Nanomaterial-Based Fluorescence Resonance Energy Transfer (FRET) and Metal-Enhanced Fluorescence (MEF) to Detect Nucleic Acid in Cancer Diagnosis
Abstract
1. Introduction
2. Nucleic Acid Targets
2.1. Genomic DNA (gDNA)
2.2. Other DNAs
2.3. Messenger RNA (mRNA)
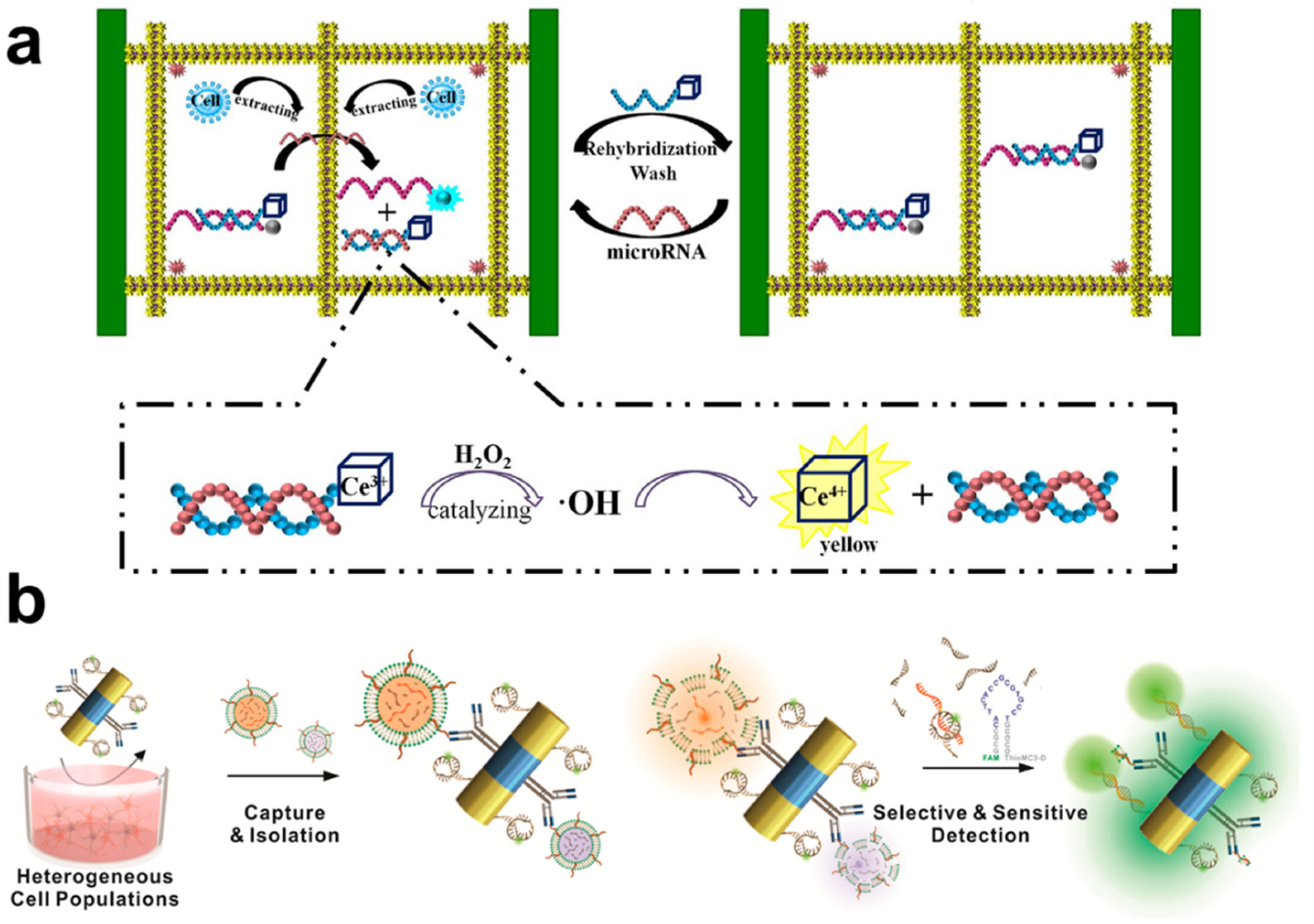
2.4. Non-Coding RNA (ncRNA)
3. FRET-Based Nucleic Acid Biosensors for Cancer Diagnosis
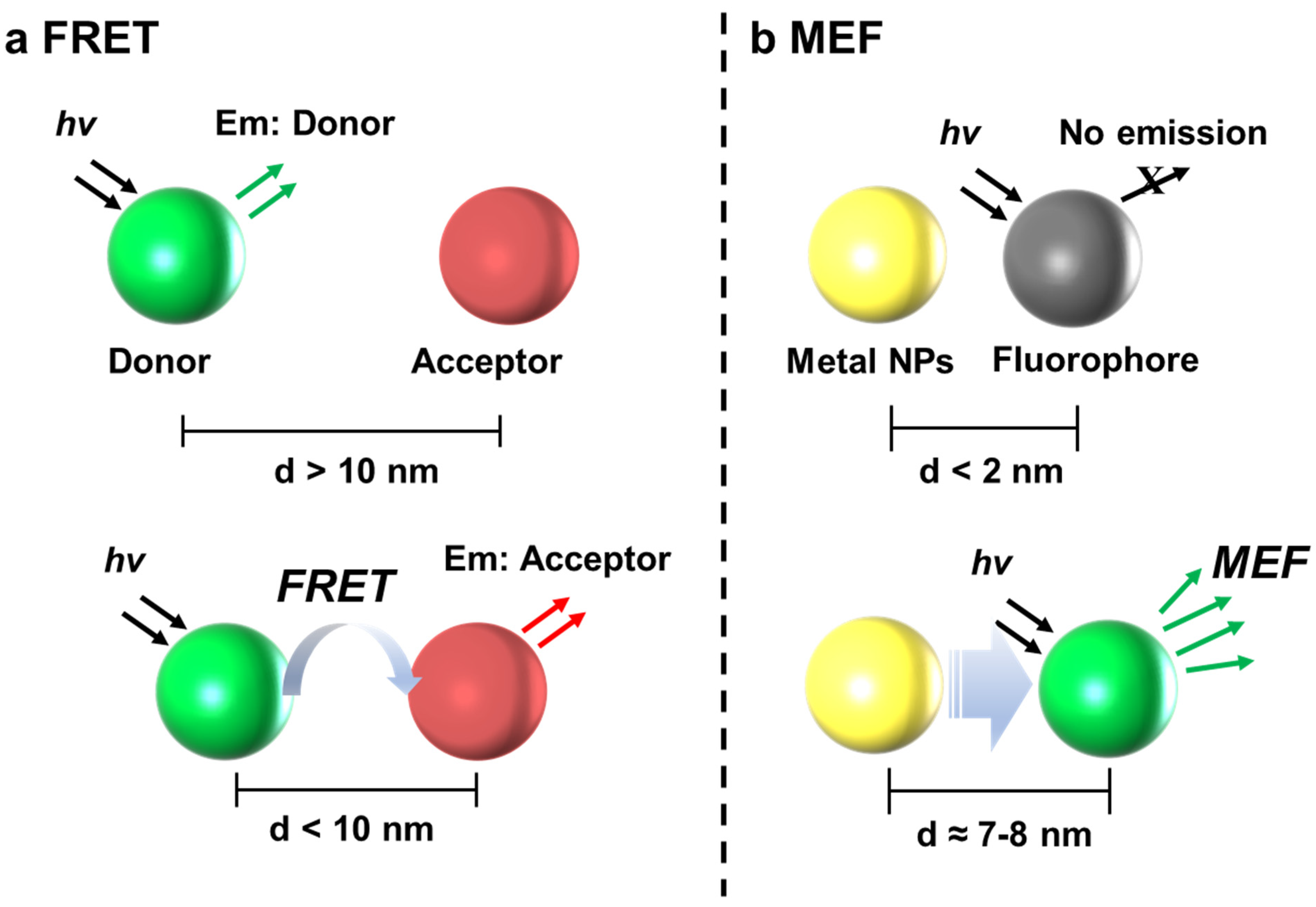
3.1. FRET
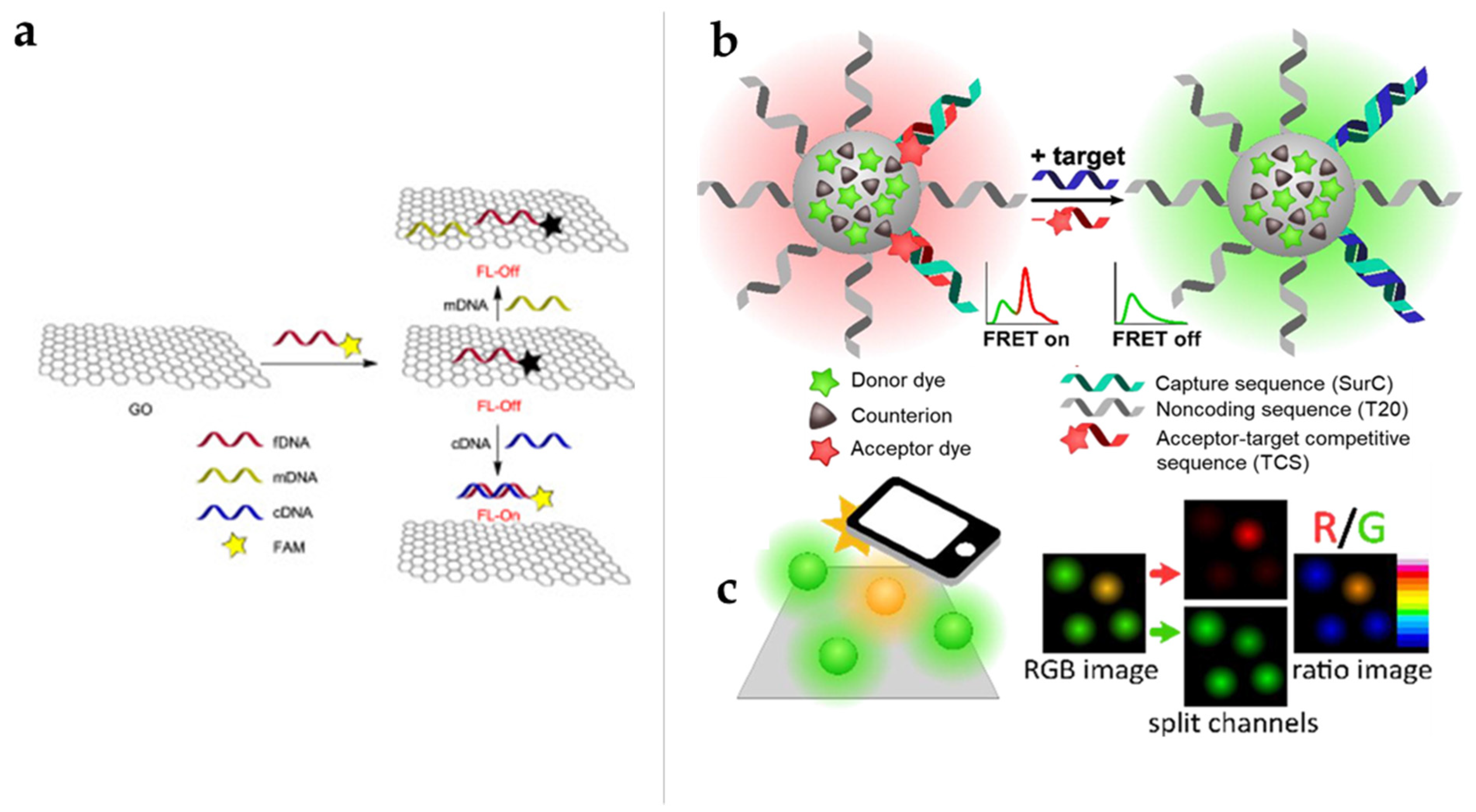
3.2. FRET-Based Biosensors for Detection of DNA Targets
3.3. FRET-Based Biosensors for Detection of RNA Targets
4. MEF-Based Nucleic Acid Biosensors for Cancer Diagnosis
4.1. MEF
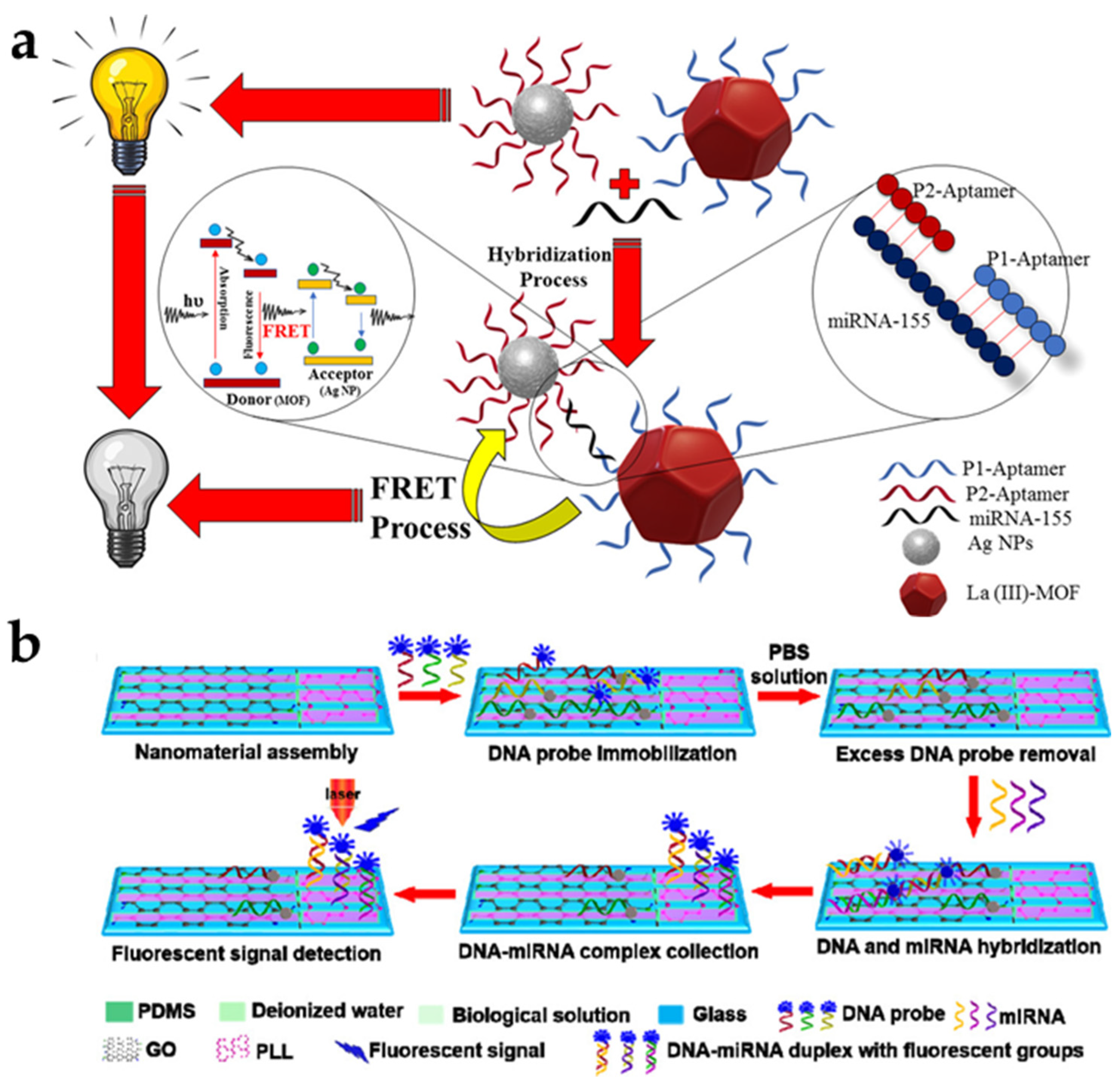
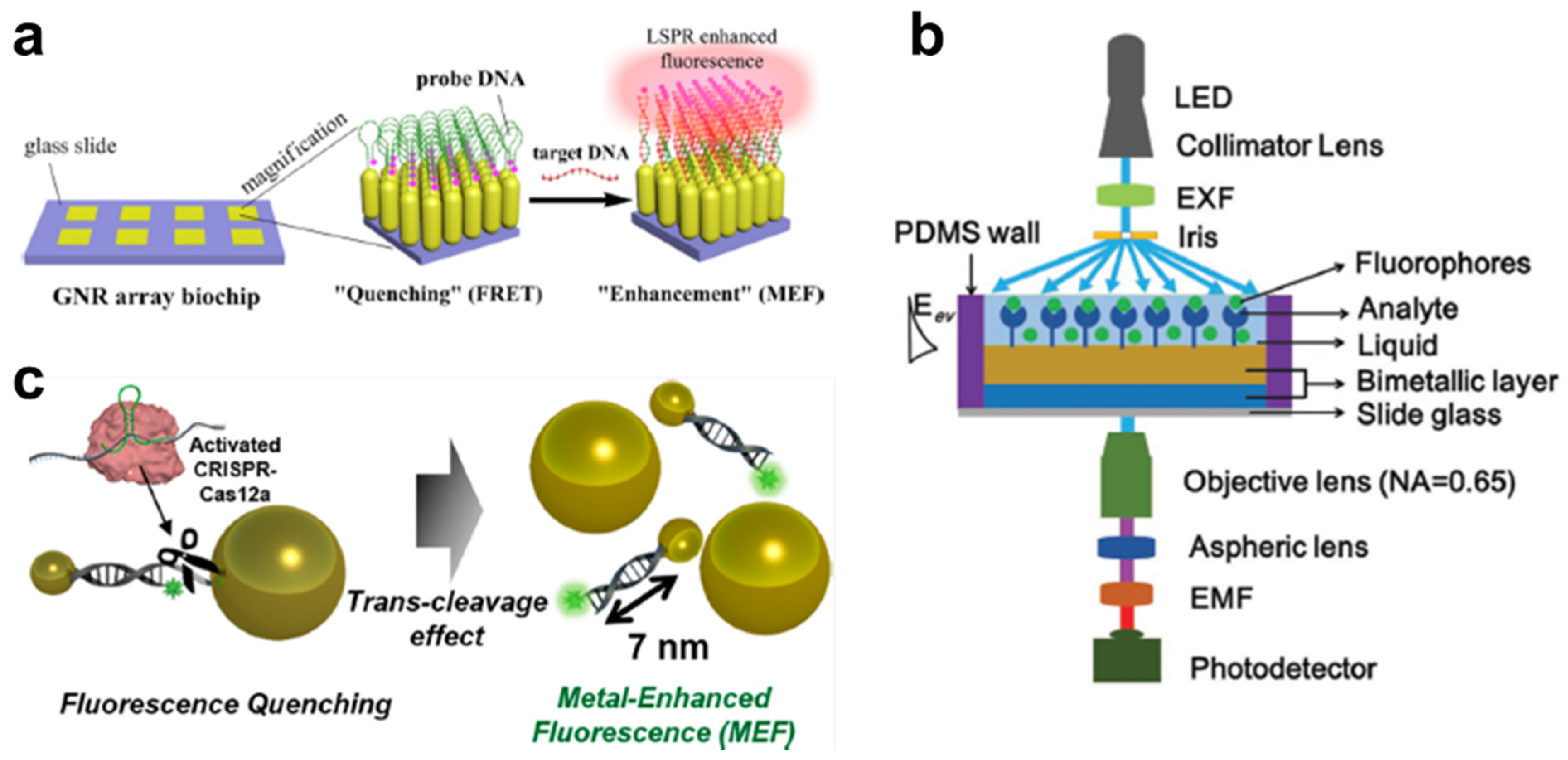
4.2. MEF-Based Biosensors for Detection of DNA Targets
4.3. MEF-Based Biosensors for Detection of RNA Targets
5. Outlook and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Armstrong, A.J.; Lin, P.; Tombal, B.; Saad, F.; Higano, C.S.; Joshua, A.M.; Parli, T.; Rosbrook, B.; van Os, S.; Beer, T.M. Five-year Survival Prediction and Safety Outcomes with Enzalutamide in Men with Chemotherapy-naive Metastatic Castration-resistant Prostate Cancer from the PREVAIL Trial. Eur. Urol. 2020, 78, 347–357. [Google Scholar] [CrossRef]
- Hong, Y.; Kim, J.; Choi, Y.J.; Kang, J.G. Clinical study of colorectal cancer operation: Survival analysis. Korean J. Clin. Oncol. 2020, 16, 3–8. [Google Scholar] [CrossRef]
- van Rijt, S.H.; Sadler, P.J. Current applications and future potential for bioinorganic chemistry in the development of anticancer drugs. Drug Discov. Today 2009, 14, 1089–1097. [Google Scholar] [CrossRef]
- Kashyap, D.; Kaur, H. Cell-free miRNAs as non-invasive biomarkers in breast cancer: Significance in early diagnosis and metastasis prediction. Life Sci. 2020, 246, 117417. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, M.; Dong, Z.; Zhang, Y.; Yao, D. Circulating specific biomarkers in diagnosis of hepatocellular carcinoma and its metastasis monitoring. Tumour Biol. 2014, 35, 9–20. [Google Scholar] [CrossRef] [PubMed]
- deVos, T.; Tetzner, R.; Model, F.; Weiss, G.; Schuster, M.; Distler, J.; Steiger, K.V.; Grutzmann, R.; Pilarsky, C.; Habermann, J.K.; et al. Circulating Methylated SEPT9 DNA in Plasma Is a Biomarker for Colorectal Cancer. Clin. Chem. 2009, 55, 1337–1346. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef] [PubMed]
- Borrebaeck, C.A. Precision diagnostics: Moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer 2017, 17, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Davis, V.W.; Bathe, O.F.; Schiller, D.E.; Slupsky, C.M.; Sawyer, M.B. Metabolomics and surgical oncology: Potential role for small molecule biomarkers. J. Surg. Oncol. 2011, 103, 451–459. [Google Scholar] [CrossRef]
- Lin, S.Y.; Linehan, J.A.; Wilson, T.G.; Hoon, D.S.B. Emerging Utility of Urinary Cell-free Nucleic Acid Biomarkers for Prostate, Bladder, and Renal Cancers. Eur. Urol. Focus 2017, 3, 265–272. [Google Scholar] [CrossRef]
- Lim, S.H.; Becker, T.M.; Chua, W.; Caixeiro, N.J.; Ng, W.L.; Kienzle, N.; Tognela, A.; Lumba, S.; Rasko, J.E.J.; de Souza, P.; et al. Circulating tumour cells and circulating free nucleic acid as prognostic and predictive biomarkers in colorectal cancer. Cancer Lett. 2014, 346, 24–33. [Google Scholar] [CrossRef]
- Davis-Dusenbery, B.N.; Hata, A. MicroRNA in Cancer: The Involvement of Aberrant MicroRNA Biogenesis Regulatory Pathways. Genes Cancer 2010, 1, 1100–1114. [Google Scholar] [CrossRef] [PubMed]
- Lapin, M.; Oltedal, S.; Tjensvoll, K.; Buhl, T.; Smaaland, R.; Garresori, H.; Javle, M.; Glenjen, N.I.; Abelseth, B.K.; Gilje, B.; et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J. Transl. Med. 2018, 16, 300. [Google Scholar] [CrossRef]
- Bu, J.; Shim, J.E.; Lee, T.H.; Cho, Y.H. Multi-modal liquid biopsy platform for cancer screening: Screening both cancer-associated rare cells and cancer cell-derived vesicles on the fabric filters for a reliable liquid biopsy analysis. Nano Converg. 2019, 6, 1–8. [Google Scholar] [CrossRef]
