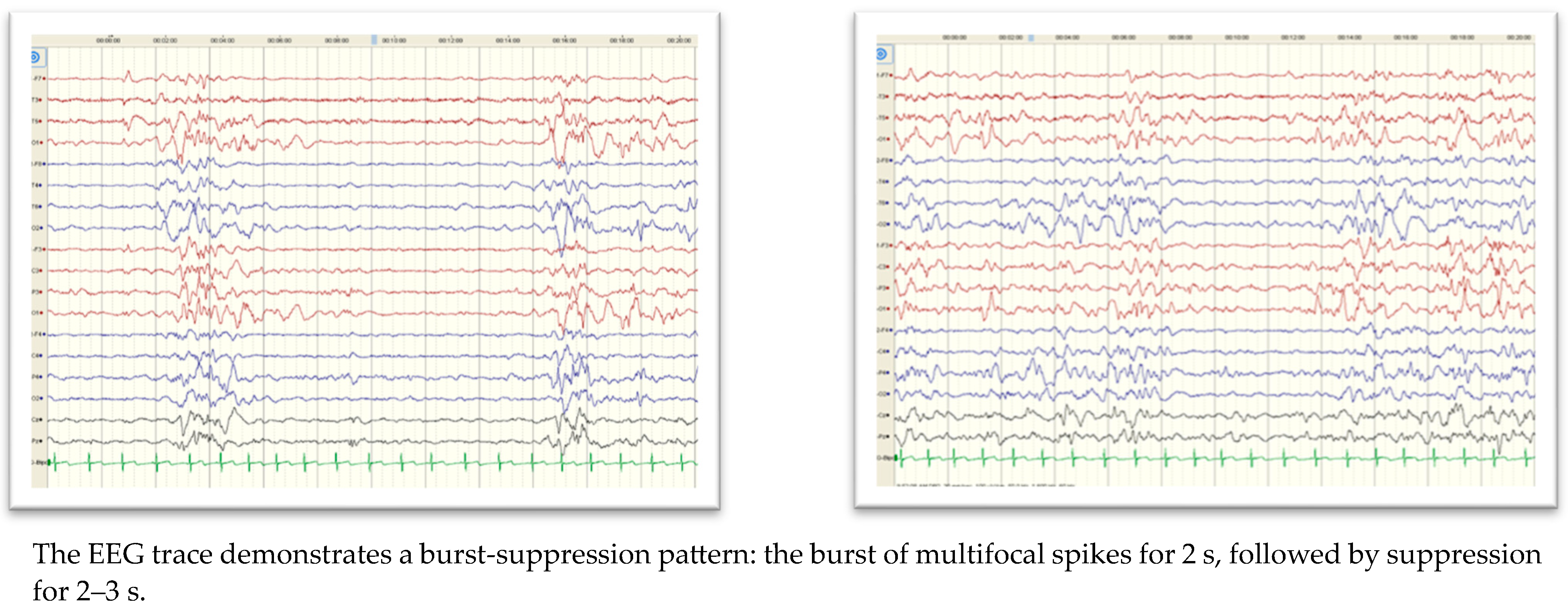
| SCN2A | 2 (4.4%) | 8 days to 3 months | Blinking of eyes and twitching of the mouth (1/2) Apnea with cyanosis and up rolling of eyes with tonic movement (1/2) Burst-suppression pattern in the EEG (1/2) | 1/2 | 0/2 | 1/2 | 0/2 | 0/2 | 1/2 |
| PCDH19 | 2 (4.4%) | 9 months to 9 years | Focal seizures (N = 1) Drop-like attacks (N = 1) | 2/2 | 2/2 | 1/2 | 0/2 | 0/2 | 1/2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashiri, F.A.; AlSheikh, R.; Hamad, M.H.; Alsheikh, H.; Alsheikh, R.A.; Kentab, A.; AlTheeb, N.; Alghamdi, M. Genotype–Phenotype Analysis of Children with Epilepsy Referred for Whole-Exome Sequencing at a Tertiary Care University Hospital. Children 2023, 10, 1334. https://doi.org/10.3390/children10081334
Bashiri FA, AlSheikh R, Hamad MH, Alsheikh H, Alsheikh RA, Kentab A, AlTheeb N, Alghamdi M. Genotype–Phenotype Analysis of Children with Epilepsy Referred for Whole-Exome Sequencing at a Tertiary Care University Hospital. Children. 2023; 10(8):1334. https://doi.org/10.3390/children10081334
Chicago/Turabian StyleBashiri, Fahad A., Rawan AlSheikh, Muddathir H. Hamad, Hamad Alsheikh, Rana Abdullah Alsheikh, Amal Kentab, Najd AlTheeb, and Malak Alghamdi. 2023. "Genotype–Phenotype Analysis of Children with Epilepsy Referred for Whole-Exome Sequencing at a Tertiary Care University Hospital" Children 10, no. 8: 1334. https://doi.org/10.3390/children10081334
APA StyleBashiri, F. A., AlSheikh, R., Hamad, M. H., Alsheikh, H., Alsheikh, R. A., Kentab, A., AlTheeb, N., & Alghamdi, M. (2023). Genotype–Phenotype Analysis of Children with Epilepsy Referred for Whole-Exome Sequencing at a Tertiary Care University Hospital. Children, 10(8), 1334. https://doi.org/10.3390/children10081334






