Abstract
Epithelial ovarian cancers (EOCs) are fatal and obstinate among gynecological malignancies in advanced stage or relapsed status, with serous carcinomas accounting for the vast majority. Unlike EOCs, borderline ovarian tumors (BOTs), including serous BOTs, maintain a semimalignant appearance. Using gene ontology (GO)-based integrative analysis, we analyzed gene set databases of serous BOTs and serous ovarian carcinomas for dysregulated GO terms and pathways and identified multiple differentially expressed genes (DEGs) in various aspects. The SRC (SRC proto-oncogene, non-receptor tyrosine kinase) gene and dysfunctional aryl hydrocarbon receptor (AHR) binding pathway consistently influenced progression-free survival and overall survival, and immunohistochemical staining revealed elevated expression of related biomarkers (SRC, ARNT, and TBP) in serous BOT and ovarian carcinoma samples. Epithelial–mesenchymal transition (EMT) is important during tumorigenesis, and we confirmed the SNAI2 (Snail family transcriptional repressor 2, SLUG) gene showing significantly high performance by immunohistochemistry. During serous ovarian tumor formation, activated AHR in the cytoplasm could cooperate with SRC, enter cell nuclei, bind to AHR nuclear translocator (ARNT) together with TATA-Box Binding Protein (TBP), and act on DNA to initiate AHR-responsive genes to cause tumor or cancer initiation. Additionally, SNAI2 in the tumor microenvironment can facilitate EMT accompanied by tumorigenesis. Although it has not been possible to classify serous BOTs and serous ovarian carcinomas as the same EOC subtype, the key determinants of relevant DEGs (SRC, ARNT, TBP, and SNAI2) found here had a crucial role in the pathogenetic mechanism of both tumor types, implying gradual evolutionary tendencies from serous BOTs to ovarian carcinomas. In the future, targeted therapy could focus on these revealed targets together with precise detection to improve therapeutic effects and patient survival rates.
1. Introduction
Ovarian tumors occupy a certain place among gynecological diseases and most cases are benign in clinical and pathological features such as follicular cysts, corpus luteum cysts, serous or mucinous cystadenomas, endometriomas, and teratomas [1,2]. Comparatively, ovarian cancer is the most lethal gynecological malignancy worldwide although the proportion is relatively low [3]. Epithelial ovarian cancers (EOCs) are the leading cause of death among patients with gynecologic cancers accounting for the vast majority of all ovarian cancers [4,5]; furthermore, serous carcinoma (SC) accounts for the most common of EOCs, with a poor prognosis and a five-year survival rate of only 25% with metastases [6,7]. SC is less likely to be found in the early stages (International Federation of Gynecology and Obstetrics (FIGO) stages I and II), which have higher survival rates because they are easier to treat, whereas patients at advanced stages (FIGO stages III and IV) have poor prognosis and high recurrence rates even after complete debulking surgery combined with chemotherapy (carboplatin and paclitaxel) due to resistance to chemotherapy [6,8,9].
Borderline ovarian tumors (BOTs), a specific subtype of EOCs, consist of disparate groups of neoplasms based on histopathological features, molecular characteristics, and clinical behaviors and BOTs can generally be classified into serous, mucinous, and other subtypes according to clinical and histopathological features [10,11]. Besides, BOTs account for approximately 10–15% of EOCs and usually occur in younger women, resulting in an excellent prognosis [12]. Compared with ovarian cancer patients, who almost always require chemotherapy after a debulking operation, patients with BOTs usually have better prognoses after adequate surgery with an extremely low probability of recurrence or metastasis [13,14]. Serous BOTs, comprising approximately 65% of BOTs, occur mostly in North America, the Middle East, and most of Europe [15]. To date, surgery is still the ideal method to treat BOTs, while adjuvant chemotherapy and radiotherapy are not usually considered as standard therapies [14,16]. Recent studies have inferred several assumptions, including the incessant ovulation, gonadotropin, hormonal, and inflammation hypotheses, to explain the tumorigenesis of serous BOTs [17,18,19]. Serous BOTs are characterized by mutations in the KRAS, BRAF, and ERBB2 genes and overexpression of the p53 and Claudin-1 genes; furthermore, the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) pathway, PI3K/AKT/mTOR pathway, Hedgehog pathway, and angiogenesis pathway are frequently activated in serous BOTs [13,14,20,21,22,23,24,25].
As a complex disease, several genetic and environmental factors contribute to SC development with a complicated carcinogenesis pathway, and the carcinogenesis of SC evolves through several aberrant functions, which fluctuate with disease progression based on findings through the widely utilized FIGO system [13,26,27,28,29,30,31,32]. It is widely known that most serous ovarian carcinomas are associated with TP53 mutations [30,33,34,35,36]; about half of them have undergone abnormal DNA repair processes through homologous recombination due to epigenetic or genetic alterations of BRCA1, BRCA2, or other DNA repair molecules [37,38]; and some show gene mutations, such as in BRAF and KRAS [20]. In addition to debulking surgery and subsequent adjuvant chemotherapy, targeted therapy and systemic immunotherapy can also be utilized to enhance the therapeutic effects. Poly-adenosine diphosphate (ADP) ribose polymerase inhibitors (PARPis), the first approved cancer drugs, were widely used targeted therapies for BRCA1/2-mutated breast and ovarian cancers, and they specifically target DNA damage and repair responses, especially for patients with homologous recombination deficiencies, resulting in increased survival [39,40,41]. However, resistance to PARPi has recently become an emerging issue and breast-related cancer antigens (BRCA) and homologous recombination deficiency (HRD) status can be considered novel predictive biomarkers of response [42,43,44,45]. Therefore, identifying potential crucial biomarkers for monitoring drug resistance and formulating new drug combination strategies are efficacious methods to resolve PARPi resistance along with precision medicine [46,47].
Furthermore, it is well known that serous ovarian carcinomas have a poor clinical prognosis because they are usually diagnosed too late, while advanced stages usually result in frequent emergence of chemoresistance [4,5,8,48,49]. Recent growing evidence suggests that epithelial–mesenchymal transition (EMT) may contribute to tumor invasion and metastasis and promote chemotherapeutic resistance, especially to cisplatin, by converting the motionless epithelial cells into mobile mesenchymal cells, escaping cell adhesion, and altering the cellular extracellular matrix [50,51,52]. EMT is a reversible process in which many crucial components, such as E-cadherin, EpCAM, vimentin, fibronectin, neural cadherin, matrix metalloproteinases, various integrins, and different cytokeratins, are regulated by a complex functional network of transcription factors, including the zinc-finger E-box-binding homeobox factors (Zeb1 and Zeb2), Snail (SNAI1), Slug (SNAI2), and the basic helix–loop–helix factors (Twist1 and Twist2) [53,54,55]. Loss of breast cancer type 1 susceptibility protein (BRCA1), a tumor suppressor that plays a role in mending double-stranded DNA breaks, is also associated with EMT and tumor initiation [50,56]. The expression of EMT signaling pathways has been correlated with poor prognosis in various epithelial cancers, including breast, pancreas, prostate, and ovarian cancer, and the role of EMT in ovarian cancer progression and therapy resistance is highlighted in current studies [57]; however, the role of EMT plasticity in serous ovarian tumors has not been comprehensively investigated.
As mentioned above, although both are named the “serous” subtype in terms of classification, serous BOTs and serous ovarian carcinomas still have decisive differences in genetic mechanisms, pathological characteristics, and clinical manifestations [13,32]. Various functions can be investigated using differentially expressed genes (DEGs) detected by microarrays. In contrast to DEGs, we established a gene set regularity (GSR) model, which reconstructed the functionomes, that is, the GSR indices of the global functions, and then investigated the dysregulated functions and dysfunctional pathways involved in the complex disease. Constructing a functionome can provide information about the dysregulated functionomes accompanied with dysfunctional pathways of complicated illness and we had conducted several gene set-based analyses by integrating microarray gene expression profiles downloaded from publicly available databases, which revealed that comprehensive methods based on functionome defined by gene ontology (GO) are useful for successfully conducting significant research on BOTs and ovarian carcinomas of different stages and subtypes [58,59,60,61,62,63,64]. Previously, individual studies have focused on gene set analysis of serous ovarian carcinomas and serous BOTs to uncover pathogenic mechanisms during tumorigenesis. However, there is no integrated analysis to compare and discover the genomic functionome of serous BOTs and serous ovarian carcinomas. Therefore, we first utilized GO-based integrative approaches to explore expression profile datasets of serous ovarian tumors, including serous BOTs and all stages of serous ovarian carcinomas, to identify common and meaningful dysregulated functions and dysfunctional pathways between these two groups. We then selected the featured DEGs by checking the significant biomarkers related to EMT with cross comparison. In this experiment, we aimed to excavate newly discovered pathogenetic mechanisms based on previous studies that differ from previous theories and hoped to take advantage of these new findings applied in medical detection with targeted therapy and effective avoidance of recurrence for better prognosis of serous ovarian tumors and patient survival.