- Nunna, B.B.; Mandal, D.; Lee, J.U.; Singh, H.; Zhuang, S.; Misra, D.; Bhuyian, M.N.U.; Lee, E.S. Detection of cancer antigens (CA-125) using gold nano particles on interdigitated electrode-based microfluidic biosensor. Nano Converg. 2019, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Faltin, B.; Zengerle, R.; von Stetten, F. Current methods for fluorescence-based universal sequence-dependent detection of nucleic acids in homogenous assays and clinical applications. Clin. Chem. 2013, 59, 1567–1582. [Google Scholar] [CrossRef][Green Version]
- Zhang, M.Y.; Ye, J.; He, J.S.; Zhang, F.; Ping, J.F.; Qian, C.; Wu, J. Visual detection for nucleic acid-based techniques as potential on-site detection methods. A review. Anal. Chim. Acta 2020, 1099, 1–15. [Google Scholar] [CrossRef]
- Kim, D.W.; Chun, H.J.; Kim, J.H.; Yoon, H.; Yoon, H.C. A non-spectroscopic optical biosensor for the detection of pathogenic Salmonella Typhimurium based on a stem-loop DNA probe and retro-reflective signaling. Nano Converg. 2019, 6, 1–8. [Google Scholar] [CrossRef]
- Kricka, L.J.; Fortina, P. Analytical ancestry:“Firsts” in fluorescent labeling of nucleosides, nucleotides, and nucleic acids. Clin. Chem. 2009, 55, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Xing, B.L.; Song, J.Z.; Zhang, F.; Nie, C.Y.; Jiao, L.; Liu, L.B.; Lv, F.T.; Wang, S. Associated Analysis of DNA Methylation for Cancer Detection Using CCP-Based FRET Technique. Anal. Chem. 2014, 86, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, J.; Yang, X.; Quan, K.; Wang, H.; Ying, L.; Xie, N.; Ou, M.; Wang, K. FRET Nanoflares for Intracellular mRNA Detection: Avoiding False Positive Signals and Minimizing Effects of System Fluctuations. J. Am. Chem. Soc. 2015, 137, 8340–8343. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.; Bhethanabotla, V.R. Integrating Metal-Enhanced Fluorescence and Surface Acoustic Waves for Sensitive and Rapid Quantification of Cancer Biomarkers from Real Matrices. ACS Sens. 2018, 3, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kook, Y.M.; Lee, K.; Koh, W.G. Metal enhanced fluorescence (MEF) for biosensors: General approaches and a review of recent developments. Biosens. Bioelectron. 2018, 111, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Yu, Y.; Suzuki, M.; Campbell, N.; Mazdo, J.; Vasanthakumar, A.; Bhagat, T.D.; Nischal, S.; Christopeit, M.; Parekh, S. Genome-wide hydroxymethylation tested using the HELP-GT assay shows redistribution in cancer. Nucleic Acids Res. 2013, 41, 157. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Wei, J.-R.; Pan, J.-X.; Zhang, W.; Dang, F.-Q.; Zhang, Z.-Q.; Zhang, J. Spectroscopic quantification of 5-hydroxymethylcytosine in genomic DNA using boric acid-functionalized nano-microsphere fluorescent probes. Biosens. Bioelectron. 2017, 91, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jiang, W.; Liu, F.; Zhou, Y.; Yin, H.; Ai, S. A novel electrochemical immunosensor for the quantitative detection of 5-hydroxymethylcytosine in genomic DNA of breast cancer tissue. Chem. Commun. 2015, 51, 14671–14673. [Google Scholar] [CrossRef]
- Chen, S.; Dou, Y.; Zhao, Z.; Li, F.; Su, J.; Fan, C.; Song, S. High-sensitivity and high-efficiency detection of DNA hydroxymethylation in genomic DNA by multiplexing electrochemical biosensing. Anal. Chem. 2016, 88, 3476–3480. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, M.; Bai, W.; Sun, H.; Li, Y.; Deng, H. A convenient electrogenerated chemiluminescence biosensing method for selective detection of 5-hydroxymethylcytosine in genomic DNA. Sens. Actuators B Chem. 2019, 284, 236–242. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, H.; Liu, F.; Wu, Z.; Lu, S.; Jin, Q.; Zhao, J.; Zhong, X.; Mao, H. Highly sensitive detection of DNA methylation levels by using a quantum dot-based FRET method. Nanoscale 2015, 7, 17547–17555. [Google Scholar] [CrossRef]
- Gao, F.; Fan, T.; Ou, S.; Wu, J.; Zhang, X.; Luo, J.; Li, N.; Yao, Y.; Mou, Y.; Liao, X. Highly efficient electrochemical sensing platform for sensitive detection DNA methylation, and methyltransferase activity based on Ag NPs decorated carbon nanocubes. Biosens. Bioelectron. 2018, 99, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wu, Y.; Xu, Q.; Ma, F.; Zhang, C.-Y. Recent advances in biosensors for in vitro detection and in vivo imaging of DNA methylation. Biosens. Bioelectron. 2020, 112712. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, Y.; Hoque, S.; Tilley, R.D.; Gooding, J.J. Rapid and ultrasensitive electrochemical detection of circulating tumor DNA by hybridization on the network of gold-coated magnetic nanoparticles. Chem. Sci. 2021, 12, 5196–5201. [Google Scholar] [CrossRef]
