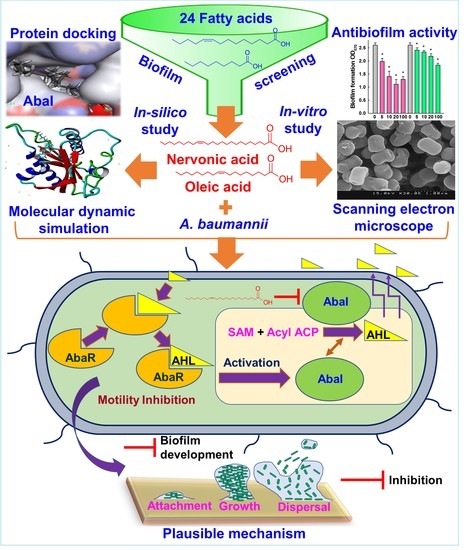
Assessment of Antibiofilm Potencies of Nervonic and Oleic Acid against Acinetobacter baumannii Using In Vitro and Computational Approaches
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Reagents, Bacterial Culture and Growth Rate Measurements
2.3. In Vitro Screening of Fatty Acids against A. baumannii Biofilm
2.4. Measurements of Surface Motility
2.5. Microscopic Architecture of A. baumannii Biofilm
2.6. Assessment of the Cytotoxicity of Fatty Acids against C. elegans
2.7. Homological Modeling of Three-Dimensional Protein Structure
2.8. Three-Dimensional Structure Validation of Model Protein
2.9. Ligands and Acyl Homoserine Lactone Synthase Protein Collection
2.10. Preparation of Ligands and Computational Screening
2.11. Estimation of ADME Property of Assigned Fatty Acids
2.12. Molecular Dynamic (MD) Simulations and Energy Calculations
2.13. Statistical Analysis
3. Results
3.1. In Vitro Validation of Antibiofilm Activity of Fatty Acids against A. baumannii
3.2. Nervonic Acid Impaired A. baumannii Motility
3.3. Microscopic Observations of Biofilm Inhibition by A. baumannii
3.4. Cytotoxicity of Fatty Acids in the Nematode Caenorhabditis elegans
3.5. Molecular Docking of Fatty Acids with AbaI and Estimation of Molecular Interactions
3.6. Conformational Stability of Fatty Acids with AbaI by Molecular Dynamic (MD) Simulation
3.7. Plausible Mechanism for the Inhibition of A. baumannii Quorum Sensing by Nervonic Acid
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rumbo-Feal, S.; Gomez, M.J.; Gayoso, C.; Alvarez-Fraga, L.; Cabral, M.P.; Aransay, A.M.; Rodriguez-Ezpeleta, N.; Fullaondo, A.; Valle, J.; Tomas, M.; et al. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS ONE 2013, 8, e72968. [Google Scholar] [CrossRef] [Green Version]
- Quintieri, L.; Caputo, L.; Monaci, L.; Cavalluzzi, M.M.; Denora, N. Lactoferrin-derived peptides as a control strategy against skinborne Staphylococcal biofilms. Biomedicines 2020, 8, 323. [Google Scholar] [CrossRef]
- Eder, A.E.; Munir, S.A.; Hobby, C.R.; Anderson, D.M.; Herndon, J.L.; Siv, A.W.; Symes, S.J.K.; Giles, D.K. Exogenous polyunsaturated fatty acids (PUFAs) alter phospholipid composition, membrane permeability, biofilm formation and motility in Acinetobacter baumannii. Microbiology 2017, 163, 1626–1636. [Google Scholar] [CrossRef] [PubMed]
- Kroger, C.; MacKenzie, K.D.; Alshabib, E.Y.; Kirzinger, M.W.B.; Suchan, D.M.; Chao, T.C.; Akulova, V.; Miranda-CasoLuengo, A.A.; Monzon, V.A.; Conway, T.; et al. The primary transcriptome, small RNAs and regulation of antimicrobial resistance in Acinetobacter baumannii ATCC 17978. Nucleic Acids Res. 2018, 46, 9684–9698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrillo, G.; Tavani, C.; Bianchi, L.; Benzi, A.; Cavalluzzi, M.M.; Salvagno, L.; Quintieri, L.; De Palma, A.; Caputo, L.; Rosato, A.; et al. Densely functionalized 2-Methylideneazetidines: Evaluation as antibacterials. Molecules 2021, 26, 3891. [Google Scholar] [CrossRef] [PubMed]
- De Silva, P.M.; Chong, P.; Fernando, D.M.; Westmacott, G.; Kumar, A. Effect of incubation temperature on antibiotic resistance and virulence factors of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 2018, 62, e01514–e01517. [Google Scholar] [CrossRef] [Green Version]