2. Materials and Methods
2.1. Workflow for the Integrative Analytic Model
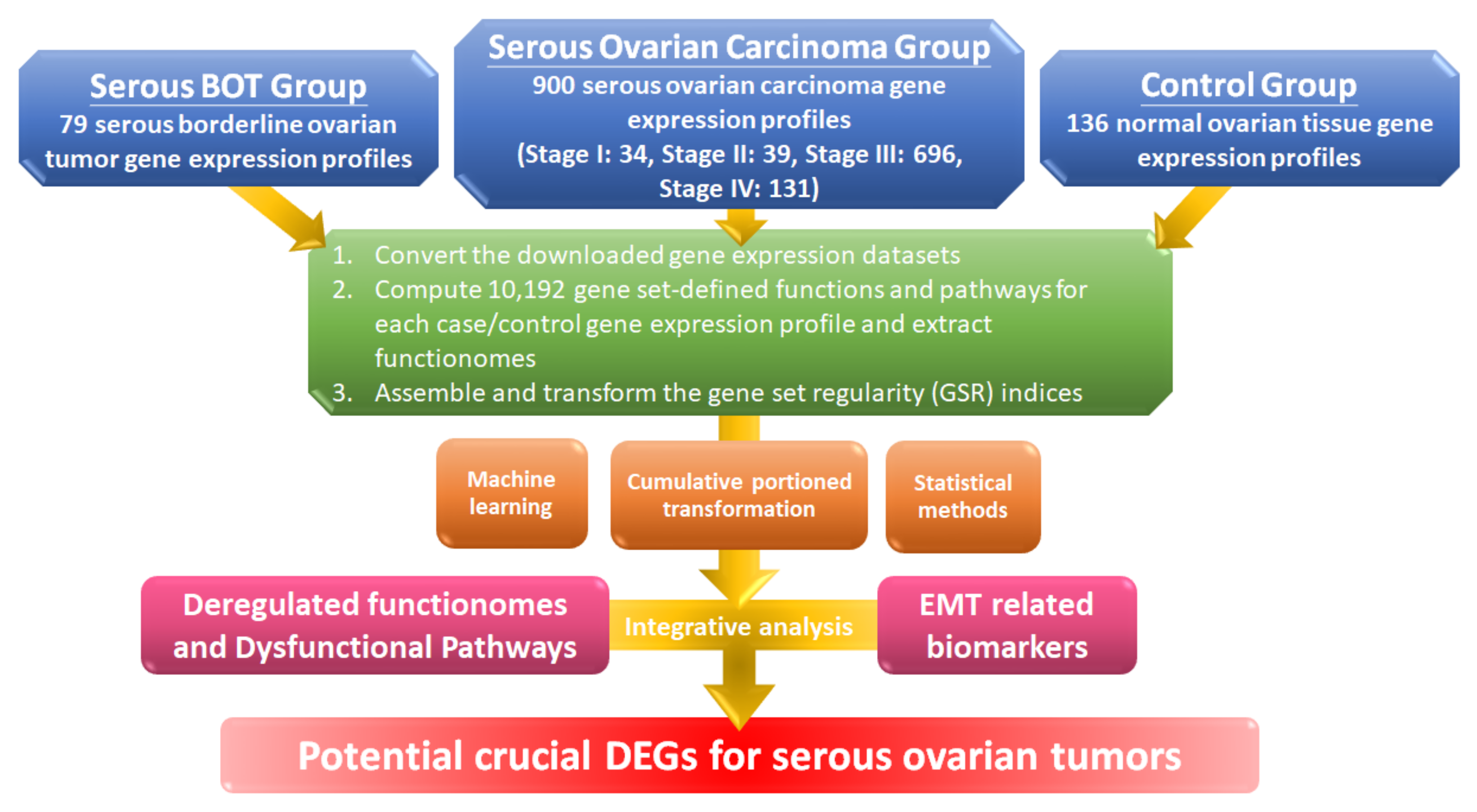
The workflow for this study is shown in the flowchart in Figure 1, and detailed information is explained below. First, we converted the gene expression profile of the extracted gene elements downloaded from the Gene Expression Omnibus (GEO) database with selection criteria for serous ovarian tumors and normal controls to ordered data and then transformed them into 10,192 quantified GO terms according to the sequential expression from the gene elements in each gene set. This process produced functionomes consisting of 10,192 GSR indices, which defined the relatively comprehensive biological and molecular functions to explore serous ovarian tumors, including serous BOTs and all stages of serous ovarian carcinomas. Next, we individually calculated the quantified functions and functional regularity patterns among serous ovarian tumors and 136 normal ovarian controls with GSR indices and established the GSR model for the functionome pattern. Then, we investigated the whole informativeness of genomic functionomes consisting of the GSR indices and constructed a functionome-based training model of classification and prediction using the support vector machine (SVM), a set of supervised mathematical commands from machine learning. The variation in the GSR indices between each serious ovarian tumor group and normal control group revealed that the biomolecular functions among serous ovarian tumors were significantly extensively dysregulated in contrast to the normal control group. Finally, we conducted whole-genome integrative analysis to identify meaningful dysfunctional pathways together with significant biomarkers of EMT involved in the progression of serous ovarian carcinomas to determine crucial DEGs that may be essential parts of the pathogenetic mechanisms for serous ovarian tumors by elucidating dysregulated functionomes using microarray analysis of gene expression profiles. The key biological functions and genes involved in the pathogenesis of serous BOTs and all stages of serous ovarian carcinomas were determined by identifying genome-wide and GO-defined functions and DEGs.
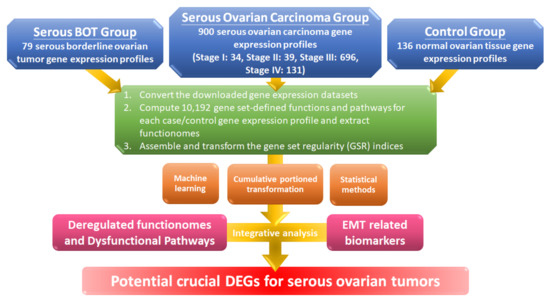
Figure 1.
Workflow of this study. The DNA microarray gene expression profiles of 79 serous borderline tumor (BOT) samples, 900 serous ovarian carcinoma specimens including all stages, and 136 normal ovarian controls were downloaded from publicly available databases with gene set regularity (GSR) indices calculated by the Gene Ontology (GO) gene set. Functionomes consisting of 10,192 GO gene sets established from the polygenic models and cumulative portion transformations with machine learning and statistical methods were utilized to identify the functionome-based patterns to investigate dysregulated GO terms, dysfunctional pathways, and biomarkers of epithelial–mesenchymal transition (EMT) together with integrative analysis and to discover potential crucial differentially expressed genes (DEGs).
2.2. Microarray Dataset Collections and the Selection Criteria
The selection criteria for the microarray gene expression datasets from the GEO database were as follows: (1) samples of normal controls and serous ovarian tumor samples, including serous BOTs and serous ovarian carcinomas, should all originate from ovarian tissues of homo sapiens; (2) datasets should offer sufficient information about the diagnosis and clinical histopathological subtypes of serous ovarian tumors and normal controls should consist of tissues or cell cultures from normal ovarian surface epithelium (NOSE) judged by histology; and (3) any extracted sample that did not meet the above-mentioned conditions was discarded and any gene expression profile in a dataset was abandoned if it contained missing data.
2.3. Computing the GSR Indices and Rebuilding the Functionomes
GSR indices were calculated and extracted from the gene expression datasets by modifying the differential rank retention (DIRAC) algorithm [65] and used to measure sequential changes among the gene elements in the gene set datasets of the gene expression profiles of serous BOTs, all stages of serous ovarian carcinomas, and the most common gene expression ordering from the normal control samples. The details and calculation process of the GSR model were described in our previous studied papers [58,59,60,61,62,63,64]. The microarray-based gene expression profiles from serous ovarian tumors and normal ovarian samples obtained from the GEO database were produced using the corresponding gene expression levels constructed according to the genetic elements in the GO-based functionome, which were then con-verted into ordered data based on each expression level. By definition, the GSR index refers to the ratio of the gene expression sequence in a gene set between the case group and the most common gene expression sequence from the normal ovarian tissue samples, ranging from 0 to 1, where 0 represents the most dysregulated state of a gene set with oppositely ordered gene set regularities between the serous ovarian tumors and the most common gene expression orderings in the normal controls, whereas 1 indicates that the genomic regularities in a gene set remain the same between the groups of serous ovarian tumors and the normal ovarian group. All GSR indices were measured using the R programming language. A functionome was defined as the complete gene set of biological functions, and we annotated and defined the human functionome using the 10,192 GO gene set-defined functions because the definitions for comprehensive biological functions are not yet available. Therefore, the functionomes used in this study were defined as a combination of 10,192 GSR indices for all samples.
2.4. Statistical Analysis
The Mann–Whitney U-test was used to test the differences in serous BOTs, all stages of serous ovarian carcinomas, and controls, and then corrected by multiple hypotheses using the false discovery rate (Benjamini–Hochberg procedure) [66]. The p-value was set at p < 0.05.
2.5. Classification and Prediction by Machine Learning with Set Analysis
An R package with the function “kvsm” provided by the “kernlab” (version 0.9–27; Comprehensive R Archive Network) and kernel-based machine learning methods were used to classify and predict patterns of GSR indices. K-fold cross-validation was used to measure the accuracy of classification and prediction of SVM. The results of ten repetitive predictions were used to evaluate the performance of the binary classification. The R package “pROC” was used to calculate the area under the curve (AUC) [67]. The R package “data.table” (version 1.12.8; Comprehensive R Archive Network) was used to display all possible logical relationships among the dysregulated gene sets of serous ovarian tumors clearly and sequentially in the tables.
2.6. Verification of Clinical Samples Using Immunohistochemical (IHC) Staining Method
Fifty clinical samples of serous ovarian tumors were collected (serous BOTs, n = 9; serous ovarian carcinomas, n = 41, including 8 stage I, 2 stage II, 23 stage III, and 8 stage IV cases). All serous ovarian tumor tissues were collected from female patients undergoing surgical treatments after signing an informed consent agreement. All patients were diagnosed and treated according to the standard therapeutic guidelines, and all tissues of patients were kept in the biobank at Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan. The Institutional Review Board of the General Hospital of the National Defense Medical Center approved the study (2-107-05-043, approved on October 26, 2018, and 2-108-05-091, approved on 20 May 2019). Informed consent was obtained from all patients and control subjects. All clinical tissue samples were confirmed via quantitative histopathological inspections and diagnosed by professional pathologists, and IHC staining results were scored as follows: the intensity (I) was multiplied by the percentage of positive cells (P) of all biomarkers utilized in this study (the formula is shown as IHC score [Q] = I × P; maximum = 300) [68,69].
3. Results
3.1. Microarrays of Sample Groups for Gene Expression Profiles and Definition for Gene Set Analysis
We performed a comprehensive bioinformatics method based on GO to explore and analyze all relational disordered functions of serous ovarian tumors, including serous BOTs and all stages of serous ovarian carcinomas [70]. The gene expression profiles of DNA microarray of serous ovarian tumors and normal controls were downloaded from the GEO repository at the National Center for Biotechnology Information (NCBI) archives. The whole-sample data were obtained from 30 datasets containing eight heterogeneous DNA microarray platforms without any missing data. There were 79 serous BOT samples and 900 serous ovarian carcinoma samples based on histopathological classification, including 34 stage I, 39 stage II, 696 stage III, and 131 stage IV cases among the 900 serous ovarian carcinoma samples according to the FIGO staging system (Table 1). In addition, 136 normal ovarian samples were collected as a control group for comparison. Table S1 provides detailed information about all obtained samples and controls. In total, 10,192 GO-based definitions for annotating all gene set-defined functionomes were also downloaded from the Molecular Signatures Database (MSigDB), the version “c5.all.v7.1.symbols.gmt” [71].