- Li, D.; Xu, Y.; Fan, L.; Shen, B.; Ding, X.; Yuan, R.; Li, X.; Chen, W. Target-driven rolling walker based electrochemical biosensor for ultrasensitive detection of circulating tumor DNA using doxorubicin@ tetrahedron-Au tags. Biosens. Bioelectron. 2020, 148, 111826. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, C.; Li, Y.; Ma, Y.; Deng, J.; Li, L.; Sun, J. Thermophoretic detection of exosomal microRNAs by nanoflares. J. Am. Chem. Soc. 2020, 142, 4996–5001. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Lim, J.; Lee, S.; Son, H.Y.; Rho, H.W.; Kim, H.; Kang, H.; Jeong, J.; Lim, E.-K.; Jung, J. Urinary exosomal mRNA detection using novel isothermal gene amplification method based on three-way junction. Biosens. Bioelectron. 2020, 167, 112474. [Google Scholar] [CrossRef]
- Ayakannu, T.; Taylor, A.H.; Konje, J.C. Selection of Endogenous Control Reference Genes for Studies on Type 1 or Type 2 Endometrial Cancer. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Chang, J.; Zhang, Y.; Li, Y.; Han, Z.; Tian, F.; Liu, C.; Feng, Q.; Wang, Y.; Sun, J.; Zhang, L. Multilayer Ratiometric Fluorescent Nanomachines for Imaging mRNA in Live Cells. Small Methods 2021, 5, 2001047. [Google Scholar] [CrossRef]
- Zhang, M.; Hong, H.; Fen-Yue, Z.; Zhong, J.-C.; Jian-Ping, L. Electrochemical luminescent DNA sensor based on polymerase-assisted signal amplification. Chin. J. Anal. Chem. 2018, 46, 203–209. [Google Scholar] [CrossRef]
- Yang, L.; Li, J.; Pan, W.; Wang, H.; Li, N.; Tang, B. Fluorescence and photoacoustic dual-mode imaging of tumor-related mRNA with a covalent linkage-based DNA nanoprobe. Chem. Commun. 2018, 54, 3656–3659. [Google Scholar] [CrossRef]
- Zhong, L.; Cai, S.; Huang, Y.; Yin, L.; Yang, Y.; Lu, C.; Yang, H. DNA octahedron-based fluorescence nanoprobe for dual tumor-related mRNAs detection and imaging. Anal. Chem. 2018, 90, 12059–12066. [Google Scholar] [CrossRef]
- Islam, M.N.; Gopalan, V.; Haque, M.H.; Masud, M.K.; Al Hossain, M.S.; Yamauchi, Y.; Nguyen, N.-T.; Lam, A.K.-Y.; Shiddiky, M.J. A PCR-free electrochemical method for messenger RNA detection in cancer tissue samples. Biosens. Bioelectron. 2017, 98, 227–233. [Google Scholar] [CrossRef]
- Rodrigues, V.C.; Soares, J.C.; Soares, A.C.; Braz, D.C.; Melendez, M.E.; Ribas, L.C.; Scabini, L.F.; Bruno, O.M.; Carvalho, A.L.; Reis, R.M. Electrochemical and optical detection and machine learning applied to images of genosensors for diagnosis of prostate cancer with the biomarker PCA3. Talanta 2021, 222, 121444. [Google Scholar] [CrossRef]
- Yang, L.; Kim, T.H.; Cho, H.Y.; Luo, J.; Lee, J.M.; Chueng, S.T.D.; Hou, Y.; Yin, P.T.T.; Han, J.; Kim, J.H. Hybrid Graphene-Gold Nanoparticle-Based Nucleic Acid Conjugates for Cancer-Specific Multimodal Imaging and Combined Therapeutics. Adv. Func. Mater. 2021, 31, 2006918. [Google Scholar] [CrossRef]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 1–19. [Google Scholar] [CrossRef]
- Shuai, H.-L.; Huang, K.-J.; Chen, Y.-X.; Fang, L.-X.; Jia, M.-P. Au nanoparticles/hollow molybdenum disulfide microcubes based biosensor for microRNA-21 detection coupled with duplex-specific nuclease and enzyme signal amplification. Biosens. Bioelectron. 2017, 89, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Shandilya, R.; Ranjan, S.; Khare, S.; Bhargava, A.; Goryacheva, I.Y.; Mishra, P.K. Point-of-care diagnostics approaches for detection of lung cancer-associated circulating miRNAs. Drug Discov. Today 2021. [Google Scholar] [CrossRef]
- Wang, C.; Peng, R.; Zeng, M.; Zhang, Z.; Liu, S.; Jiang, D.; Lu, Y.; Zou, F. An autoregulatory feedback loop of miR-21/VMP1 is responsible for the abnormal expression of miR-21 in colorectal cancer cells. Cell Death Dis. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhang, G.; Dong, H.; Ma, M.; Sun, Q. miR-203 facilitates tumor growth and metastasis by targeting fibroblast growth factor 2 in breast cancer. Onco Targets Ther. 2016, 9, 6203. [Google Scholar] [CrossRef]
