Inherited Metabolic Diseases from Past to Present: A Bibliometric Analysis (1968–2023)
Abstract
1. Introduction
2. Materials and Methods
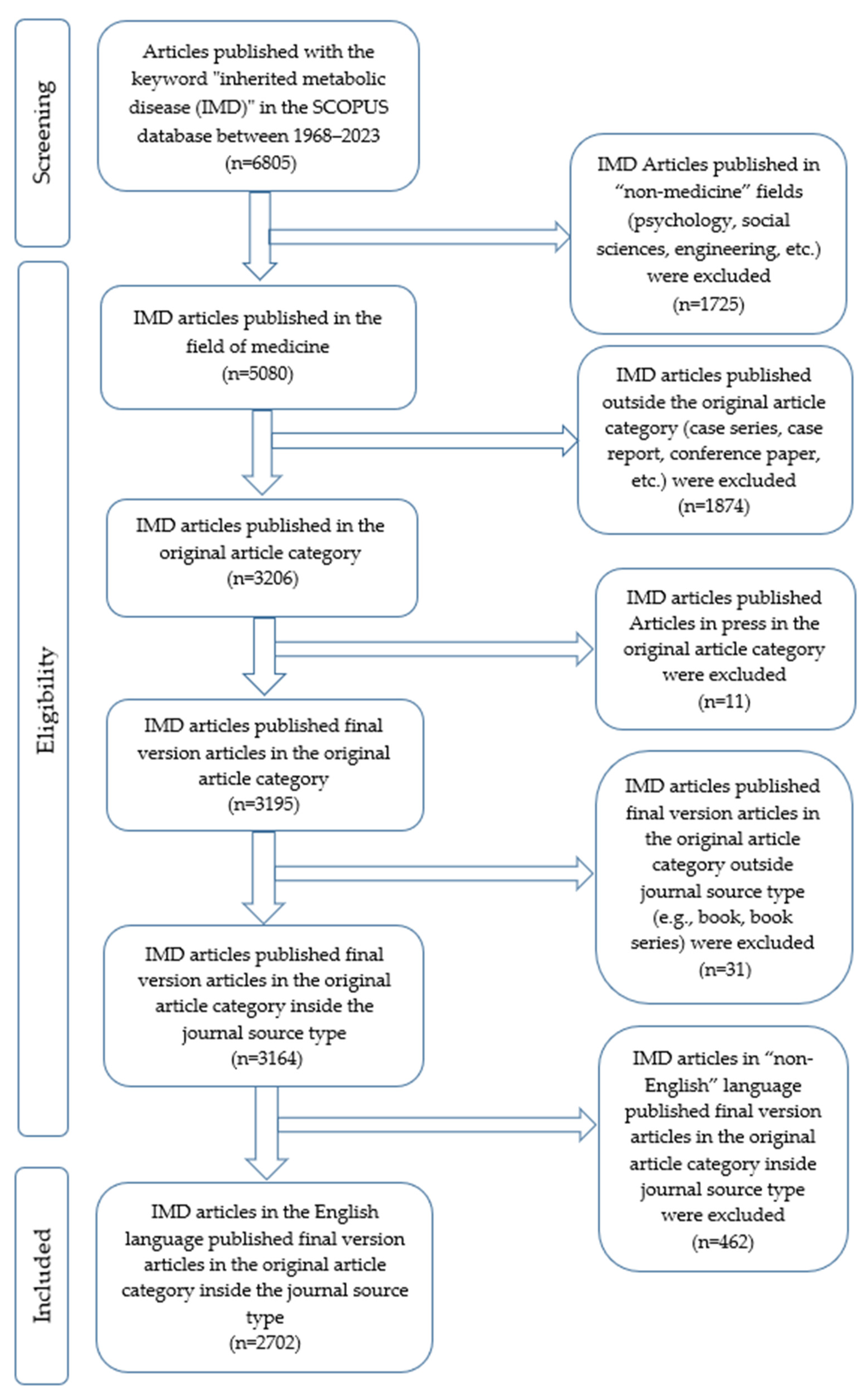
2.1. Study Design and Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Data Collection and Statistics
3. Results
3.1. Analysis of Data
3.1.1. General Analysis of Published Articles on Inherited Metabolic Diseases
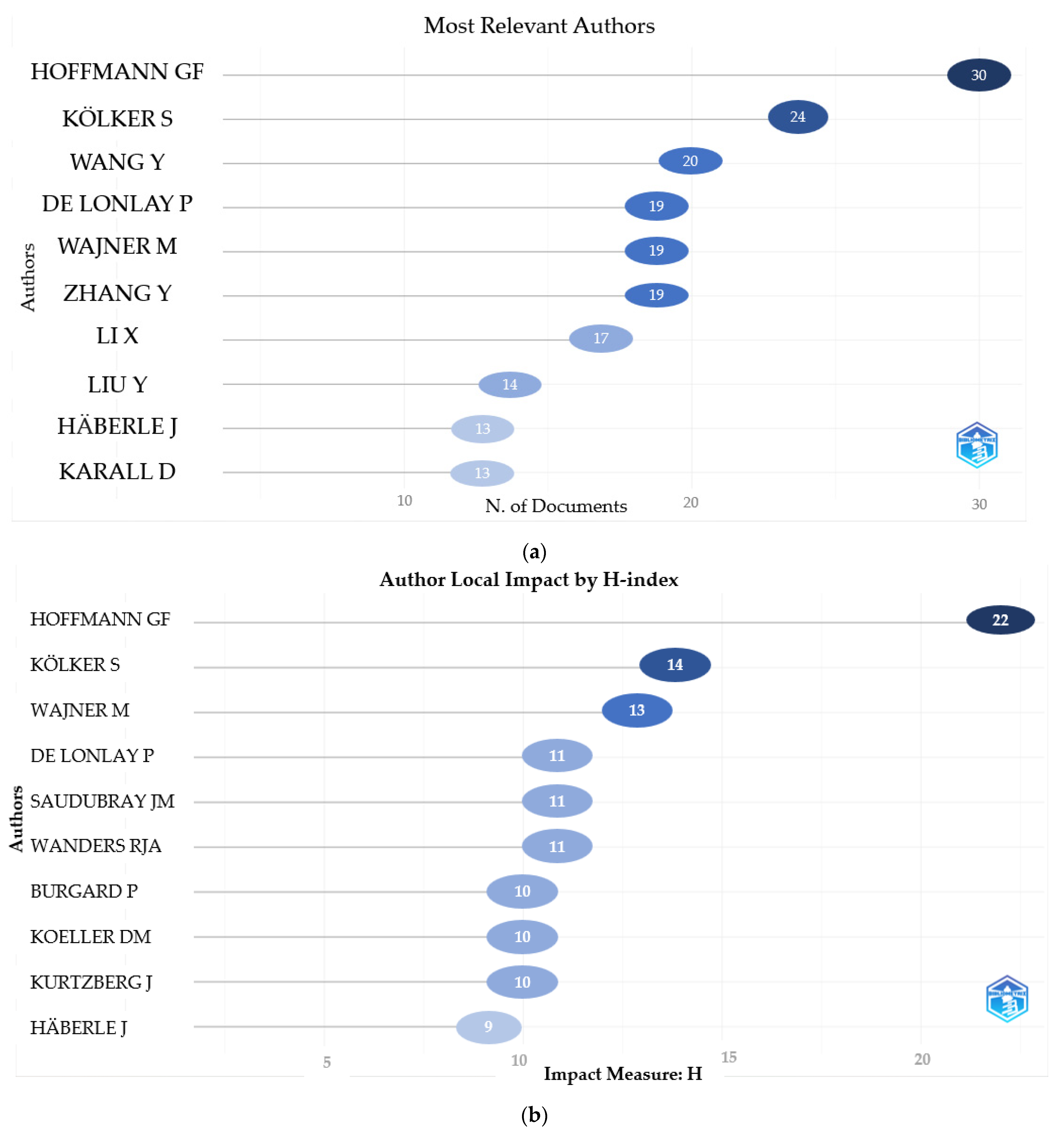
3.1.2. Distribution of Main Authors
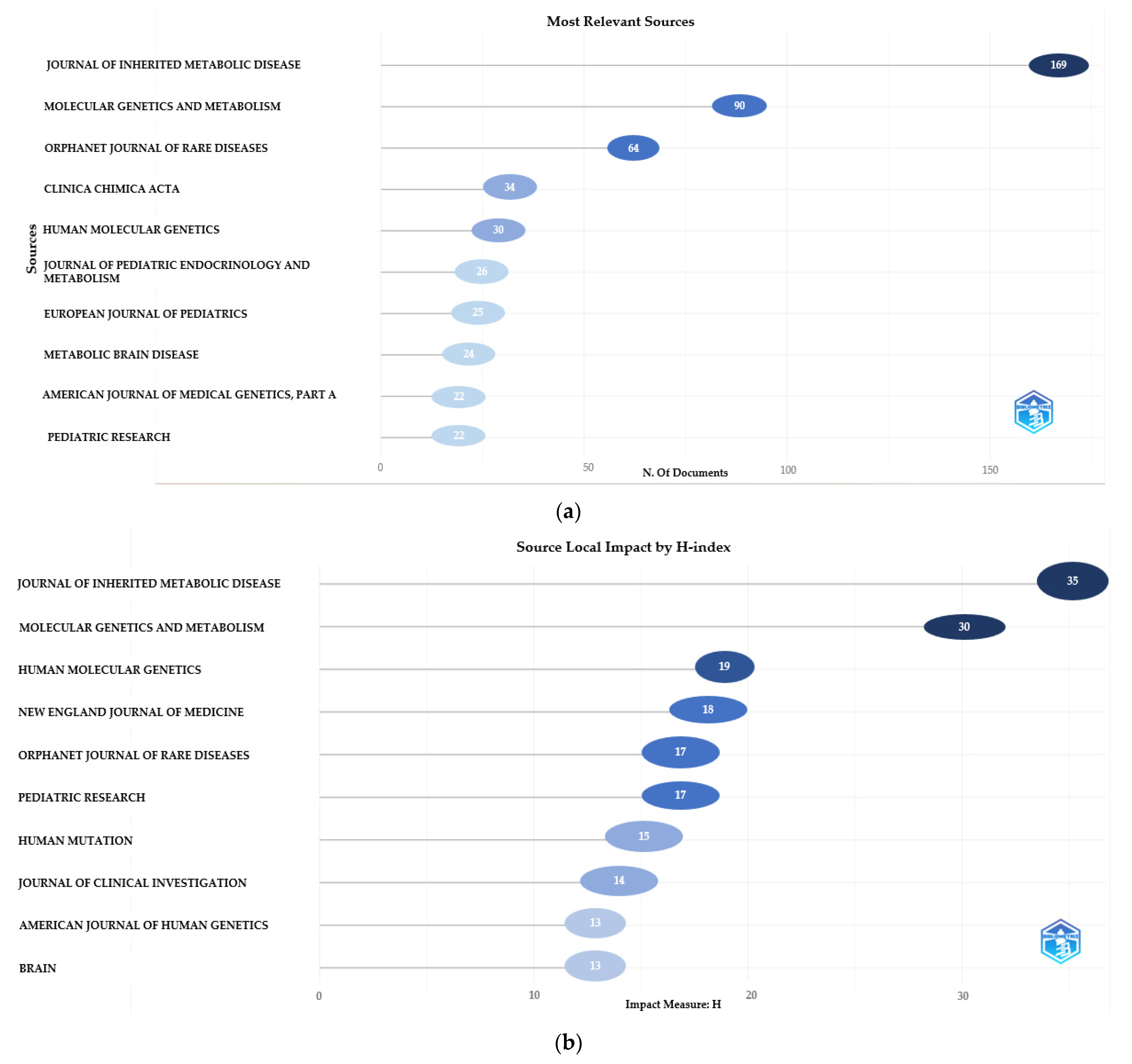
3.1.3. Distribution of Main Journals
3.1.4. Analysis of Corresponding Authors’ Countries
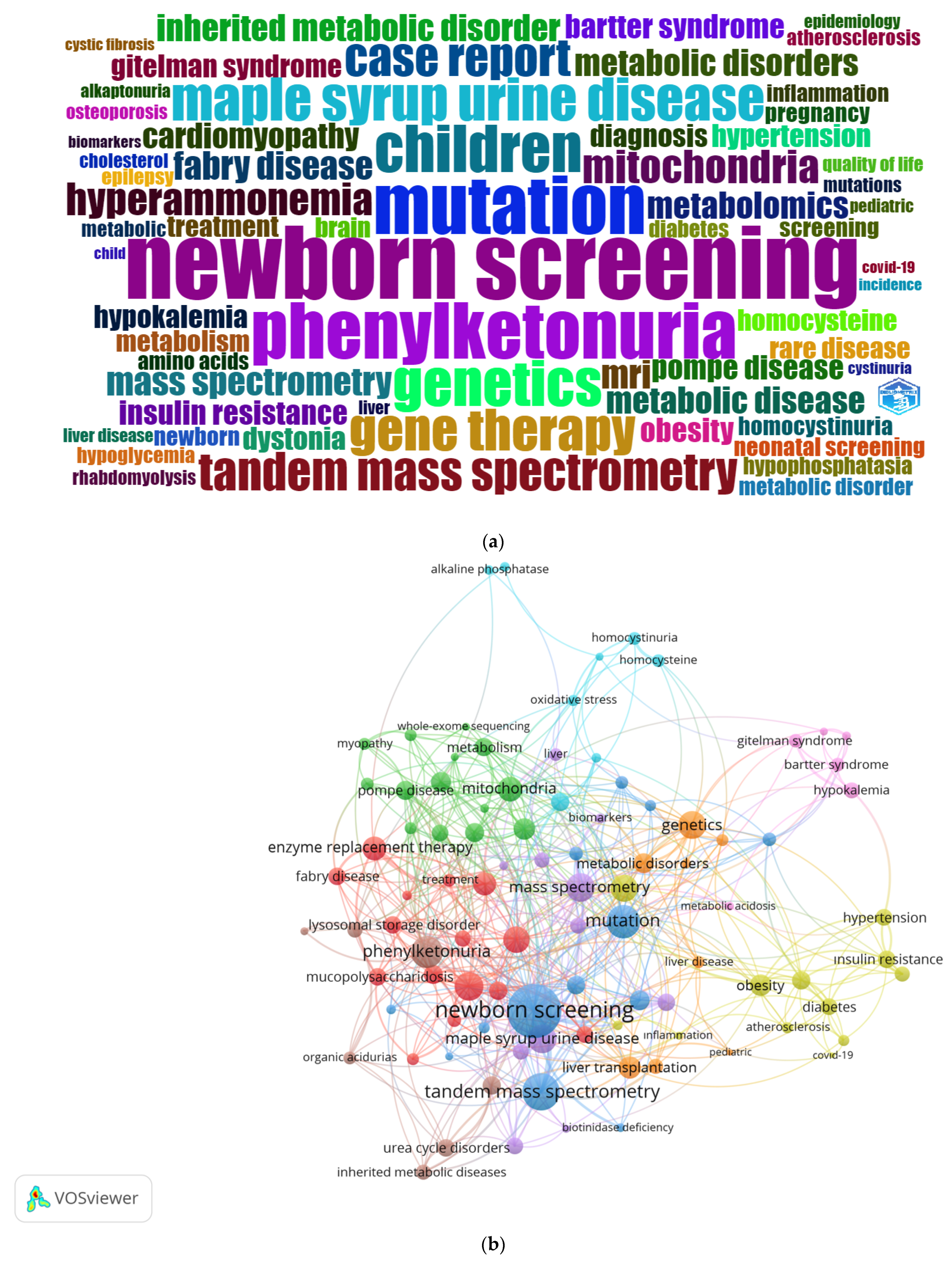
3.1.5. Analysis of Keywords
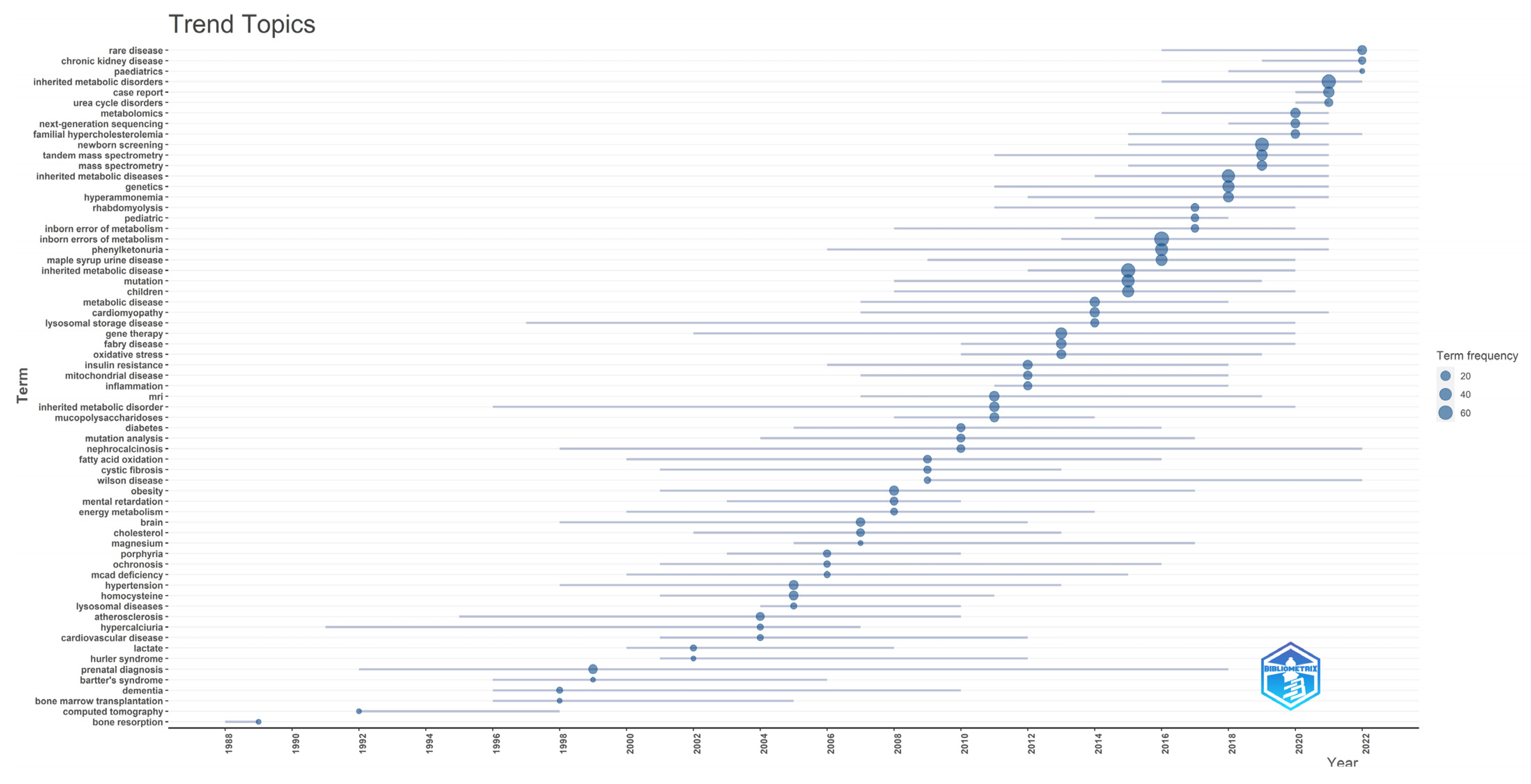
3.1.6. Trending Topics
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.J.; Kan, X. Recent research on inherited metabolic diseases in children. Zhongguo Dang Dai Er Ke Za Zhi 2022, 24, 326–331. [Google Scholar] [PubMed]
- Ferreira, C.R.; Rahman, S.; Keller, M.; Zschocke, J.; ICIMD Advisory Group. An international classification of inherited metabolic disorders (ICIMD). J. Inherit. Metab. Dis. 2021, 44, 164–177. [Google Scholar] [CrossRef]
- Schweitzer-Krantz, S. Early diagnosis of inherited metabolic disorders towards improving outcome: The controversial issue of galactosaemia. Eur. J. Pediatr. 2003, 162, 50–53. [Google Scholar] [CrossRef]
- van Konijnenburg, E.M.M.H.; Wortmann, S.B.; Koelewijn, M.J.; Tseng, L.A.; Houben, R.; Stöckler-Ipsiroglu, S.; Ferreira, C.R.; van Karnebeek, C.D.M. Treatable inherited metabolic disorders causing intellectual disability: 2021 review and digital app. Orphanet. J. Rare Dis. 2021, 16, 170. [Google Scholar] [CrossRef]