Table 1.
Number of samples and statistics for the groups of serous BOTs and all FIGO stages of serous ovarian carcinomas.
3.2. Histograms of GSR Indices of Functionomes among Each Group with Diverse Differences
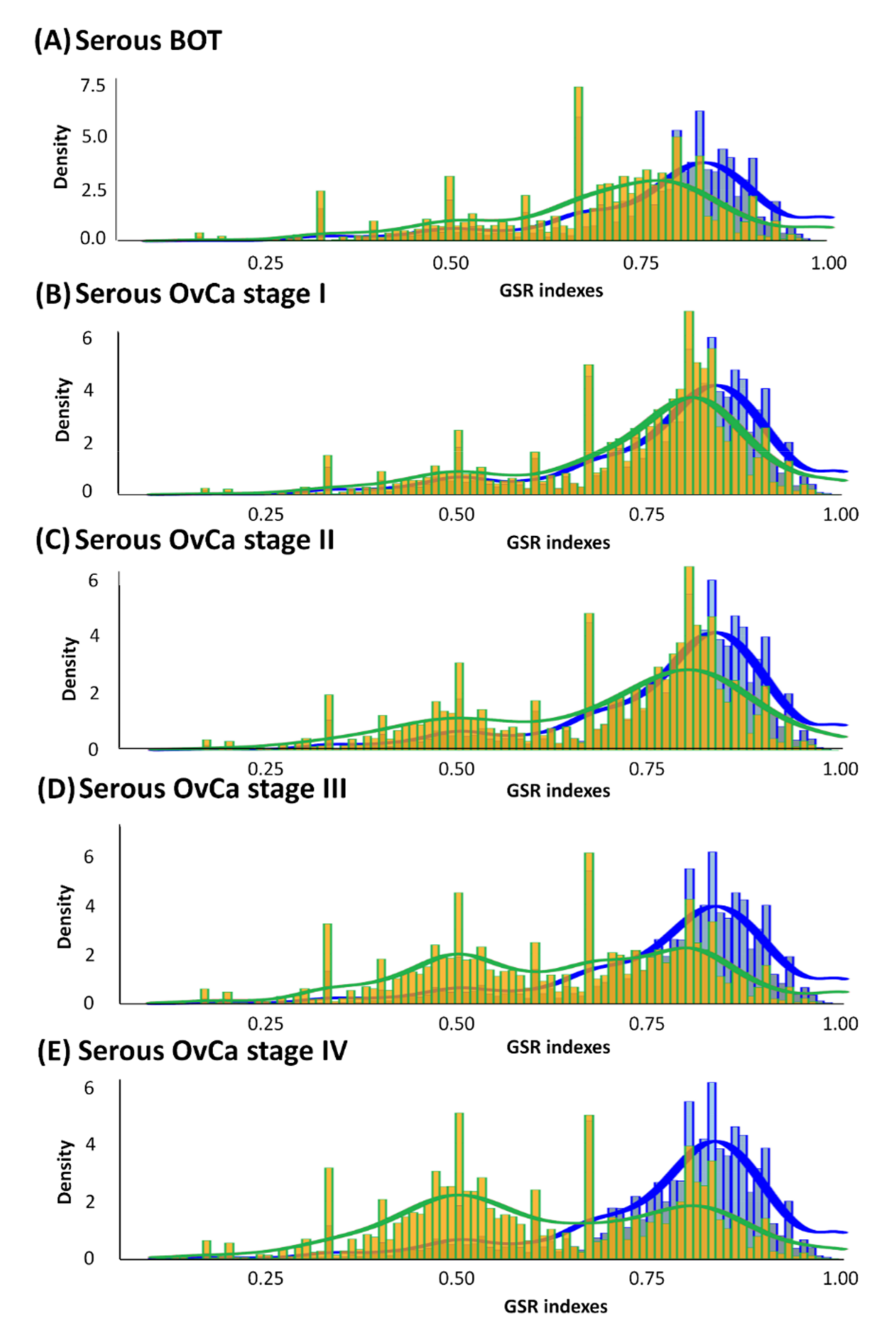
According to the divergence in ranking within a gene set between the case and control groups characterized by GO terms, GSR indices were individually calculated by quantifying alterations in the ranking of gene expression in a gene set or functionome. As displayed in Figure 2, all averages of GSR indices for the functions of serous ovarian tumors were computed and then rectified by the mean values of the control group. Divergences in GSR indices between serous ovarian tumor and normal control groups were statistically significant (p < 0.05). We found that in the serous ovarian carcinoma group, as the FIGO stage progressed from early stages (I and II, yellow-green grids in Figure 2B,C, respectively) to advanced stages (III and IV, yellow-green grids in Figure 2D,E, respectively), the differences from corresponding normal controls (blue grids in Figure 2) became increasingly distinct. Furthermore, differences in mutations among serous BOTs (yellow-green grids in Figure 2A) and normal controls seemed to be more irregular than those at the early stages (FIGO stage I and II) but less aberrant than at the late stages (FIGO stage III and IV) of serous ovarian carcinomas. The modulation of dysregulated functionomes was quantified using the means of the total GSR indices of each functionome of serous ovarian tumors and control groups with adjustments compared to the control group. The average corrected GSR indices for serous BOTs and serous ovarian carcinomas from stage I to IV were 0.7036, 0.7230, 0.6976, 0.6355, and 0.6147, respectively.
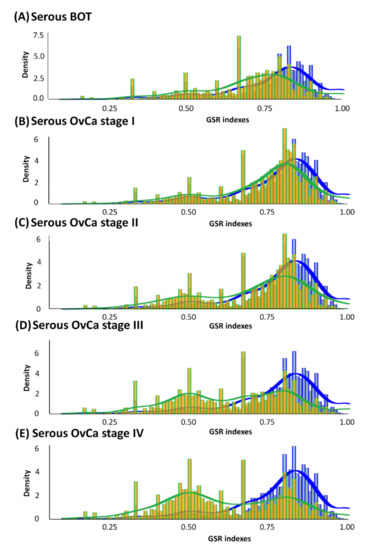
Figure 2.
Histograms of global GSR indices of functionomes for serous BOTs, all stages of serous ovarian carcinomas (yellow green), and control groups (blue). Different distributions of the functionomes among five case sample groups and control groups are shown with statistical significance (p < 0.05). The normal control group (blue, right) was used as the control and is the same in all panels. Peaks in distribution were observed (yellow green), indicating dysregulated biomolecular functionomes of serous BOTs and all stages of serous ovarian carcinomas. (A–E) Corrected GSR indices of serous BOTs: 0.7036 (A); FIGO stage I: 0.7230 (B); FIGO stage II: 0.6976 (C); FIGO stage III: 0.6355 (D); and FIGO stage IV: 0.6147 (E).
3.3. Regularity Patterns of Functionomes Classified and Predicted by Supervised Machine Learning with High Sensitivity, Specificity, and Accuracy
As displayed in Figure 2, the regularity patterns of functionomes among the five serous ovarian tumor groups were compared with those of the normal control group, and the functional regularity patterns of the five case groups (serous BOTs and all four stages of serous ovarian carcinomas) showed significant divergence. We then identified, classified, and predicted different functions of various gene sets defined by GO using SVM, a powerful technological algorithm for supervised machine learning. The accumulated data of assessed performances were operated with ten consecutive binary classifications and checked with forecasting approaches by five-fold cross-validation; all the calculated results with high sensitivity, specificity, and accuracy are listed in Table S2. The sensitivity, specificity, and accuracy of binary classification for gene set databases among serous ovarian tumor and control groups were approximately 95.13–100.00%, 99.85–100.00%, and 98.97–100.00%, respectively. The AUCs for the performance of each case group ranged from 0.9771 to 1.0000. The evaluated performances by binary classifications between serous ovarian carcinoma, FIGO stage IV, and normal control groups had the best overall effects. The results revealed that the quantified functional regularity patterns with the GSR indices transformed from the DNA microarray gene expression profiles could offer sufficient and credible information to the SVM for accurate identification and classification. These results also indicated that all the functional regularity patterns of serous ovarian tumors were demarcated and suitable for integrated genetic and molecular classifications interpreted in this study.
3.4. The Most Dysregulated and Common GO Terms among Serous Ovarian Tumors
We used the cluster weight index (CWI) with SVM to uncover 655, 662, 643, 828, and 841 GO terms among serous BOTs and serous ovarian carcinomas at FIGO stages I–IV, respectively. CWI, a calculated exponent based on the p-values with statistical significance, is defined as the weighted ratio of the single weight of each clustered GO term divided by the total weights of the whole clusters, and it is used to measure the representative weight and express the mutual correlation for every cluster in the GO trees. All identified GO terms were meaningful and could represent dysregulated functionomes in each group of serous ovarian tumors. We used the calculated CWI to quantify and judge the value of each dysregulated GO cluster among the pathogenetic mechanisms of serous ovarian tumors. We ranked the 50 most dysregulated GO terms judged by CWI for serous ovarian tumors, as shown in Table 2. The first dysregulated GO terms for each group were “regulation of immune system process (GO:0002682)” for serous BOT; “transporter activity (GO:0005215)” for serous ovarian carcinoma, FIGO stage I; “small molecule metabolic process (GO:0044281)” for serous ovarian carcinoma, FIGO stage II; “regulation of immune system process (GO:0002682)” for serous ovarian carcinoma, FIGO stage III; and “small molecule metabolic process (GO:0044281)” for serous ovarian carcinoma, FIGO stage IV. Details on dysregulated GO terms for all disease groups of serous ovarian tumors are listed in Table S3. We then selected and reorganized the top 25 from the 50 most dysregulated GO terms among the five groups by comprehensively comparing weighted CWIs with their original rankings in each group, as listed in Table S4. Next, we summarized the 25 most common dysregulated GO terms among the five disease groups of serous ovarian tumors with representative biological and molecular effects and reclassified them into three major categories: cellular cycle and signaling-related effects, membrane and transport-related effects, and metabolic, immunological, and other effects.

Table 2.