- Mohammadi, A.; Mansoori, B.; Duijf, P.H.; Safarzadeh, E.; Tebbi, L.; Najafi, S.; Shokouhi, B.; Sorensen, G.L.; Holmskov, U.; Baradaran, B. Restoration of miR-330 expression suppresses lung cancer cell viability, proliferation, and migration. J. Cell. Physiol. 2021, 236, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, S.; Mohammadi, S.; Salimi, A. A 3D hydrogel based on chitosan and carbon dots for sensitive fluorescence detection of microRNA-21 in breast cancer cells. Talanta 2021, 224, 121895. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, T.; Bai, Y.; Li, Y.; Qiu, J.; Yu, W.; Zhuo, Y. Dual-amplified strategy for ultrasensitive electrochemical biosensor based on click chemistry-mediated enzyme-assisted target recycling and functionalized fullerene nanoparticles in the detection of microRNA-141. Biosens. Bioelectron. 2020, 150, 111964. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yang, F.; Tu, T.-T.; Liao, N.; Chai, Y.-Q.; Yuan, R.; Zhuo, Y. A synergistic promotion strategy remarkably accelerated electrochemiluminescence of SnO2 QDs for MicroRNA detection using 3D DNA walker amplification. Biosens. Bioelectron. 2021, 173, 112820. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-F.; Chou, Y.-T.; Cheng, C.-Y.; Hsu, J.-F.; Su, L.-C.; Ho, J.-A.A. Amplification-free Detection of Cytomegalovirus miRNA Using a Modification-free Surface Plasmon Resonance Biosensor. Anal. Chem. 2021. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, F.; Zhang, H.; Zhang, Y.; Liu, M.; Liu, Y. Universal Ti3C2 MXenes based self-standard ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal. Chem. 2018, 90, 12737–12744. [Google Scholar] [CrossRef]
- Kaur, A.; Dhakal, S. Recent applications of FRET-based multiplexed techniques. Trends Anal. Chem. 2020, 123, 115777. [Google Scholar] [CrossRef]
- Pollock, A.J.; Zaver, S.A.; Woodward, J.J. A STING-based biosensor affords broad cyclic dinucleotide detection within single living eukaryotic cells. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Melnychuk, N.; Egloff, S.; Runser, A.; Reisch, A.; Klymchenko, A.S. Light-Harvesting Nanoparticle Probes for FRET-Based Detection of Oligonucleotides with Single-Molecule Sensitivity. Angew. Chem. 2020, 132, 6878–6885. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, Y.; Yang, X.; Tang, Y.; Han, S.; Kang, A.; Deng, H.; Chi, Y.; Zhu, D.; Lu, Y. FÖrster resonance energy transfer (FRET)-based biosensors for biological applications. Biosens. Bioelectron. 2019, 138, 111314. [Google Scholar] [CrossRef]
- Selvin, P.R. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Mol. Biol. 2000, 7, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, Z.; Jiang, D.; Wang, H.; Lai, W.-F.; Lv, Y.; Zhai, Y. A FRET biosensor based on MnO2 nanosphere/copper nanocluster complex: From photoluminescence quenching to recovery and magnification. Sens. Actuators B Chem. 2019, 290, 535–543. [Google Scholar] [CrossRef]
- Karimi, M.A.; Dadmehr, M.; Hosseini, M.; Korouzhdehi, B.; Oroojalian, F. Sensitive detection of methylated DNA and methyltransferase activity based on the lighting up of FAM-labeled DNA quenched fluorescence by gold nanoparticles. RSC Adv. 2019, 9, 12063–12069. [Google Scholar] [CrossRef]
- Tyagi, A.; Chu, K.L.; Abidi, I.H.; Cagang, A.A.; Zhang, Q.; Leung, N.L.; Zhao, E.; Tang, B.Z.; Luo, Z. Single-probe multistate detection of DNA via aggregation-induced emission on a graphene oxide platform. Acta Biomater. 2017, 50, 334–343. [Google Scholar] [CrossRef]
- Eftekhari-Sis, B.; Aliabad, M.A.; Karimi, F. Graphene oxide based nano-biosensor for the detection of deletion mutation in exon 19 of EGFR gene, leading to lung cancer. Mater. Lett. 2016, 183, 441–443. [Google Scholar] [CrossRef]
- Peng, X.; Zhang, Y.; Lu, D.; Guo, Y.; Guo, S. Ultrathin Ti3C2 nanosheets based “off-on” fluorescent nanoprobe for rapid and sensitive detection of HPV infection. Sens. Actuators B Chem. 2019, 286, 222–229. [Google Scholar] [CrossRef]
- Severi, C.; Melnychuk, N.; Klymchenko, A.S. Smartphone-assisted detection of nucleic acids by light-harvesting FRET-based nanoprobe. Biosens. Bioelectron. 2020, 168, 112515. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wei, S.; Wang, S.; Zhu, X.; Lei, C.; Huang, Y.; Nie, Z.; Yao, S. Chimeric DNA-functionalized titanium carbide MXenes for simultaneous mapping of dual cancer biomarkers in living cells. Anal. Chem. 2018, 91, 1651–1658. [Google Scholar] [CrossRef] [PubMed]