- Lenzini, L.; Carraro, G.; Avogaro, A.; Vitturi, N. Genetic Diagnosis in a Cohort of Adult Patients with Inherited Metabolic Diseases: A Single-Center Experience. Biomolecules 2022, 12, 920. [Google Scholar] [CrossRef]
- Wasim, M.; Khan, H.N.; Ayesha, H.; Awan, F.R. Need and Challenges in Establishing Newborn Screening Programs for Inherited Metabolic Disorders in Developing Countries. Adv. Biol. 2023, e2200318, online ahead of print. [Google Scholar] [CrossRef]
- Villoria, J.G.; Pajares, S.; López, R.M.; Marin, J.L.; Ribes, A. Neonatal Screening for Inherited Metabolic Diseases in 2016. Semin. Pediatr. Neurol. 2016, 23, 257–272. [Google Scholar] [CrossRef]
- Lin, Y.; Zheng, Q.; Zheng, T.; Zheng, Z.; Lin, W.; Fu, Q. Expanded newborn screening for inherited metabolic disorders and genetic characteristics in a southern Chinese population. Clin. Chim. Acta 2019, 494, 106–111. [Google Scholar] [CrossRef]
- Verma, J.; Thomas, D.C.; Sharma, S.; Jhingan, G.; Saxena, R.; Kohli, S.; Puri, R.D.; Bijarnia, S.; Verma, I.C. Inherited metabolic disorders: Prenatal diagnosis of lysosomal storage disorders. Prenat. Diagn. 2015, 35, 1137–1147. [Google Scholar] [CrossRef]
- Arnold, G.L. Inborn errors of metabolism in the 21st century: Past to present. Ann. Transl. Med. 2018, 6, 467. [Google Scholar] [CrossRef]
- Gonzalez, J.; Willis, M.S. Ivar Asbjörn Følling: Discovered Phenylketonuria (PKU). Lab. Med. 2010, 41, 118–119. [Google Scholar] [CrossRef]
- Blau, N.; van Spronsen, F.J.; Levy, H.L. Phenylketonuria. Lancet 2010, 376, 1417–1427. [Google Scholar] [CrossRef]
- Scriver, C.R. The PAH gene, phenylketonuria, and a paradigm shift. Hum. Mutat. 2007, 28, 831–845. [Google Scholar] [CrossRef]
- Chace, D.H.; Kalas, T.A.; Naylor, E.W. Use of tandem mass spectrometry for multianalyte screening of dried blood specimens from newborns. Clin. Chem. 2003, 49, 1797–1817. [Google Scholar] [CrossRef]