- Kadam, S.; Shai, S.; Shahane, A.; Kaushik, K.S. Recent advances in non-conventional antimicrobial approaches for chronic wound biofilms: Have we found the ‘chink in the armor’? Biomedicines 2019, 7, 35. [Google Scholar] [CrossRef] [Green Version]
- Bahamondez-Canas, T.F.; Heersema, L.A.; Smyth, H.D.C. Current status of in vitro models and assays for susceptibility testing for wound biofilm infections. Biomedicines 2019, 7, 34. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, V.; Patel, V.; Tiwari, M. In-silico screening and experimental validation reveal L-Adrenaline as anti-biofilm molecule against biofilm-associated protein (Bap) producing Acinetobacter baumannii. Int. J. Biol. Macromol. 2018, 107, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.M.; Tracy, E.N.; Carruthers, M.D.; Rather, P.N.; Actis, L.A.; Munson, R.S., Jr. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 2013, 4, e00360-13. [Google Scholar] [CrossRef] [Green Version]
- Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 2011, 157, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Eijkelkamp, B.A.; Stroeher, U.H.; Hassan, K.A.; Elbourne, L.D.; Paulsen, I.T.; Brown, M.H. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect. Immun. 2013, 81, 2574–2583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, C.; Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008, 190, 3386–3392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Chen, Y.; Wang, X.; Ding, Y.; Sun, X.; Ni, Z. Contribution of the AbaI/AbaR quorum sensing system to resistance and virulence of Acinetobacter baumannii clinical strains. Infect. Drug Resist. 2020, 13, 4273–4281. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, N.; Sharma, P.; Capalash, N. Quorum sensing in Acinetobacter: An emerging pathogen. Crit. Rev. Microbiol. 2010, 36, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Han, K. AbaR is a LuxR type regulator essential for motility and the formation of biofilm and pellicle in Acinetobacter baumannii. Genes Genom. 2020, 42, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ni, Z.; Tang, J.; Ding, Y.; Wang, X.; Li, F. The abaI/abaR quorum sensing system effects on pathogenicity in Acinetobacter baumannii. Front. Microbiol. 2021, 12, 679241. [Google Scholar] [CrossRef]
- Saipriya, K.; Swathi, C.H.; Ratnakar, K.S.; Sritharan, V. Quorum-sensing system in Acinetobacter baumannii: A potential target for new drug development. J. Appl. Microbiol. 2020, 128, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kim, Y.G.; Khadke, S.K.; Lee, J. Antibiofilm and antifungal activities of medium-chain fatty acids against Candida albicans via mimicking of the quorum-sensing molecule farnesol. Microb. Biotechnol. 2020, 14, 1353–1366. [Google Scholar] [CrossRef]
- Kumar, P.; Lee, J.H.; Beyenal, H.; Lee, J. Fatty acids as antibiofilm and antivirulence agents. Trends Microbiol. 2020, 28, 753–768. [Google Scholar] [CrossRef]
- Kim, Y.G.; Lee, J.H.; Raorane, C.J.; Oh, S.T.; Park, J.G.; Lee, J. Herring oil and omega fatty acids inhibit Staphylococcus aureus biofilm formation and virulence. Front. Microbiol. 2018, 9, 1241. [Google Scholar] [CrossRef] [Green Version]