- Oudeng, G.; Au, M.; Shi, J.; Wen, C.; Yang, M. One-step in situ detection of miRNA-21 expression in single cancer cells based on biofunctionalized MoS2 nanosheets. ACS Appl. Mater. Interfaces 2018, 10, 350–360. [Google Scholar] [CrossRef]
- Afzalinia, A.; Mirzaee, M. Ultrasensitive fluorescent miRNA biosensor based on a “sandwich” oligonucleotide hybridization and fluorescence resonance energy transfer process using an Ln (III)-MOF and Ag nanoparticles for early cancer diagnosis: Application of central composite design. ACS Appl. Mater. Interfaces 2020, 12, 16076–16087. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Gao, Y.; Tang, W.; Qiang, L.; Han, Y.; Gao, J.; Zhang, Y.; Liu, H.; Han, L. Attomolar-Level Ultrasensitive and Multiplex microRNA Detection Enabled by a Nanomaterial Locally Assembled Microfluidic Biochip for Cancer Diagnosis. Anal. Chem. 2021, 93, 5129–5136. [Google Scholar] [CrossRef]
- Sekhon, S.S.; Kaur, P.; Kim, Y.-H.; Sekhon, S.S. 2D graphene oxide–aptamer conjugate materials for cancer diagnosis. npj 2D Mater. Appl. 2021, 5, 1–19. [Google Scholar] [CrossRef]
- Yadav, V.; Roy, S.; Singh, P.; Khan, Z.; Jaiswal, A. 2D MoS2-based nanomaterials for therapeutic, bioimaging, and biosensing applications. Small 2019, 15, 1803706. [Google Scholar] [CrossRef] [PubMed]
- Dhenadhayalan, N.; Yadav, K.; Sriram, M.I.; Lee, H.-L.; Lin, K.-C. Ultra-sensitive DNA sensing of a prostate-specific antigen based on 2D nanosheets in live cells. Nanoscale 2017, 9, 12087–12095. [Google Scholar] [CrossRef] [PubMed]
- Badshah, M.A.; Koh, N.Y.; Zia, A.W.; Abbas, N.; Zahra, Z.; Saleem, M.W. Recent Developments in Plasmonic Nanostructures for Metal Enhanced Fluorescence-Based Biosensing. Nanomaterials 2020, 10, 1749. [Google Scholar] [CrossRef] [PubMed]
- Geddes, C.D. Metal-enhanced fluorescence. Phys. Chem. Chem. Phys. PCCP 2013, 15, 19537. [Google Scholar] [CrossRef]
- Bauch, M.; Toma, K.; Toma, M.; Zhang, Q.; Dostalek, J. Plasmon-Enhanced Fluorescence Biosensors: A Review. Plasmonics 2014, 9, 781–799. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Meng, L.; Zhang, W.; Zheng, Y.; Wang, S.; Ren, B.; Yang, Z.; Yan, X. High-throughput single-particle analysis of metal-enhanced fluorescence in free solution using Ag@ SiO2 core–shell nanoparticles. ACS Sens. 2017, 2, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.D.; Liu, C.; Li, C.Y.; Song, C.Y.; Kang, Y.F.; Qi, C.B.; Lin, Y.; Pang, D.W.; Tang, H.W. Dual Amplification Fluorescence Assay for Alpha Fetal Protein Utilizing Immunohybridization Chain Reaction and Metal-Enhanced Fluorescence of Carbon Nanodots. ACS Appl. Mater. Interfaces 2017, 9, 37606–37614. [Google Scholar] [CrossRef]
- Zhou, R.; Sun, S.; Li, C.; Wu, L.; Hou, X.; Wu, P. Enriching Mn-Doped ZnSe Quantum Dots onto Mesoporous Silica Nanoparticles for Enhanced Fluorescence/Magnetic Resonance Imaging Dual-Modal Bio-Imaging. ACS Appl. Mater. Interfaces 2018, 10, 34060–34067. [Google Scholar] [CrossRef]
- Manurung, R.V.; Wu, C.T.; Roy, P.K.; Chattopadhyay, S. A plasmon-tuned ‘gold sandwich’for metal enhanced fluorescence in silica coated NaYF 4: Yb, Er upconversion nanoparticles. RSC Adv. 2016, 6, 87088–87095. [Google Scholar] [CrossRef]
- Choi, J.H.; Choi, J.W. Metal-Enhanced Fluorescence by Bifunctional Au Nanoparticles for Highly Sensitive and Simple Detection of Proteolytic Enzyme. Nano Lett. 2020, 20, 7100–7107. [Google Scholar] [CrossRef] [PubMed]
- Miranda, B.; Chu, K.Y.; Maffettone, P.L.; Shen, A.Q.; Funari, R. Metal-Enhanced Fluorescence Immunosensor Based on Plasmonic Arrays of Gold Nanoislands on an Etched Glass Substrate. ACS Appl. Nano Mater. 2020, 3, 10470–10478. [Google Scholar] [CrossRef]
- Sabanayagam, C.R.; Lakowicz, J.R. Increasing the sensitivity of DNA microarrays by metal-enhanced fluorescence using surface-bound silver nanoparticles. Nucleic Acids Res. 2007, 35, e13. [Google Scholar] [CrossRef]
- Ji, X.; Xiao, C.; Lau, W.F.; Li, J.; Fu, J. Metal enhanced fluorescence improved protein and DNA detection by zigzag Ag nanorod arrays. Biosens. Bioelectron. 2016, 82, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.; Tang, L. Surface-Plasmon-Coupled Fluorescence Enhancement Based on Ordered Gold Nanorod Array Biochip for Ultrasensitive DNA Analysis. Anal. Chem. 2017, 89, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Badshah, M.A.; Ju, J.; Lu, X.; Abbas, N.; Kim, S.M. Enhancing the sensitivity of DNA microarrays by metal-enhanced fluorescence using vertical nanorod structures. Sens. Actuator B Chem. 2018, 274, 451–457. [Google Scholar] [CrossRef]