- Therrell, B.L.; Padilla, C.D.; Loeber, J.G.; Kneisser, I.; Saadallah, A.; Borrajo, G.J.; Adams, J. Current status of newborn screening worldwide: 2015. Semin. Perinatol. 2015, 39, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Gao, J.; Kang, J.; Wang, X.; Niu, Q.; Liu, J.; Zhang, L. Knowledge Mapping of Exosomes in Autoimmune Diseases: A Bibliometric Analysis (2002–2021). Front. Immunol. 2022, 13, 939433. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, O.J.; Da Silva, F.F.; Juliani, F.; Barbosa, L.C.F.M.; Nunhes, T.V. Bibliometric Method for Mapping the State-of-the-Art and Identifying Research Gaps and Trends in Literature: An Essential Instrument to Support the Development of Scientific Projects. In Scientometrics Recent Advances; Intech: London, UK, 2019. [Google Scholar] [CrossRef]
- Ke, L.; Lu, C.; Shen, R.; Lu, T.; Ma, B.; Hua, Y. Knowledge Mapping of Drug-Induced Liver Injury: A Scientometric Investigation (2010–2019). Front. Pharmacol. 2020, 11, 842. [Google Scholar] [CrossRef]
- Synnestvedt, M.B.; Chen, C.; Holmes, J.H. CiteSpace II: Visualization and Knowledge Discovery in Bibliographic Databases. AMIA Annu. Symp. Proc. 2005, 2005, 724–728. [Google Scholar]
- Yeung, A.W.K.; Tzvetkov, N.T.; Balacheva, A.A.; Georgieva, M.G.; Gan, R.-Y.; Jozwik, A.; Pyzel, B.; Horbańczuk, J.O.; Novellino, E.; Durazzo, A.; et al. Lignans: Quantitative Analysis of the Research Literature. Front. Pharmacol. 2020, 11, 37. [Google Scholar] [CrossRef]
- Li, C.; Ojeda-Thies, C.; Renz, N.; Margaryan, D.; Perka, C.; Trampuz, A. The Global State of Clinical Research and Trends in Periprosthetic Joint Infection: A Bibliometric Analysis. Int. J. Infect. Dis. 2020, 96, 696–709. [Google Scholar] [CrossRef]