- Inoue, T.; Shingaki, R.; Fukui, K. Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS. Microbiol. Lett. 2008, 281, 81–86. [Google Scholar] [CrossRef]
- Murzyn, A.; Krasowska, A.; Stefanowicz, P.; Dziadkowiec, D.; Lukaszewicz, M. Capric acid secreted by S. boulardii inhibits C. albicans filamentous growth, adhesion and biofilm formation. PLoS ONE 2010, 5, e12050. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, S.; Ravindran, D.; Arunachalam, K.; Arumugam, V.R. Inhibition of quorum sensing-dependent biofilm and virulence genes expression in environmental pathogen Serratia marcescens by petroselinic acid. Antonie Van Leeuwenhoek 2018, 111, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Song, S.; Yang, C.; Sun, X.; Huang, Y.; Li, K.; Zhao, S.; Zhang, Y.; Deng, Y. Disruption of quorum sensing and virulence in Burkholderia cenocepacia by a structural analogue of the cis-2-dodecenoic acid signal. Appl. Environ. Microbiol. 2019, 85, e00105–e00119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Impallomeni, G.; Ballistreri, A.; Carnemolla, G.M.; Guglielmino, S.P.; Nicolo, M.S.; Cambria, M.G. Synthesis and characterization of poly(3-hydroxyalkanoates) from Brassica carinata oil with high content of erucic acid and from very long chain fatty acids. Int. J. Biol. Macromol. 2011, 48, 137–145. [Google Scholar] [CrossRef]
- Coraça-Huber, D.C.; Steixner, S.; Wurm, A.; Nogler, M. Antibacterial and anti-biofilm activity of omega-3 polyunsaturated fatty acids against periprosthetic joint infections-isolated multi-drug resistant strains. Biomedicines 2021, 9, 334. [Google Scholar] [CrossRef]
- Chanda, W.; Joseph, T.P.; Padhiar, A.A.; Guo, X.; Min, L.; Wang, W.; Lolokote, S.; Ning, A.; Cao, J.; Huang, M.; et al. Combined effect of linolenic acid and tobramycin on Pseudomonas aeruginosa biofilm formation and quorum sensing. Exp. Ther. Med. 2017, 14, 4328–4338. [Google Scholar] [CrossRef] [PubMed]
- Amri, Z.; Zaouay, F.; Lazreg-Aref, H.; Soltana, H.; Mneri, A.; Mars, M.; Hammami, M. Phytochemical content, fatty acids composition and antioxidant potential of different pomegranate parts: Comparison between edible and non edible varieties grown in Tunisia. Int. J. Biol. Macromol. 2017, 104, 274–280. [Google Scholar] [CrossRef]
- Van Thuoc, D.; My, D.N.; Loan, T.T.; Sudesh, K. Utilization of waste fish oil and glycerol as carbon sources for polyhydroxyalkanoate production by Salinivibrio sp. M318. Int. J. Biol. Macromol. 2019, 141, 885–892. [Google Scholar] [CrossRef]
- Davies, D.G.; Marques, C.N. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicol, M.; Alexandre, S.; Luizet, J.B.; Skogman, M.; Jouenne, T.; Salcedo, S.P.; De, E. Unsaturated fatty acids affect quorum sensing communication system and inhibit motility and biofilm formation of Acinetobacter baumannii. Int. J. Mol. Sci. 2018, 19, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Muñiz, M.Y.; López-Jacome, L.E.; Hernández-Durán, M.; Franco-Cendejas, R.; Licona-Limón, P.; Ramos-Balderas, J.L.; Martinéz-Vázquez, M.; Belmont-Díaz, J.A.; Wood, T.K.; García-Contreras, R. Repurposing the anticancer drug mitomycin C for the treatment of persistent Acinetobacter baumannii infections. Int. J. Antimicrob. Agents 2017, 49, 88–92. [Google Scholar] [CrossRef]