- Tran, N.H.T.; Trinh, K.T.L.; Lee, J.H.; Yoon, W.J.; Ju, H. Reproducible Enhancement of Fluorescence by Bimetal Mediated Surface Plasmon Coupled Emission for Highly Sensitive Quantitative Diagnosis of Double-Stranded DNA. Small 2018, 14, 1801385. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, T.; Huang, H.; Chen, Y.; Liu, F.; Huang, C.; Li, N. A dual amplification strategy for DNA detection combining bio-barcode assay and metal-enhanced fluorescence modality. Chem. Commun. (Camb.) 2014, 50, 13373–13376. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.F.; Wu, Y.S.; Zhang, L.Z.; Liu, Y.C.; Li, Y.; Yan, Y.L.; Wu, D.C. Hybrid magnetic nanoparticle/nanogold clusters and their distance-dependent metal-enhanced fluorescence effect via DNA hybridization. Nanoscale 2014, 6, 8681–8693. [Google Scholar] [CrossRef]
- Wu, S.; Li, C.; Shi, H.; Huang, Y.; Li, G. Design of metal–organic framework-based nanoprobes for multicolor detection of DNA targets with improved sensitivity. Anal. Chem. 2018, 90, 9929–9935. [Google Scholar] [CrossRef]
- Zhu, Z.J.; Yuan, P.Y.; Li, S.; Garai, M.; Hong, M.H.; Xu, Q.H. Plasmon-Enhanced Fluorescence in Coupled Nanostructures and Applications in DNA Detection. Acs Appl. Bio Mater. 2018, 1, 118–124. [Google Scholar] [CrossRef]
- Choi, J.H.; Lim, J.; Shin, M.; Paek, S.H.; Choi, J.W. CRISPR-Cas12a-Based Nucleic Acid Amplification-Free DNA Biosensor via Au Nanoparticle-Assisted Metal-Enhanced Fluorescence and Colorimetric Analysis. Nano Lett. 2021, 21, 693–699. [Google Scholar] [CrossRef]
- Aslan, K.; Huang, J.; Wilson, G.M.; Geddes, C.D. Metal-enhanced fluorescence-based RNA sensing. J. Am. Chem. Soc. 2006, 128, 4206–4207. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Lan, F.; Yin, X.; Ge, S.; Yu, J.; Yan, M. Metal-enhanced fluorescence/visual bimodal platform for multiplexed ultrasensitive detection of microRNA with reusable paper analytical devices. Biosens. Bioelectron. 2017, 95, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Zong, S.F.; Wang, Z.Y.; Wu, L.; Chen, P.; Yun, B.F.; Cui, Y.P. Microfluidic chip based micro RNA detection through the combination of fluorescence and surface enhanced Raman scattering techniques. Nanotechnology 2017, 28, 105501. [Google Scholar] [CrossRef]
- Masterson, A.N.; Liyanage, T.; Berman, C.; Kaimakliotis, H.; Johnson, M.; Sardar, R. A novel liquid biopsy-based approach for highly specific cancer diagnostics: Mitigating false responses in assaying patient plasma-derived circulating microRNAs through combined SERS and plasmon-enhanced fluorescence analyses. Analyst 2020, 145, 4173–4180. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.Y.; Tu, D.T.; Liu, Y.; Zhou, S.Y.; Zheng, W.; Chen, X.Y. Ultrasensitive detection of cancer biomarker microRNA by amplification of fluorescence of lanthanide nanoprobes. Nano Res. 2018, 11, 264–273. [Google Scholar] [CrossRef]
- Lee, J.H.; Choi, J.H.; Chueng, S.D.; Pongkulapa, T.; Yang, L.; Cho, H.Y.; Choi, J.W.; Lee, K.B. Nondestructive Characterization of Stem Cell Neurogenesis by a Magneto-Plasmonic Nanomaterial-Based Exosomal miRNA Detection. ACS Nano 2019, 13, 8793–8803. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, J.; Zhang, R. Four microRNAs Signature for Survival Prognosis in Colon Cancer using TCGA Data. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yu, L.; Lin, X.; Zheng, Q.; Zhang, S.; Chen, D.; Pan, X.; Huang, Y. Combination of Serum miRNAs with Serum Exosomal miRNAs in Early Diagnosis for Non-Small-Cell Lung Cancer. Cancer Manag. Res. 2020, 12, 485–495. [Google Scholar] [CrossRef]
- Song, C.; Chen, W.H.; Kuang, J.Y.; Yao, Y.; Tang, S.; Zhao, Z.; Guo, X.J.; Shen, W.; Lee, H.K. Recent advances in the detection of multiple microRNAs. Trends Anal. Chem. 2021, 139, 116269. [Google Scholar] [CrossRef]
| Feature | Donor/Acceptor | Wavelengths (Ex,Em) | Target | Required Time | Detection Limit | Ref |
|---|---|---|---|---|---|---|