The 50 most dysregulated GO terms for serous BOT and all stages of serous ovarian carcinoma ranked by cluster weight index (CWI).
3.5. Three Reclassified Categories of the Top 25 Common Dysregulated GO Terms and the Most Relevant Corresponding DEGs
As displayed in Table 3, we reclassified the top 25 most common dysregulated GO terms among serous ovarian tumors into three major categories based on each representative function. There were 9, 8, and 8 GO terms belonging to “cellular cycle and signaling-related effects”, “membrane and transport-related effects”, and “metabolic, immunological, and other effects”, respectively. We sorted the potential genes annotated for all regrouped GO terms among each category with definitions (http://geneontology.org/, accessed on 5 June 2021) and selected the most relevant DEGs with the highest repetitive frequencies determined statistically with cross comparisons. Nine GO terms were reclassified to cellular cycle- and signaling-related effects and four most relevant DEGs were identified with the highest repetition: EDN1 (endothelin 1), AKT1 (AKT serine/threonine kinase 1), IL1B (interleukin 1 beta), and INS (insulin). Eight GO terms were reclassified to membrane- and transport-related effects and the two most relevant DEGs were identified with the highest repetition: CDK5 (cyclin dependent kinase 5) and ATP1B1 (sodium/potassium-transporting ATPase subunit beta-1). Eight GO terms were reclassified to metabolic, immunological, and other effects and the seven most relevant DEGs were identified with the highest repetition: PTK2B (protein tyrosine kinase 2 beta), MTOR (mechanistic target of rapamycin kinase), APP (amyloid beta precursor protein), KIT (tyrosine-protein kinase KIT), LEP (leptin), MAPK3 (mitogen-activated protein kinase 3), and SRC (proto-oncogene tyrosine-protein kinase Src).

Table 3.
Categorized lists of the top 25 common dysregulated GO terms among serous ovarian tumors reclassified by biological functions and the most relevant corresponding DEGs in each group.
3.6. The Significant Common Dysfunctional GO-Defined Pathways and Corresponding DEGs
We firstly discovered that there were 5346, 4047, 6779, 7985, and 8251 dysfunctional pathways defined with GO terms in the serous BOT and serous ovarian carcinoma stage I–IV groups, respectively. Then, we placed these pathways in order of correlation for each group according to statistically significant p-values. Next, we selected the top 50 most dysfunctional pathways ranked by p-value for each disease group to investigate meaningful correlations, as listed in Table 4 and the detailed GO-defined pathways for serous ovarian tumors are listed in Table S5. “Negative regulation of isotype switching (GO:0045829)” ranked first in the serous BOT group; “modified amino acid transmembrane transporter activity (GO:0072349)” ranked first in serous ovarian carcinoma, FIGO stage I; “DNA double-strand break processing involved in repair via single-strand annealing (GO:0010792)” ranked first in serous ovarian carcinoma, FIGO stage II; “DNA double-strand break processing involved in repair via single-strand annealing (GO:0010792)” ranked first in serous ovarian carcinoma, FIGO stage III; and “aryl hydrocarbon receptor binding (GO:0017162)” ranked first in serous ovarian carcinoma, FIGO stage IV. Moreover, we found only one common dysfunctional pathway among the five disease groups of serous ovarian tumors: “aryl hydrocarbon receptor binding (GO:0017162),” which is ranked at 42, 2, 2, 29, and 1 in the groups of serous BOTs and serous ovarian carcinomas of stages I–IV, respectively. Meanwhile, we also found ten corresponding genes, AHR (aryl-hydrocarbon receptor), AIP (aryl-hydrocarbon receptor-interacting protein), ARNT (aryl hydrocarbon receptor nuclear translocator), ARNT2 (aryl hydrocarbon receptor nuclear translocator 2), ARNTL (aryl hydrocarbon receptor nuclear translocator-like), NCOA1 (nuclear receptor coactivator 1), NCOA2 (nuclear receptor coactivator 2), TAF4 (TATA-box binding protein associated factor 4), TAF6 (TATA-box binding protein associated factor 6), and TBP (TATA box binding protein), with their representative proteins annotated for these GO term-defined dysfunctional pathways acquired from the GO gene set database (http://geneontology.org/, accessed on 5 June 2021).

Table 4.
The top 50 most dysfunctional GO-defined pathways among serous BOT and all stages of serous ovarian carcinoma ranked by p-values.
3.7. The Influences of Distinct Valuable DEGs with Corresponding Biomarkers Expressed in Serous Ovarian Tumors
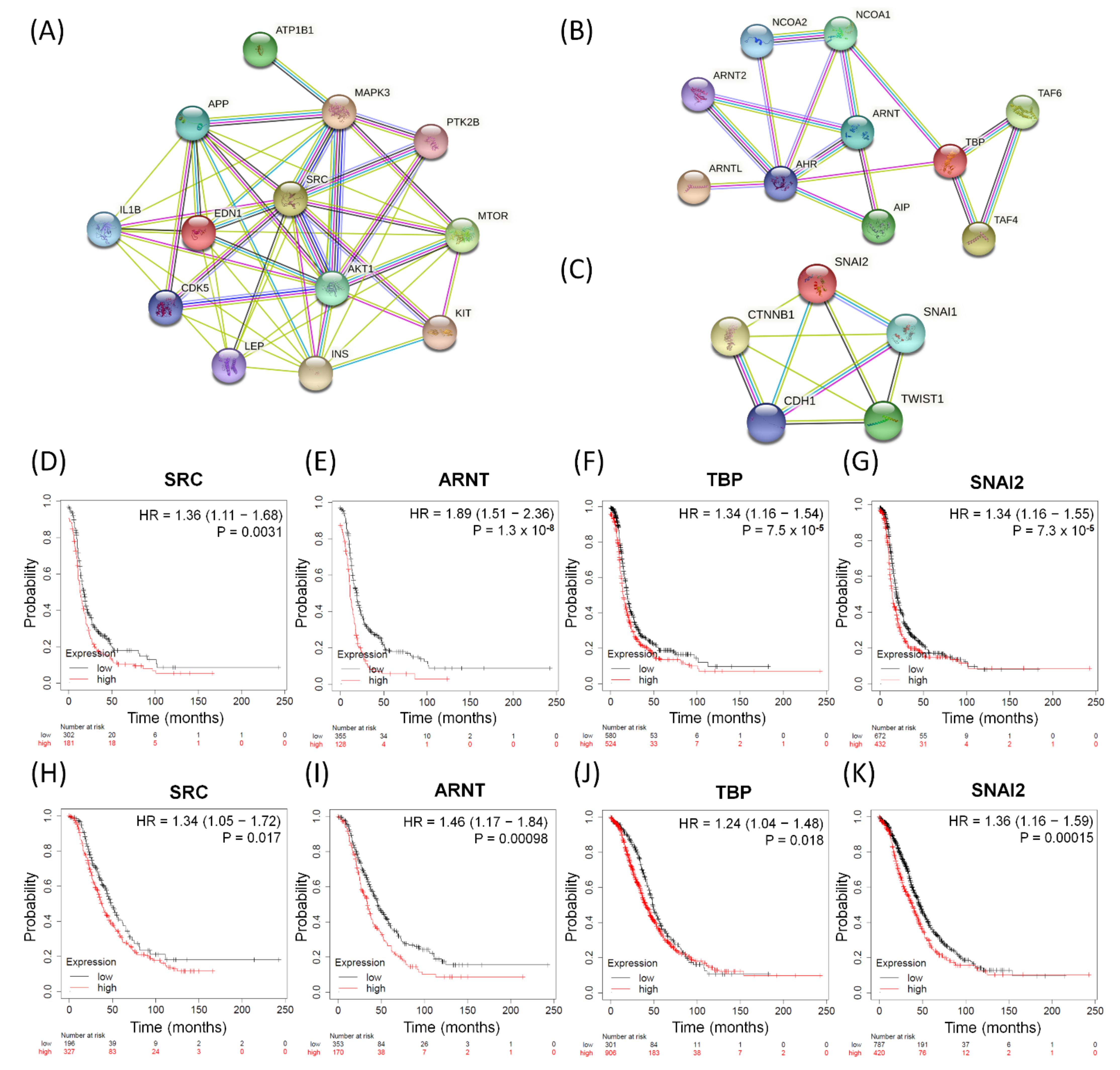
So far, we have performed GO-based integrative methods to analyze, discover, and reclassify the 25 most important common dysregulated functions among the serous ovarian tumor groups into distinct effective categories and obtained 13 corresponding DEGs in total. We also found one common dysfunctional pathway among the five disease groups and the corresponding ten DEGs. Next, we searched several important biomarkers and relevant genes with close relationships with EMT among ovarian cancers from previous research [50,57,72] and compared them with the DEGs of the top 25 meaningful dysregulated functionomes in this experiment by checking for repetitions and cross comparisons. Five featured DEGs were selected: CDH1 (cadherin 1), CTNNB1 (catenin beta 1), SNAI1 (snail family transcriptional repressor 1, SNAIL), SNAI2 (snail family transcriptional repressor 2, SLUG), and TWIST1 (twist-related protein 1). In addition, we have individually established three functional protein–protein interaction networks using the STRING database (https://string-db.org, accessed on 5 June 2021) based on the relevant DEGs and their corresponding proteins as biomarkers. These networks comprised the following: one, all relevant DEGs sorted from the top 25 common dysregulated functionomes (Figure 3A); two, the 10 DEGs involved in the dysfunctional AHR binding pathway (Figure 3B); and three, the featured DEGs among relevant biomarkers associated with EMT (Figure 3C). All these biomarkers revealed intensive interactions with regulatory cross effects in each network. Simultaneously, we searched the GEO (http://www.ncbi.nlm.nih.gov/geo/, accessed on 5 June 2021) and The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov, accessed on 5 June 2021) repositories, including datasets downloaded from three major microarray platforms, GPL96 (Affymetrix HG-U133A), GPL570 (Affymetrix HG-U133 Plus 2.0), and GPL571/GPL3921 (Affymetrix HG-U133A 2.0), which contained extracted and corrected raw data of 1232 patients with serous ovarian carcinoma. We entered these datasets with gene expression into the PostgreSQL relational database and compared 28 meaningful DEGs (EDN1, AKT1, IL1B, INS, CDK5, ATP1B1, PTK2B, MTOR, APP, KIT, LEP, MAPK3, SRC, AHR, AIP, ARNT, ARNT2, ARNTL, NCOA1, NCOA2, TAF4, TAF6, TBP, CDH1, CTNNB1, SNAI1, SNAI2, and TWIST1) selected from the above steps. Then, we calculated and investigated the DEG expression levels, progression-free survival (PFS), and overall survival (OS) among serous ovarian carcinoma patients using the Mann–Whitney test and the receiver operating characteristic test in the R statistical environment (http://www.r-project.org, accessed on 5 June 2021) with Bioconductor libraries (http://www.bioconductor.org, accessed on 5 June 2021) followed by a second normalization to set the average expression of the 22,277 identical probes (http://kmplot.com/analysis/index.php?p=service&cancer=ovar, accessed on 5 June 2021) [73]. Combining all the methods mentioned above, we found that only four DEGs (SRC, ARNT, TBP, and SNAI2) showed stronger and closer relationships than the other biomarkers in each functional protein–protein interaction network (Figure 3A–C) and had consistent synchronous poor effects on PFS and OS among patients with serous ovarian carcinomas with statistical significance (Figure 3D–K). The high expression levels of the four potentially crucial genes (SRC, ARNT, TBP, and SNAI2) were significantly correlated with poor prognosis and survival, and the hazard ratios of PFS and OS are shown in each graph below (Figure 3D–K).