- Lu, C.; Liu, M.; Shang, W.; Yuan, Y.; Li, M.; Deng, X.; Li, H.; Yang, K. Knowledge Mapping of Angelica Sinensis (Oliv.) Diels (Danggui) Research: A Scientometric Study. Front. Pharmacol. 2020, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.; Sampson, M.; Barrowman, N.; Doja, A. Bibliometric Analysis of Neurology Articles Published in General Medicine Journals. JAMA Netw. Open 2021, 4, e215840. [Google Scholar] [CrossRef]
- Liu, B.; He, X.; Wang, Y.; Huang, J.-W.; Zheng, Y.-B.; Li, Y.; Lu, L.-G. Bibliometric Analysis of γδ T Cells as Immune Regulators in Cancer Prognosis. Front. Immunol. 2022, 13, 874640. [Google Scholar] [CrossRef]
- Li, C.; Wang, L.; Perka, C.; Trampuz, A. Clinical application of robotic orthopedic surgery: A bibliometric study. BMC Musculoskelet. Disord. 2021, 22, 968. [Google Scholar] [CrossRef]
- Vural, S.; Kaya, H.B.; Coskun, F. A bibliometric study on the publication errors in emergency medicine journals from 2000 to 2020. Am. J. Emerg. Med. 2022, 60, 140–144. [Google Scholar] [CrossRef]
- Bharadwaj, A.; Wahi, N.; Saxena, A. Occurrence of Inborn Errors of Metabolism in Newborns, Diagnosis and Prophylaxis. Endocr. Metab. Immune Disord. Drug Targets 2021, 21, 592–616. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, J.; Shi, S.; Bo, R.; Zhang, X.; Hu, Y. Bibliometric Analysis on the Progress of Chronic Heart Failure. Curr. Probl. Cardiol. 2022, 47, 101213. [Google Scholar] [CrossRef]
- Akmal, M.; Hasnain, N.; Rehan, A.; Iqbal, U.; Hashmi, S.; Fatima, K.; Farooq, M.Z.; Khosa, F.; Siddiqi, J.; Khan, M.K. Glioblastome Multiforme: A Bibliometric Analysis. World Neurosurg. 2020, 136, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Koo, M. Systemic Lupus Erythematosus Research: A Bibliometric Analysis over a 50-Year Period. Int. J. Environ. Res. Public Health 2021, 18, 7095. [Google Scholar] [CrossRef]