| FAM-labeled DNA probe adsorption on AuNPs via thiol-modified probes; When hybridizing with target DNA, fluorescence occurrence due to reduced interaction and increased distance. | AuNPs/FAM-labeled DNA probe | Ex: 545 nm Em: 580 nm | methylated DNA | >2 h | 2.2 pM | [62] |
| Aggregation-induced release (AIE) molecules and cDNA adsorption on GO via π-π stacking; Increased distance by reduced interaction when hybridizing with target DNA eliminates quenching effect | GO/AIE labeled DNA probe | Ex: 370 nm Em: 465 nm | DNA | 3 min | 3.1 pM | [63] |
| FAM-labeled DNA adsorption on GO via π-π stacking; FAM-labeled ssDNA desorption and fluorescence recovery from GO through addition of target DNA | GO/FAM-labeled DNA probe | Ex: 488 nm Em: 520 nm | Exon 10 (EGFR gene) | 10 min | 35 fmol/μL | [64] |
| FAM-labeled DNA adsorption on Ti3C2 NSs via π-π stacking; Detection of PCR-amplified HPV-18 DNA from cervical scrape samples | Ti3C2 NSs/FAM-DNA probe | Ex: 495 nm Em: 520 nm | HPV-18 | 5 min | 100 pM | [65] |
| Target DNA detection by Rho 110 and 6G-PMMA using a smartphone RGB camera using a two-color fluorescence microscope; Prevention dye self-quenching of polymer nanoparticles encapsulating green fluorescent donor (Rho 110 and 6G) and hydrophobic counterions (ATTO647N) as acceptors | Rho 110/6G-PMMA/ATTO647N | Ex: 488 nm Em: 662 nm | DNA | 3 h | 10 fM | [66] |
| Covalent binding of TAMRA-labeled MUC1 aptamer and TAMRA-labeled miR-21 to the surface of chimeric DNA-functionalized Ti3C2; Red and green fluorescence recovery by nanoprobe and target MUC1, miR-21 hybridization. | Functionalized Ti3C2/TAMRA- miR-21, TAMRA-MUC1 | Ex: 488 nm Em: 525 nm | miR-21 MUC1 | 2 h | 0.8 nM | [67] |
| Isolation and green fluorescence recovery of dye-labeled ssDNA on the surface of MoS2 nanosheets due to hybridization between the probe and the target miR. | MoS2/FAM-miR 21 probe | Ex: 488 nm Em: 520 nm | miR-21 | > 2 h | 10–50 nM | [68] |
| Detection of UV-vis and gel electrophoresis from 1-aptamers attached to the probe surface using high surface area MOFs. | La (III) MOFs/AgNPs | Ex: 220 nm Em: 430 nm | miR-155 | 45 min | 5.5 fM | [69] |
| FAM-labeled DNA adsorption on GO via π-π stacking; Collection and detection of DNA-miR complexes after the target miRNA hybridization with the probe and detaches from the reaction channel. | GO/FAM-labeled DNA probes | Ex: 488 nm Em: 520 nm | miR-125, miR-126, miR-191, miR-155, miR-21, | 35 min | 0.146 aM | [70] |
| Detection Strategy | Fluorescent Dye/Enhancer | Wavelengths (Ex, Em) | Target | Required Time | Detection Limit | Ref |
|---|---|---|---|---|---|---|
| Detection of DNA by using engineered substrates to increase the emissivity of the fluorophore (proximity of surface plasmons (<100 Å)) | Cy3, Cy5/Ag-nanostructure | Ex: 549 nm, 646 nm Em: 562 nm, 663 nm | DNA | <3 h | - | [83] |
| By applying neutravidin-coated fluorescent nanospheres to biotinylated Au-nanorods, the signal enhancement under the fluorescence microscope using Ag zigzag nanorods (ZNR) arrays | Alexa 488/Ag zigzag nanorod | Ex: 488 nm Em: 525 nm | DNA | <3 h | 0.01 pM | [84] |
| Induction of localized surface plasmon resonance effect (LSPR) of highly ordered monolayer Au-nanorods; Distance-dependent MEF effect between extinction and enhancement | Quasar670/Au-nanorod | Ex: 664 nm Em: 670 nm | ssDNA | <20 min | 10 pM | [85] |
| Detection of DNA by using Ag vertical nanorod (VNR) arrays fabricated via the glancing angle deposition (GLAD) method | Cy5/Ag vertical nanorods | Ex: 635 nm Em: 670 nm | Kallikrein (KLK7) | >12 h | - | [86] |
| Bimetallic structures (containing Au and Ag 2 nm and 50 nm thick) are used to maximize the MEF effect; Detection of amplified target DNA using polymerase chain reaction (PCR). | SYBR Green I/Au-Ag | Ex: 470 nm Em: 535 nm | dsDNA | - | 400 fg/μL | [87] |
| Fix with capture DNA of magnetic nanoparticles implanted with silver nanoparticles; After introduction of target DNA, sandwich structure formation by hybridization reaction and isolation of barcode DNA from silver nanoparticles and fluorescence enhancement | FAM/NMP-AgNP | Ex: 495 nm Em: 518 nm | DNA | >10 min | 1 pM | [88] |