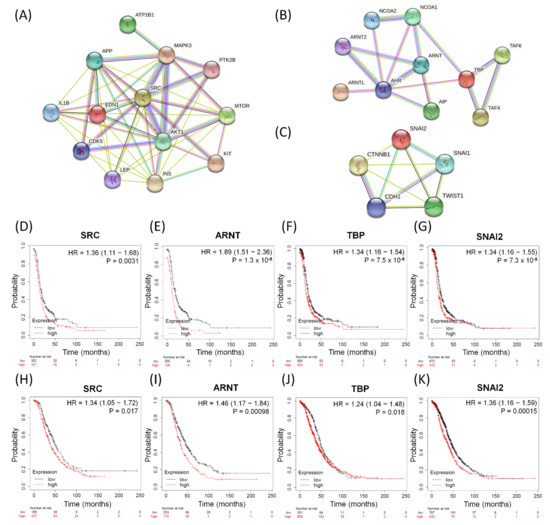
Figure 3.
The significant biomarkers influencing serous ovarian tumors. (A–C) Panels display identified potential involving DEGs subjected to protein–protein interaction (PPI) analysis with interactive network from the STRING database (https://string-db.org (accessed on 21 July 2021)) with intensive interactions. (A) All 13 relevant DEGs sorted from the top 25 common dysregulated GO terms, (B) all ten DEGs in-volved in dysfunctional aryl hydrocarbon receptor (AHR) binding pathway, and (C) five featured DEGs among relevant biomarkers associated with EMT. (D–K) The four meaningful DEGs (SRC, ARNT, TBP and SNAI2) associated with poor survival outcomes. PFS: (D) SRC, (E) ARNT, (F) TBP, (G) SNAI2; OS: (H) SRC, (I) ARNT, (J) TBP, and (K) SNAI2 in serous ovarian carcinomas. The hazard ratios of the PFS of SRC, ARNT, TBP, and SNAI2 were 1.36 (1.11–1.68, p = 0.0031), 1.89 (1.51–2.36, p = 1.3 × 10−8), 1.34 (1.16–1.54, p = 7.5 × 10−5), and 1.34 (1.16–1.55, p = 7.3 × 10−5), respectively. The hazard ratios of the OS of SRC, ARNT, TBP, and SNAI2 were 1.34 (1.05–1.72, p = 0.017), 1.46 (1.17–1.84, p = 0.00098), 1.24 (1.04–1.48, p = 0.018), 1.36 (1.16–1.59, p = 0.00015), respectively.
3.8. Immunohistochemical Validation of Expression Levels for SRC, ARNT, TBP, and SNAI2 among Serous Ovarian Tumors
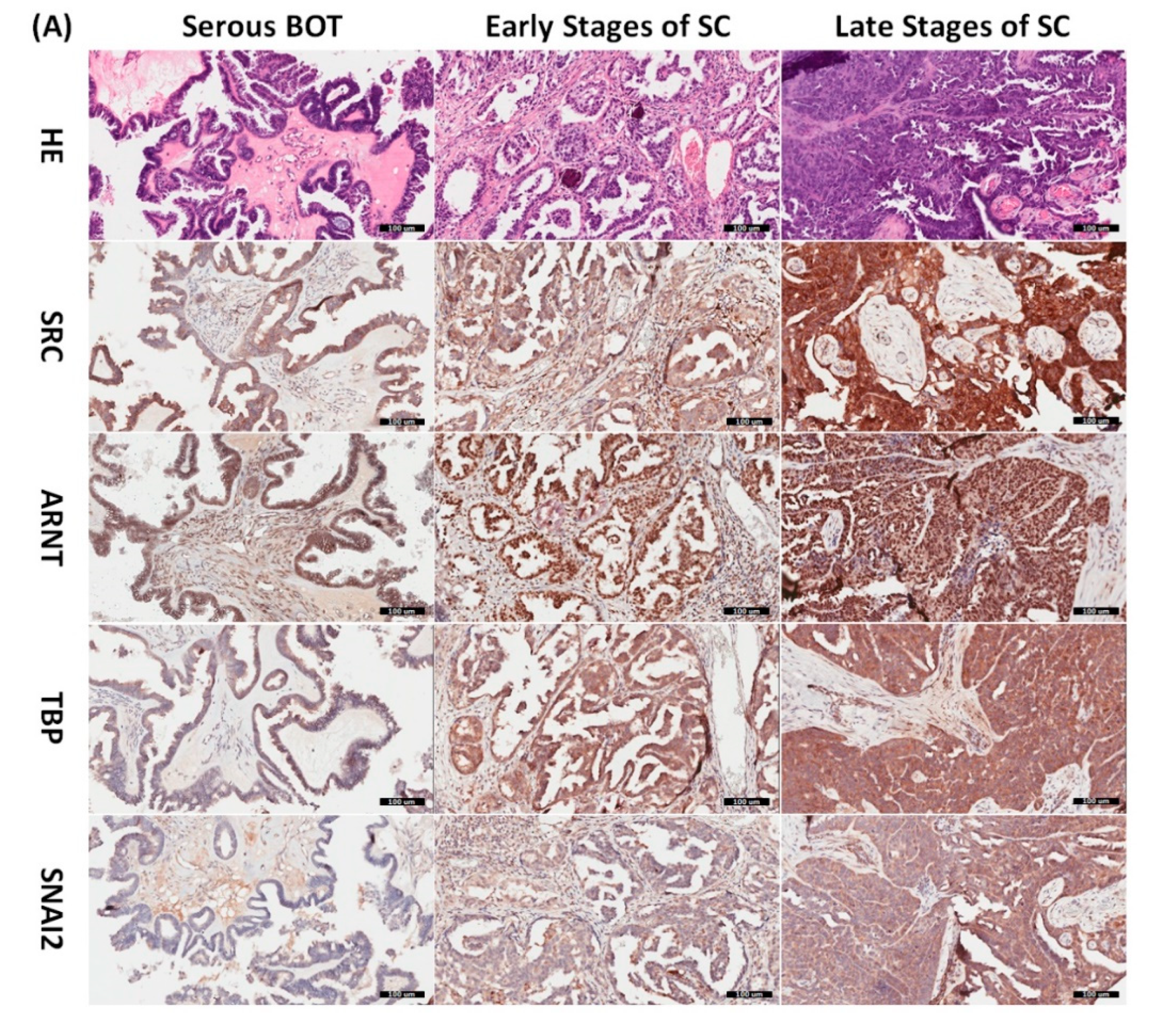
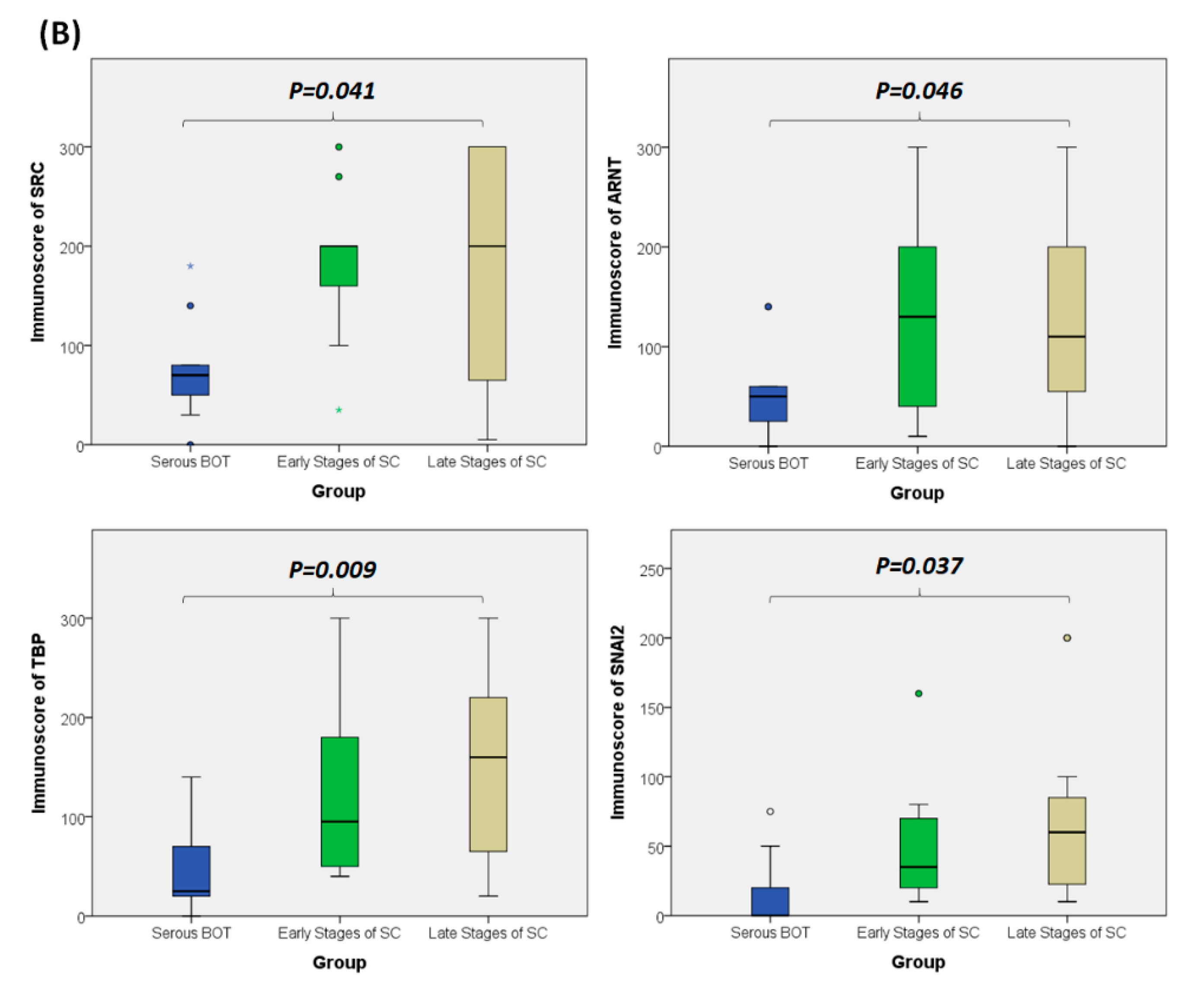
Since the inferred biomarkers including SRC, ARNT, TBP, and SNAI2 from the previous analysis were assumed to be influential in the tumorigenesis of serous ovarian tumors, we gathered relevant clinical samples from a cohort of patients (serous BOT, n = 9; serous ovarian carcinoma, n = 41, including n = 8, 2, 23, and 8 for FIGO stages I–IV, respectively) to explore the clinical characteristics and verify the specific manifestations of the four abovementioned selected DEGs that were determined to participate in the pathogenetic mechanisms of serous ovarian tumors. Because the number of samples in each group was inconsistent, we combined groups as follows to facilitate verification and comparison: serous BOTs, early-stage serous ovarian carcinomas (FIGO stages I and II), and late-stage serous ovarian carcinomas (FIGO stages III and IV). We then performed IHC staining of anti-SRC, anti-ARNT, anti-TBP, and anti-SNAI2 antibodies separately among the three modified disease groups to clinically assess the significant manifestation of SRC, ARNT, TBP, and SNAI2. Professional pathologists verified and interpreted the results evenly and repeatedly throughout the whole diagnostic process using SPSS software (IBM SPSS Statistics version 22.0 for Windows, IBM Corp., Armonk, NY, USA) to quantify the immunoscores of SRC, ARNT, TBP, and SNAI2. The organized results clearly showed that the highest biomarker expression levels tended to occur in the group of late-stage serous ovarian carcinoma, followed by the early-stage group, and lastly the serous BOT group (Figure 4A). We also found that the highest mean values of expression levels for all these biomarkers (SRC, ARNT, TBP, and SNAI2) belonged to the late-stage serous ovarian carcinoma group, with clear increasing trends from the serous BOT group to the late-stage group, and the calculated mean values of the relevant biomarkers were statistically significant (Figure 4B). The detailed results of all scores for relevant featured biomarkers of clinical samples and detailed clinical characteristics of the patients (grade, menopausal status, the presence of BRCA1, BRCA2 mutation, overall survival, and Ca125 level) are listed in Table S6. These results were in accordance with our inferences, implying that many dysregulated functionomes deduced from the integrative GO-based enrichment analysis are dedicated to the pathogenetic mechanisms of serous ovarian tumors. Similarly, these validated results also demonstrated that the dysfunctional AHR binding pathway played a role in the tumorigenesis of serous ovarian tumors. Furthermore, this verification supported the association between EMT and tumor progression. All these significant results confirmed the importance of the previously proposed DEGs and related pathogenic tumorigenesis for serous ovarian tumors.
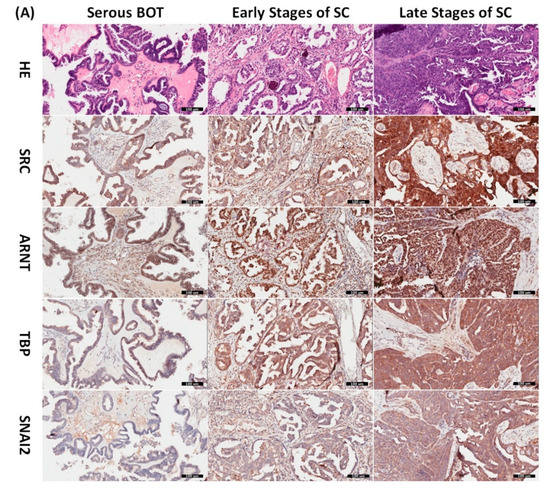
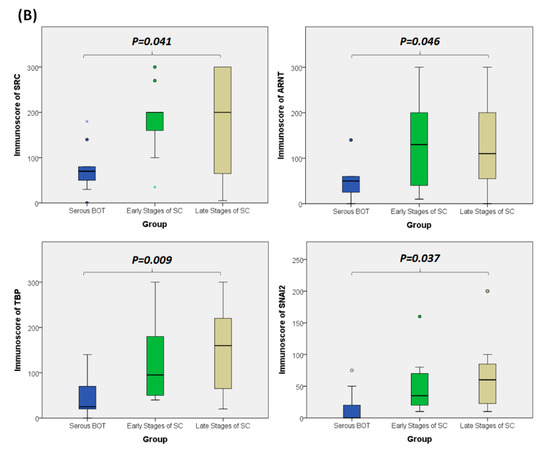
Figure 4.
Verified analysis of biomarkers among serous ovarian tumors by IHC staining. (A) Clinical samples from patients with serous BOTs (n = 9, left column), early stages of serous ovarian carcinomas (n = 10, middle column), and late stages of serous ovarian carcinomas (n = 31, right column) were immunostained with hematoxylin and eosin (first row), anti-SRC antibody (second row), anti-ARNT antibody (third row), anti-TBP antibody (fourth row), and anti-SNAI2 antibody (fifth row). (B) Box plots for expressed biomarkers including SRC, ARNT, TBP, and SNAI2 among groups of serous BOTs (blue), early stages of serous ovarian carcinomas (green), and late stages of serous ovarian carcinomas (light brown). All the expression levels of these meaningful biomarkers were quantified and clearly revealed an increasing trend of mean values from serous BOTs to late stages of serous ovarian carcinomas with statistical significance.