- Wayne, P. Methods for Dilultion Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically in: Approved Standard, 10th ed.; CLSI document; M07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; Volume 35. [Google Scholar]
- Khadke, S.K.; Lee, J.H.; Woo, J.T.; Lee, J. Inhibitory effects of honokiol and magnolol on biofilm formation by Acinetobacter baumannii. Biotechnol. Bioprocess. Eng. 2019, 24, 359–365. [Google Scholar] [CrossRef]
- Raorane, C.J.; Lee, J.H.; Kim, Y.G.; Rajasekharan, S.K.; Garcia-Contreras, R.; Lee, J. Antibiofilm and antivirulence efficacies of flavonoids and curcumin against Acinetobacter baumannii. Front. Microbiol. 2019, 10, 990. [Google Scholar] [CrossRef] [PubMed]
- Subhani, S.; Jayaraman, A.; Jamil, K. Homology modelling and molecular docking of MDR1 with chemotherapeutic agents in non-small cell lung cancer. Biomed. Pharmacother. 2015, 71, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.G.; Gianoulis, T.A.; Pukatzki, S.; Mekalanos, J.J.; Ornston, L.N.; Gerstein, M.; Snyder, M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21, 601–614. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger, L. Schrödinger Suite; Schrödinger LLC: New York, NY, USA, 2016. [Google Scholar]
- Parr, R.G.; Yang, W. Density functional approach to the frontier-electron theory of chemical reactivity. J. Am. Chem. Soc. 2002, 106, 4049–4050. [Google Scholar] [CrossRef]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput. 2010, 6, 1509–1519. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.Y.; Zou, X. Advances and challenges in protein-ligand docking. Int. J. Mol. Sci. 2010, 11, 3016–3034. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Xiao, X.; Cheng, W.; Yang, W.; Yu, P.; Song, Z.; Yarov-Yarovoy, V.; Zheng, J. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol. 2015, 11, 518–524. [Google Scholar] [CrossRef] [Green Version]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Raj, V.; Park, J.G.; Cho, K.H.; Choi, P.; Kim, T.; Ham, J.; Lee, J. Assessment of antiviral potencies of cannabinoids against SARS-CoV-2 using computational and in vitro approaches. Int. J. Biol. Macromol. 2021, 168, 474–485. [Google Scholar] [CrossRef]
- Krieger, E.; Dunbrack, R.L., Jr.; Hooft, R.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012, 819, 405–421. [Google Scholar] [CrossRef]
- Dickson, C.J.; Madej, B.D.; Skjevik, A.A.; Betz, R.M.; Teigen, K.; Gould, I.R.; Walker, R.C. Lipid14: The amber lipid force field. J. Chem. Theory Comput. 2014, 10, 865–879. [Google Scholar] [CrossRef]
- Piotto, S.; Sessa, L.; Iannelli, P.; Concilio, S. Computational study on human sphingomyelin synthase 1 (hSMS1). Biochim. Biophys. Acta Biomembr. 2017, 1859, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, N.A.; Barnard, A.M.; Slater, H.; Simpson, N.J.; Salmond, G.P. Quorum-sensing in gram-negative bacteria. FEMS. Microbiol. Rev. 2001, 25, 365–404. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, M.M.; Khodi, S.; Mirhosseini, A. Quorum sensing in bacteria and a glance on Pseudomonas aeruginosa. Clin. Microbiol. 2014, 3, 156. [Google Scholar] [CrossRef]