| Increase the signal-to-background ratio by using the high quenching efficiency of AuNPs; Distance-dependent MEF effect and high specificity of target fluorescein isothiocyanate-tagged DNA-HMNC and target DNA hybridization | FITC/Magnetic NP/nanogold clusters | Ex: 488 nm Em: 520 nm | DNA | >2 h | 0.01 μM | [89] |
| Amine-functionalized Fe3O4 nanoparticles bind with amine-gold bonding reaction and form magnetic Au-nanoclusters; After binding of target DNA, distance-dependent MEF effect between AuNP and FITC | Flu, Rho 6G, Cy5/MOF-based nanoprobes | Ex: 494, 525, and 646 nm Em: 513 nm, 553 nm, 664 nm | DNA | <30 min | 20 fM | [90] |
| Quenching of AuNPconjugated Cy5 by AuNP, complementary binding of target DNA with capture DNA on AuNP and other DNA on Au-nanorod, AuNP or Au@AgNP, and enhancement of fluorescence signal. | Au-NP-conjugated Cy5/Au@AgNP | Ex: 410 nm Em: 660 nm | DNA | >12 h | 3.1 pM | [91] |
| Length optimization of dsDNA and attachment of Au-nanoparticles (AuNPs) to the surface; Activation of CRISPR-Cas12a complex by target cfDNA, degradation of AuNP and fluorophore ssDNA, and generation of red-violet fluorescence | FITC/DNA-functionalized AuNP | Ex: 490 nm Em: 520 nm | breast cancer gene-1 (BRCA-1) | <30 min | 0.34 fM | [92] |
| Annealing of target RNA and DNA anchor probes tagged with silver nanoparticles and fluorophores on a solid surface and enhancement of fluorescence signal | TAMRA/AgNP | Ex: 532 nm Em: 585 nm | β-globin mRNA | 30–60 min | 25 fM | [93] |
| Fabrication of sandwich structure using 484-mer RNA and 15-mer complementary RNA attached to Flower-like silver film; Enhanced fluorescence emission of TAMRA-tagged 15-mer RNA; Shortening the spacing distance between N-CD and CeO2 and significantly quenching the fluorescence of N-CD; Fluorescence recovery of quenched probes in the presence of target miRNAs | (DNA1-NCDs)/Flower-like Ag | Ex: 390 nm Em: 462 nm | miR-210, miR-21 | 15 min | 0.03 fM, 0.06 fM | [94] |
| FAM-MB formed a hairpin structure and the fluorescence reduction of 6-FAM and AgNPs; Upon target miRNA hybridization, increasing the distance between 6-FAM and AgNPs and Fluorescence enhancement | FAM-tagged MB/AgNRs | Ex: 488 nm Em: 520 nm | miR-21 | 60 min | 1 pM | [95] |
| Fluorescent dye (FAM) tagging of chemically synthesized gold triangular nanoprisms (Au TNPs); When the target miRNA hybridizes with the molecular beacon, the distance between the FAM and AgNP increases and the fluorescence intensity of the FAM increases. | FAM/Au TNPs | Ex: 488 nm Em: 520 nm | miR-10b, miR-96 | 120 min | 1.13 pM, 30 fM | [96] |
| Fluorescence enhancement using biotin-functionalized lanthanide nanoparticles as signal enhancers and capture of target miRNAs of surface-modified molecular labels; Detection of Biotin-NPs-probe hybridized with miR in a sandwich structure. | Biotin-NPs/Ln3+-nanoprobe | Ex: 340 nm Em: 617 nm | miR-21 | >2 h | 1.38 fM | [97] |
| The miR-124 specific MB (molecular beacon) attachment on magnetic plasmonic nanorods; Immunomagnetic isolation and enrichment of exosomes in cell culture medium collection, non-destructive analysis of exosome miR-124 expression. | FAM/plasmonic AuNRs | Ex: 490 nm Em: 520 nm | miR-124 | 30 min | 1 pM | [98] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.-H.; Ha, T.; Shin, M.; Lee, S.-N.; Choi, J.-W. Nanomaterial-Based Fluorescence Resonance Energy Transfer (FRET) and Metal-Enhanced Fluorescence (MEF) to Detect Nucleic Acid in Cancer Diagnosis. Biomedicines 2021, 9, 928. https://doi.org/10.3390/biomedicines9080928
Choi J-H, Ha T, Shin M, Lee S-N, Choi J-W. Nanomaterial-Based Fluorescence Resonance Energy Transfer (FRET) and Metal-Enhanced Fluorescence (MEF) to Detect Nucleic Acid in Cancer Diagnosis. Biomedicines. 2021; 9(8):928. https://doi.org/10.3390/biomedicines9080928
Chicago/Turabian StyleChoi, Jin-Ha, Taehyeong Ha, Minkyu Shin, Sang-Nam Lee, and Jeong-Woo Choi. 2021. "Nanomaterial-Based Fluorescence Resonance Energy Transfer (FRET) and Metal-Enhanced Fluorescence (MEF) to Detect Nucleic Acid in Cancer Diagnosis" Biomedicines 9, no. 8: 928. https://doi.org/10.3390/biomedicines9080928
APA StyleChoi, J.-H., Ha, T., Shin, M., Lee, S.-N., & Choi, J.-W. (2021). Nanomaterial-Based Fluorescence Resonance Energy Transfer (FRET) and Metal-Enhanced Fluorescence (MEF) to Detect Nucleic Acid in Cancer Diagnosis. Biomedicines, 9(8), 928. https://doi.org/10.3390/biomedicines9080928