4. Discussion
In this study, we implemented a comprehensive GO-based multi-genome interpretative model using gene set defined functionomes and GSR indices calculated based on gene expression profiles and levels downloaded from public gene set databases to further investigate the complicated and divergent molecular and genetic events of serous ovarian tumors, including serous BOTs and serous ovarian carcinomas at all stages. All results obtained using SVM were statistically significant with high sensitivity, specificity, and accuracy. The GSR indices of all groups of serous ovarian tumors compared to the control groups revealed obvious deviations. The most apparent divergence detected was in the group of serous ovarian carcinoma, FIGO stage IV, and the deviation of serous BOT was just between the early and late stages of serous ovarian carcinomas. Among all groups of serous ovarian tumors, we first identified the top 25 significant common dysregulated functionomes with 13 relevant DEGs, then found one common dysfunctional pathway, AHR binding (GO:0017162), containing 10 corresponding DEGs and excavated five applicable EMT-related DEGs that were related with ovarian neoplasms. Recently, EMT, a reversible process in which epithelial cells acquire mesenchymal cell characteristics due to the loss of cellular polarity and adhesion with increasing cellular migration, has become an important concept in research on tumorigenesis, progression, and chemoresistance of ovarian neoplasms; thus, we included biomarkers of EMT for ovarian tumors in this research. After integrative analysis, including comparison of functional protein–protein interactions and patient survival (PFS and OS) of serous ovarian carcinomas, we obtained four potentially important DEGs: SRC, ARNT, TBP, and SNAI2. Finally, IHC validation of these four biomarkers revealed that they significantly increased in samples incrementally from serous BOT to early stages and then to late stages of serous ovarian carcinomas. Since the results obtained in this study are extraordinarily rich and complex, we mainly explained and discussed the crucial dysfunctional AHR binding pathways accompanied by four consequential DEGs that were statistically verified. However, other related meaningful results deserve further exploration and investigation.
Among the preliminary results of GO-based analysis for each group of serous ovarian tumors, we noticed significant differences between serous BOT and serous ovarian carcinomas, that is similar to the divergences of clinical manifestations and histopathological characteristics between the two groups; furthermore, there were also discrepancies even in the four stages of serous ovarian carcinomas. This experiment thus revealed that serous BOTs and serous ovarian carcinomas are basically inconsistent, although all histopathological classifications are confirmed as “serous”. Even so, we identified the top 25 dysregulated functionomes from the first 50 GO-defined terms among the five groups and reclassified them into three categories according to their representative functions. After statistical comparison, we noticed that the category of metabolic and immunological effects had the greatest influence on serous ovarian tumors, followed by membrane and transport-related effects, and lastly, cellular cycle and signaling-related effects. Therefore, we can reasonably infer the importance of the metabolome and immunome in the tumorigenesis of serous ovarian tumors, which require investigation in the future together with the other two effects. In our experiments, we also identified 13 highly relevant DEGs. Many related studies have examined how these DEGs affected the formation of serous ovarian tumors, such as tyrosine kinase related DEGs (PTK2B, KIT, and SRC) [74,75,76], crucial factors known to be related to tumorigenesis (AKT1, MTOR, MAPK3) [64,77,78,79,80], DEGs related to cellular metabolism and immunity (EDN1, IL1B, INS, APP, LEP) [29,56,60,81,82,83,84,85,86], and agents for signal transmission and channels of cell membranes (CDK5 and ATP1B1) [59,87,88]. Among these DEGs, we found that SRC has consistently poor effects on the survival of serous ovarian carcinoma patients with a poor prognosis of PFS and OS. SRC, a non-receptor protein tyrosine kinase known as a proto-oncogene, participates in the regulation of embryonic development and cell growth [89]. SRC has been found to be activated and overexpressed in association with HER-2/neu overexpression in a high percentage of ovarian cancers, especially in the late stage, and to increase proliferation, angiogenesis, and invasion during tumor development [90]. Silencing of SRC could enhance the cytotoxicity of taxol in ovarian cancer cells to improve the efficacy of chemotherapy [91].