- de Diego, I.M.; González-Fernández, C.; Fernández-Isabel, A.; Fernández, R.R.; Cabezas, J. System for evaluating the reliability and novelty of medical scientific papers. J. Inf. 2021, 15, 101188. [Google Scholar]
- Rajabi, F. Updates in Newborn Screening. Pediatr. Ann. 2018, 47, e187–e190. [Google Scholar] [CrossRef] [PubMed]
- Wasserstein, M.P.; Andriola, M.; Arnold, G.; Aron, A.; Duffner, P.; Erbe, R.W.; Escolar, M.L.; Estrella, L.; Galvin-Parton, P.; Iglesias, A.; et al. Clinical outcomes of children with abnormal newborn screening results for Krabbe disease in New York State. Genet. Med. 2016, 18, 1235–1243. [Google Scholar] [CrossRef] [PubMed]
- van Spronsen, F.J.; Enns, G.M. Future treatment strategies in phenylketonuria. Mol. Genet. Metab. 2010, 99, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, A.; Oussalah, A.; Jeannesson, É.; Guéant, J.L.; Feillet, F. La phénylcétonurie—De la diététique à la thérapie génique [Phenylketonuria, from diet to gene therapy]. Med. Sci. 2020, 36, 725–734. [Google Scholar]
- Pronicka, E.; Piekutowska-Abramczuk, D.; Ciara, E.; Trubicka, J.; Rokicki, D.; Karkucińska-Więckowska, A.; Pajdowska, M.; Jurkiewicz, E.; Halat, P.; Kosińska, J.; et al. New perspective in diagnostics of mitochondrial disorders: Two years’ experience with whole-exome sequencing at a national paediatric centre. J. Transl. Med. 2016, 14, 174. [Google Scholar] [CrossRef]
- Chen, P.S.; Chao, C.C.; Tsai, L.K.; Huang, H.-Y.; Chien, Y.-H.; Huang, P.-H.; Hwu, W.-L.; Hsieh, S.-T.; Lee, N.-C.; Hsueh, H.-W.; et al. Diagnostic Challenges of Neuromuscular Disorders after Whole Exome Sequencing. J. Neuromuscul. Dis. 2023, 10, 667–684. [Google Scholar] [CrossRef]