- Subhadra, B.; Oh, M.H.; Choi, C.H. Quorum sensing in Acinetobacter: With special emphasis on antibiotic resistance, biofilm formation and quorum quenching. AIMS Microbiol. 2016, 2, 27–41. [Google Scholar] [CrossRef]
- Chen, R.; Lv, R.; Xiao, L.; Wang, M.; Du, Z.; Tan, Y.; Cui, Y.; Yan, Y.; Luo, Y.; Yang, R.; et al. A1S_2811, a CheA/Y-like hybrid two-component regulator from Acinetobacter baumannii ATCC 17978, is involved in surface motility and biofilm formation in this bacterium. Microbiologyopen 2017, 6, e00510. [Google Scholar] [CrossRef]
- Mussi, M.A.; Gaddy, J.A.; Cabruja, M.; Arivett, B.A.; Viale, A.M.; Rasia, R.; Actis, L.A. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J. Bacteriol. 2010, 192, 6336–6345. [Google Scholar] [CrossRef] [Green Version]
- McBride, M.J. Shining a light on an opportunistic pathogen. J. Bacteriol. 2010, 192, 6325–6326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seleem, N.M.; Abd El Latif, H.K.; Shaldam, M.A.; El-Ganiny, A. Drugs with new lease of life as quorum sensing inhibitors: For combating MDR Acinetobacter baumannii infections. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1687–1702. [Google Scholar] [CrossRef] [PubMed]






| Compounds | Receptor | Binding Energy (Kcal/mol) | Binding Energy (Kcal/mol) | Indicating Amino Acids | Bonds |
|---|---|---|---|---|---|
| VINA | AUTODOCK | ||||
| Nervonic acid | AbaI | −5.3 | −5.3 | Tyr30, Leu31, Trp33, Val120, Pro149, Tyr175, Met177 | 7π–π |
| Oleic acid | AbaI | −5.07 | −5.07 | Trp33, Ile172, Tyr175, Met177 | 4π–π |
| Myristoleic acid | AbaI | −5.9 | −5.9 | Val26, Tyr30, Phe109, Ser117 | 2π–π, 2H |
| S-adenosyl methionine | AbaI | −5.9 | −6.7 | Leu31, Ile65, Ser105, Val107, Ser121, Ile128, Pro149, Leu150, Met170, Met171, Ile172, Gly174, Tyr175, Ser176, Met177 | 2π–π, 13H |
| Linolenic acid | AbaI | −4.9 | −4.7 | Leu31, Ala106, Val107, Pro149 | 2π–π, 2H |
| Tricosanoic acid | AbaI | −4.6 | −4.8 | Tyr30 | 2π–π |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khadke, S.K.; Lee, J.-H.; Kim, Y.-G.; Raj, V.; Lee, J. Assessment of Antibiofilm Potencies of Nervonic and Oleic Acid against Acinetobacter baumannii Using In Vitro and Computational Approaches. Biomedicines 2021, 9, 1133. https://doi.org/10.3390/biomedicines9091133
Khadke SK, Lee J-H, Kim Y-G, Raj V, Lee J. Assessment of Antibiofilm Potencies of Nervonic and Oleic Acid against Acinetobacter baumannii Using In Vitro and Computational Approaches. Biomedicines. 2021; 9(9):1133. https://doi.org/10.3390/biomedicines9091133
Chicago/Turabian StyleKhadke, Sagar Kiran, Jin-Hyung Lee, Yong-Guy Kim, Vinit Raj, and Jintae Lee. 2021. "Assessment of Antibiofilm Potencies of Nervonic and Oleic Acid against Acinetobacter baumannii Using In Vitro and Computational Approaches" Biomedicines 9, no. 9: 1133. https://doi.org/10.3390/biomedicines9091133
APA StyleKhadke, S. K., Lee, J.-H., Kim, Y.-G., Raj, V., & Lee, J. (2021). Assessment of Antibiofilm Potencies of Nervonic and Oleic Acid against Acinetobacter baumannii Using In Vitro and Computational Approaches. Biomedicines, 9(9), 1133. https://doi.org/10.3390/biomedicines9091133