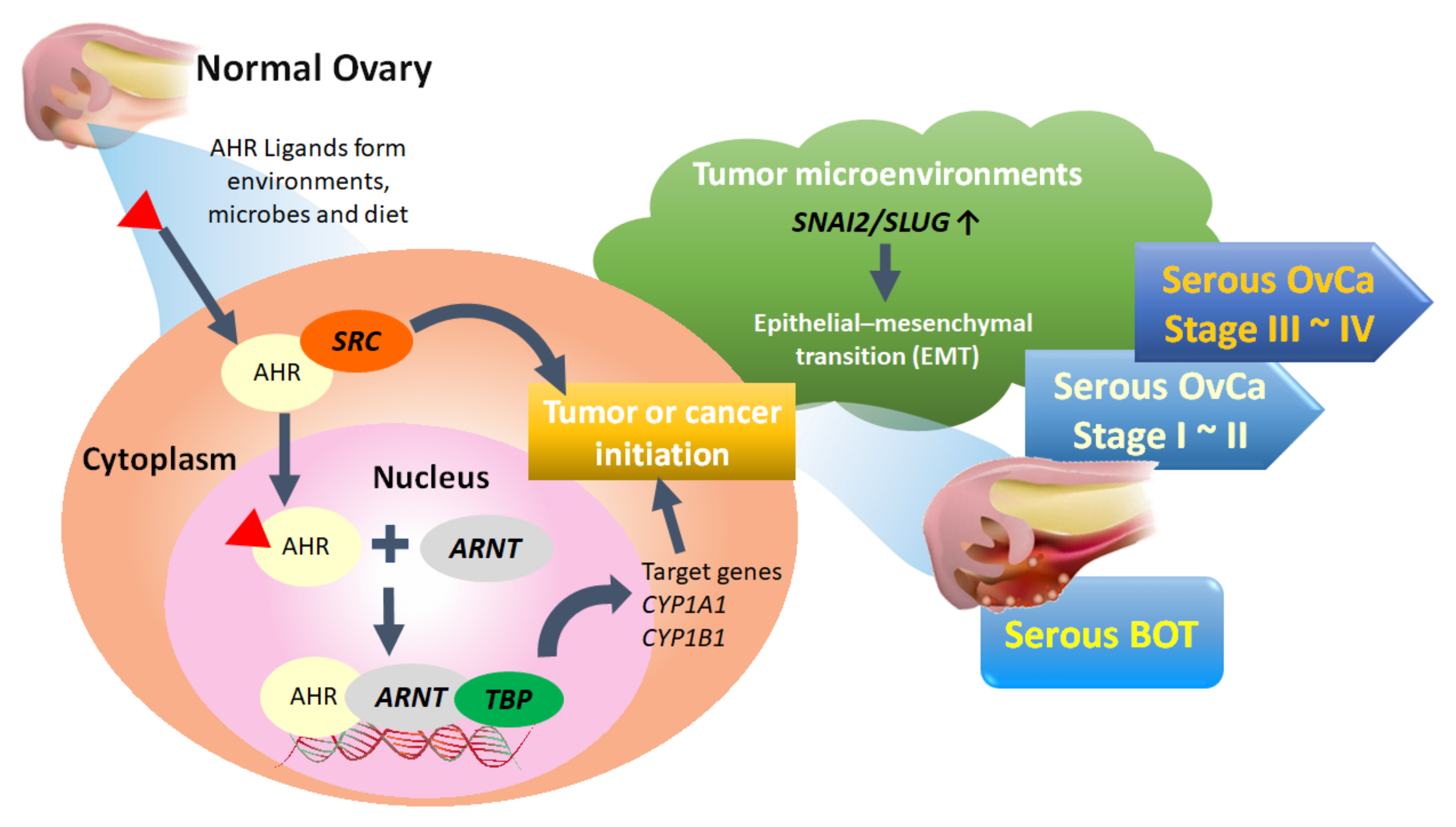
Of the top 50 GO-defined dysfunctional pathways, we found only one meaningful common pathway (AHR binding, GO:0017162) among the five disease groups. We conducted integrated analysis to comprehensively discover the pivotal role of the AHR binding pathway in the tumorigenesis of serous ovarian tumors for the first time. However, in addition to the dysfunctional AHR binding pathway, we also found two common disordered pathways, including positive regulation of keratinocyte differentiation (GO:0045618) and adiponectin secretion (GO:0070162), in all stages of serous ovarian carcinomas. Although not in the top 50 pathways of serous BOTs, these two disordered pathways may be potential problems to be investigated further for the pathogenesis of serous ovarian carcinoma. Through comprehensive analysis, it was revealed that ARNT and TBP have consistently poor effects on PFS and OS. AHR, a ligand-activated transcription factor, is notable for its role in environmental chemical toxicity [92,93,94]; however, in recent studies, AHR was also recognized to play a critical role in tumorigenesis through complex epigenetic and pathogenetic mechanisms encompassing both pro- and anti-tumorigenic activities [95,96]. AHR exists in the cytoplasm and is induced and activated by linking with a group of environmental pollutants as well as other AHR ligands from microbes and diet, and it undergoes certain conformational transformations together with SRC and other cofactors in the cytoplasm to translocate to the nucleus in a dissociated form [95,97,98,99]. AHR can heterodimerize with ARNT, a nuclear translocator, to compose the AhR-ARNT complex, which subsequently binds with specific DNA sequences and xenobiotic response element (XRE) in the enhancer region of certain genes associated with TBP, leading to transcriptional activation of enzymes, such as the cytochrome P450 (CYP) enzymes 1A1 (CYP1A1), CYP1A2, and CYP1B1, for xenobiotic metabolism to induce carcinogenicity of cancer stem cells as tumors or initiate cancer (Figure 5) [100,101,102,103]. Although the current research on the AHR binding pathway and serous ovarian tumors is still limited, it can be roughly understood that the AHR binding pathway influences the formation and occurrence of serous ovarian malignancy through the deep deletion and amplification of AHR transcription factors [95,96,104,105]. Moreover, localization of AHR in the nucleus of tumor cells has been associated with a worse outcome in patients with ovarian cancer, and the role of the AHR/ARNT/CYP-enzyme pathway [106,107] and AHR-driven TBP gene expression in carcinogenesis and cancer initiation, as well as its potential use, have been considered as therapeutic targets for better outcomes [108]. In addition, AHR and NCOA1 discovered in this experiment may also be targets warranting further discussion [98].
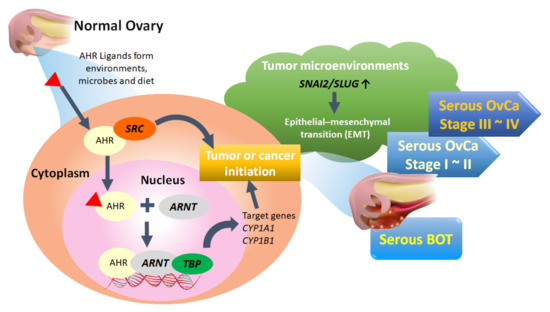
Figure 5.
Proposed pathogenetic mechanism of the AHR binding pathway combined with EMT-related factors for tumorigenesis of serous ovarian tumors. BOT: borderline ovarian tumor; OvCa: ovarian carcinoma.
Approximately 80% of patients with ovarian cancer suffer from recurrence of metastasis within five years after the initial therapy with debulking operation and chemotherapy due to the development of resistance [109,110]. Accumulating findings have recently demonstrated that EMT may induce chemotherapy resistance and cancer cell stemness by regulating EMT transcription factors, such as Zeb1, Zeb2, Snail, Slug, and Twist1, in a complicated network, and all functional EMT in the tumor microenvironment could exchange tumor cell morphology to upgrade metastatic abilities via migration and invasion [72,111,112,113]. Because avoidance of EMT may be crucial for evaluating and managing tumor metastasis and recurrence [114], we selected five featured DEGs by proofreading and collation with all meaningful DEGs from the top 25 common functionomes of all groups of serous ovarian tumors, and we found that SNAI2 was the most influential DEG due to the concordant results of patient survival. IHC analysis showed an increasing trend from borderline tumors to the late stage of ovarian malignancy. The transcriptional factor SNAI2, also known as SLUG, is considered important for cell migration, differentiation, and metastasis [115,116]. Our study identified the expression and role of SNAI2 in serous ovarian tumors, indicating the progression of serous ovarian tumors possibly through EMT. So far, the association between the AHR binding pathway and EMT among serous ovarian tumors remains unexplored thoroughly, and this experiment provides the opportunity to solve this problem. Aromatic hydrocarbon substances, such as phthalates, di(2-ethylhexyl)phthalate (DEHP), or bisphenol A, are recognized as aggravators, as they upregulate and promote cell proliferation and tumor progression [117,118,119]. In contrast, dietary phytoestrogen and kaempferol could exert anti-carcinogenic and anti-proliferative effects through AHR-related pathways to inhibit the EMT process [120]. Our results showed that there is indeed a tight correlation between AHR and EMT as the degree of malignancy develops in serous ovarian tumors, just like other malignancy [121]. However, how the AHR binding pathway and EMT interact and influence each other in tumor progression and resistance to chemotherapy warrants further research.
This study had several limitations. First, we noticed some limitations in the integrative analytic methods utilized in this study, because the gene set databases of GO terms and related biomolecular pathways did not completely contain or fully define all functionomes of humans. False positivity was attributed to the heterogenicity of disparate cellular histopathological compositions and the indistinguishable elements of different gene sets among the chosen tumor and control samples, and detection by the GSR model was uncertain due to missed errors and untransformed GSR indices if the expression levels were undetectable when converting levels for ordering gene expression. However, these disadvantages may not be obvious in the overall results coupled with the statistically significant high sensitivity, specificity, and accuracy of this experiment. To eliminate these problems in the future, a more precise programming syntax design and more specified sample screening are required. The second limitation is the uneven distribution of case groups. The numbers of serous BOTs, serous ovarian carcinomas, and normal control samples are quite different, and even in the largest population of serous ovarian carcinomas, the numbers of tumors in each stage are quite different. According to the known proportions of serous ovarian tumors, serous ovarian carcinomas account for more EOCs than BOTs, and early stages of serous ovarian carcinomas are usually difficult to diagnose, resulting in fewer diagnoses than at advanced stages. The number of specimens collected for subsequent clinical verification also fits this situation. Although the number of clinical samples is small, with the support of the support vector machine (SVM) used in this study, the preliminary results of this multidisciplinary comprehensive analysis are reliable with high sensitivity, specificity, and accuracy. Even a relatively small number could obtain statistically significant results through IHC verification. Perhaps a more intact gene expression profile database could be constructed to decrease individual discrepancies among ethnic groups in retrospective or prospective cohort studies conducted on a larger scale globally. Third, this study only investigated the common pathogenetic mechanisms of serous ovarian tumors. However, due to the current lack of research, the small number of clinical specimens, and limited funds, data gathered from the GO term database are somewhat obstructed, especially data of serous BOT. Nevertheless, the results are clear and statistically significant, as determined by clinical verification with the immunostaining method. In the future, it may be necessary to gather more specimens, examine more global academic research, and utilize databases of various subtypes to compare and investigate more profoundly and comprehensively the pathogenetic mechanisms with the aid of large-scale experimental tests and funding.