- Meng, L.; Pammi, M.; Saronwala, A.; Magoulas, P.; Ghazi, A.R.; Vetrini, F.; Zhang, J.; Theodore, C.; Dharmadhikari, A.; Qu, C.; et al. Use of Exome Sequencing for Infants in Intensive Care Units: Ascertainment of Severe Single-Gene Disorders and Effect on Medical Management. JAMA Pediatr. 2017, 171, e173438. [Google Scholar] [CrossRef]
- Roman, T.S.; Crowley, S.B.; Roche, M.I.; Foreman, A.K.M.; O’daniel, J.M.; Seifert, B.A.; Lee, K.; Brandt, A.; Gustafson, C.; DeCristo, D.M.; et al. Genomic Sequencing for Newborn Screening: Results of the NC NEXUS Project. Am. J. Hum. Genet. 2020, 107, 596–611. [Google Scholar] [CrossRef]
- Śmigiel, R.; Biela, M.; Szmyd, K.; Błoch, M.; Szmida, E.; Skiba, P.; Walczak, A.; Gasperowicz, P.; Kosińska, J.; Rydzanicz, M.; et al. Rapid Whole-Exome Sequencing as a Diagnostic Tool in a Neonatal/Pediatric Intensive Care Unit. J. Clin. Med. 2020, 9, 2220. [Google Scholar] [CrossRef]
- Parenti, G.; Andria, G.; Ballabio, A. Lysosomal storage diseases: From pathophysiology to therapy. Annu. Rev. Med. 2015, 66, 471–486. [Google Scholar] [CrossRef]
- Dursun, A.; Henneke, M.; Özgül, K.; Gartner, J.; Coşkun, T.; Tokatli, A.; Kalkanoğlu, H.S.; Demirkol, M.; Wendel, U.; Özalp, I. Maple syrup urine disease: Mutation analysis in Turkish patients. J. Inherit. Metab. Dis. 2002, 25, 89–97. [Google Scholar] [CrossRef]
- Carrillo-Carrasco, N.; Chandler, R.J.; Venditti, C.P. Combined methylmalonic acidemia and homocystinuria, cblC type. I. Clinical presentations, diagnosis and management. J. Inherit. Metab. Dis. 2011, 35, 91–102. [Google Scholar] [CrossRef]
- Yu, Y.; Ling, S.; Shuai, R.; Qiu, W.; Zhang, H.; Liang, L.; Ji, W.; Liu, Y.; Gu, X.; Han, L. Clinical features and outcomes of patients with cblC type methylmalonic acidemia carrying gene c.609G>A mutation. Zhejiang Da Xue Xue Bao Yi Xue Ban 2021, 50, 436–443. [Google Scholar]
- Yilmaz, B.S.; Gurung, S.; Perocheau, D.; Counsell, J.; Baruteau, J. Gene therapy for inherited metabolic diseases. J. Mother Child 2020, 24, 53–64. [Google Scholar]
- Coene, K.L.M.; Kluijtmans, L.A.J.; van der Heeft, E.; Engelke, U.F.H.; de Boer, S.; Hoegen, B.; Kwast, H.J.T.; van de Vorst, M.; Huigen, M.C.D.G.; Keularts, I.M.L.W.; et al. Next-generation metabolic screening: Targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients. J. Inherit. Metab. Dis. 2018, 41, 337–353. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Heiles, S. Advanced tandem mass spectrometry in metabolomics and lipidomics-methods and applications. Anal. Bioanal. Chem. 2021, 413, 5927–5948. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef]
- Brooks, E.D.; Landau, D.J.; Everitt, J.I.; Brown, T.T.; Grady, K.M.; Waskowicz, L.; Bass, C.R.; D’Angelo, J.; Asfaw, Y.G.; Williams, K.; et al. Long-term complications of glycogen storage disease type Ia in the canine model treated with gene replacement therapy. J. Inherit. Metab. Dis. 2018, 41, 965–976. [Google Scholar] [CrossRef]
- Bryson, T.E.; Anglin, C.M.; Bridges, P.H.; Cottle, R.N. Nucleasemediated gene therapies for inherited metabolic diseases of the liver. Yale J. Biol. Med. 2017, 90, 553–566. [Google Scholar]
- Lee, Y.M.; Conlon, T.J.; Specht, A.; Coleman, K.E.; Brown, L.M.; Estrella, A.M.; Dambska, M.; Dahlberg, K.R.; Weinstein, D.A. Long-term safety and efficacy of AAV gene therapy in the canine model of glycogen storage disease type Ia. J. Inherit. Metab. Dis. 2018, 41, 977–984. [Google Scholar] [CrossRef] [PubMed]




| Description | Results |
|---|---|
| Main Information about Data | |
| Timespan | 1968:2023 |
| Sources (Journals, Books, etc.) | 1012 |
| Documents | 2702 |
| Annual Growth Rate % | 3.69 |
| Document Average Age | 14.2 |
| Average citations per doc | 26.4 |
| References | 88,136 |
| Document Contents | |
| Keywords Plus (ID) | 16,542 |
| Author’s Keywords (DE) | 5380 |
| Authors Collaboration | |
| Authors | 14,782 |
| Single-authored docs | 207 |
| Co-Authors per Doc | 6.97 |
| International co-authorships % | 21.06 |
| Document Types | |
| Research article | 2702 |
| Keywords | Number of Articles |
|---|---|
| newborn screening | 54 |
| Mutation | 43 |
| Phenylketonuria | 42 |
| Children | 35 |
| Genetics | 34 |
| maple syrup urine disease | 32 |
| gene therapy | 31 |
| case report | 28 |
| tandem mass spectrometry | 28 |
| glycogen storage disease | 27 |
| Mucopolysaccharidosis | 27 |
| liver transplantation | 25 |
| Mitochondria | 25 |
| Hyperammonemia | 24 |
| enzyme replacement therapy | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadıoğlu Yılmaz, B.; Akgül, A.H. Inherited Metabolic Diseases from Past to Present: A Bibliometric Analysis (1968–2023). Children 2023, 10, 1205. https://doi.org/10.3390/children10071205
Kadıoğlu Yılmaz B, Akgül AH. Inherited Metabolic Diseases from Past to Present: A Bibliometric Analysis (1968–2023). Children. 2023; 10(7):1205. https://doi.org/10.3390/children10071205
Chicago/Turabian StyleKadıoğlu Yılmaz, Banu, and Ayşe Hümeyra Akgül. 2023. "Inherited Metabolic Diseases from Past to Present: A Bibliometric Analysis (1968–2023)" Children 10, no. 7: 1205. https://doi.org/10.3390/children10071205
APA StyleKadıoğlu Yılmaz, B., & Akgül, A. H. (2023). Inherited Metabolic Diseases from Past to Present: A Bibliometric Analysis (1968–2023). Children, 10(7), 1205. https://doi.org/10.3390/children10071205