In summary, to investigate potential crucial pathogenetic mechanisms, we performed an integrated GO-based analysis to obtain global genome-wide expression profiles individually and explore meaningful dysregulated functionomes, dysfunctional pathways, and relevant biomarkers of EMT among assorted groups of serous ovarian tumors with the support of elementary machine learning. Based on the above conclusions, we proposed the inferred hypothesis for the formative process of serous ovarian tumor that activated AHR could cooperate with SRC in the cytoplasm to enter cell nuclei and then bind to ARNT together with TBP to act on DNA for initiating targeted AHR-responsive genes to cause tumor or cancer initiation. Besides, biomarker of EMT such as SNAI2 in the tumor microenvironment could also facilitate EMT process accompanied with tumorigenesis (Figure 5). These results provided new directions for understanding the tumorigenesis of serous ovarian tumors and more potential crucial targets for the identification, treatment, monitoring, and even prevention of recurrence combined with targeted therapies as precision medicine in the future.
5. Conclusions
Serous ovarian tumors, consisting mainly of serous ovarian carcinoma and serous BOT, are epithelial tumors of the ovary with distinctive characteristics for each subtype. In this study, we made use of integrative analytic methods to select the top 25 significant common GO terms as dysregulated functionomes reclassified into three crucial categories (metabolic, immunological, and other effects; membrane and transport-related effects; and cellular cycle and signaling-related effects) and acquired 13 corresponding DEGs with high probability through cross comparison. For the first time, the dysfunctional AHR binding pathway accompanied with 10 corresponding DEGs was found significantly to be participated in tumorigenesis of both serous BOT and serous ovarian carcinoma and five vital biomarkers related to EMT were searched and gathered for this analytic study. Finally, four important DEGs (SRC, ARNT, TBP, and SNAI2) were compiled to have distinct effects on the survivals of serous ovarian tumor patients with the help of IHC staining for verification showing elevated expression among all clinical samples with increasing malignancy from serous BOT to early stages and to late stages of serous ovarian carcinomas. All acquired results initially supported the inference that dysregulated functionomes with active DEGs and relevant biomarkers could cooperate with the dysfunctional AHR binding pathway together with increased EMT effects in the tumor microenvironment to synergistically influence tumor initiation. These findings considerably contributed to elucidating the pathogenesis of serous ovarian tumors.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biomedicines9080866/s1, Table S1, Detailed informative data of all collected samples. Table S2, Sensitivity, specificity, and accuracy of binary classification and prediction by supervised machine learning. Table S3, The whole deregulated GO terms of serous ovarian tumors. Table S4. The top 25 common dysregulated GO terms among the five case groups (serous BOT and all stages of serous ovarian carcinoma). Table S5, The whole dysfunctional GO pathways for serous ovarian tumors. Table S6, All IHC scores for relevant biomarkers of clinical samples and detailed clinical characteristics of the patients.
Author Contributions
Conceptualization, K.-M.S. and C.-M.C.; methodology, K.-M.S. and Y.-F.L.; software, C.-M.C. and Y.-F.L.; validation, H.-W.G., Y.-F.L., and K.-M.S.; formal analysis, L.-C.L. and C.-C.C. (Chia-Ching Chang); investigation, K.-H.L., Y.-H.L., and L.-C.L.; resources, M.-H.Y., Y.-H.L., and C.-C.C. (Chia-Ching Chang); data curation, H.-W.G., C.-M.C., and K.-H.L.; writing—original draft preparation, K.-M.S. and Y.-F.L.; writing—review and editing, K.-M.S. and C.-C.C. (Cheng-Chang Chang); visualization, K.-M.S. and H.-W.G.; supervision, M.-H.Y. and C.-C.C. (Cheng-Chang Chang); funding acquisition, K.-M.S., Y.-F.L., and C.-C.C. (Cheng-Chang Chang). All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part and funded individually by the following grants from the Ministry of Science and Technology, R.O.C. (MOST 110-2321-B-016-002); the Tri-Service General Hospital (TSGH-D-109189, TSGH-D-109203, TSGH-D-110136, TSGH-D-110172, and TSGH-E-110230), the Teh-Tzer Study Group for Human Medical Research Foundation, the Veterans General Hospital, Tri-Service General Hospital and Academia Sinica Joint Research Programs (VTA110-T-4-1).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Tri-Service General Hospital, National Defense Medical Center for agreement of the study (2-107-05-043, approved on 26 October 2018; 2-108-05-091, approved on 20 May 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The gene set databases of microarrays expression profiles are publicly available and downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/, accessed on 5 June 2021).
Acknowledgments
We would like to thank Kuo-Chih Su and Hui-Yin Su for figure editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Russell, P. The pathological assessment of ovarian neoplasms. I: Introduction to the common ‘epithelial’tumours and analysis of benign ‘epithelial’tumours. Pathology 1979, 11, 5–26. [Google Scholar] [CrossRef] [PubMed]
- Iwabuchi, H.; Sakamoto, M.; Sakunaga, H.; Ma, Y.-Y.; Carcangiu, M.L.; Pinkel, D.; Yang-Feng, T.L.; Gray, J.W. Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res. 1995, 55, 6172–6180. [Google Scholar]
- Allemani, C.; Weir, H.K.; Carreira, H.; Harewood, R.; Spika, D.; Wang, X.-S.; Bannon, F.; Ahn, J.V.; Johnson, C.J. Bonaventure, A. Global surveillance of cancer survival 1995–2009: Analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015, 385, 977–1010. [Google Scholar] [CrossRef]
- Runnebaum, I.B.; Stickeler, E. Epidemiological and molecular aspects of ovarian cancer risk. J. Cancer Res. Clin. Oncol. 2001, 127, 73–79. [Google Scholar] [CrossRef]
- Karnezis, A.N.; Cho, K.R.; Gilks, C.B.; Pearce, C.L.; Huntsman, D.G. The disparate origins of ovarian cancers: Pathogenesis and prevention strategies. Nat. Rev. Cancer 2017, 17, 65. [Google Scholar] [CrossRef] [PubMed]
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Vargas, A.N. Natural history of ovarian cancer. Ecancermedicalscience 2014, 8, 1–10. [Google Scholar]
- Chien, J.; Poole, E.M. Ovarian cancer prevention, screening, and early detection: Report from the 11th biennial ovarian cancer research symposium. Int. J. Gynecol. Cancer 2017, 27. [Google Scholar] [CrossRef]
- Santillan, A.; Kim, Y.; Zahurak, M.; Gardner, G.; Giuntoli, R.; Shih, I.; Bristow, R. Differences of chemoresistance assay between invasive micropapillary/low-grade serous ovarian carcinoma and high-grade serous ovarian carcinoma. Int. J. Gynecol. Cancer 2007, 17, 601–606. [Google Scholar] [CrossRef]
- Song, T.; Lee, Y.-Y.; Choi, C.H.; Kim, T.-J.; Lee, J.-W.; Bae, D.-S.; Kim, B.-G. Histologic distribution of borderline ovarian tumors worldwide: A systematic review. J. Gynecol. Oncol. 2013, 24, 44–51. [Google Scholar] [CrossRef]
- Yasmeen, S.; Hannan, A.; Sheikh, F.; Syed, A.A.; Siddiqui, N. Borderline tumors of the ovary: A clinicopathological study. Pak. J. Med Sci. 2017, 33, 369. [Google Scholar] [CrossRef]
- Silverberg, S.G.; Bell, D.A.; Kurman, R.J.; Seidman, J.D.; Prat, J.; Ronnett, B.M.; Copeland, L.; Silva, E.; Gorstein, F.; Young, R.H. Borderline ovarian tumors: Key points and workshop summary. Hum. Pathol. 2004, 35, 910–917. [Google Scholar] [CrossRef]
- Hauptmann, S.; Friedrich, K.; Redline, R.; Avril, S. Ovarian borderline tumors in the 2014 WHO classification: Evolving concepts and diagnostic criteria. Virchows Arch. 2017, 470, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xu, J.; Jia, X. The Diagnosis, Treatment, Prognosis and Molecular Pathology of Borderline Ovarian Tumors: Current Status and Perspectives. Cancer Manag. Res. 2020, 12, 3651–3659. [Google Scholar] [CrossRef] [PubMed]
- Lalwani, N.; Shanbhogue, A.K.; Vikram, R.; Nagar, A.; Jagirdar, J.; Prasad, S.R. Current update on borderline ovarian neoplasms. Am. J. Roentgenol. 2010, 194, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Trillsch, F.; Mahner, S.; Ruetzel, J.; Harter, P.; Ewald-Riegler, N.; Jaenicke, F.; Du Bois, A. Clinical management of borderline ovarian tumors. Expert Rev. Anticancer Ther. 2010, 10, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Fathalla, M. Incessant ovulation—A factor in ovarian neoplasia. Lancet 1971, 2, 163. [Google Scholar] [CrossRef]
- Van Leeuwen, F.; Klip, H.; Mooij, T.M.; Van De Swaluw, A.; Lambalk, C.B.; Kortman, M.; Laven, J.; Jansen, C.; Helmerhorst, F.; Cohlen, B. Risk of borderline and invasive ovarian tumours after ovarian stimulation for in vitro fertilization in a large Dutch cohort. Hum. Reprod. 2011, 26, 3456–3465. [Google Scholar] [CrossRef]
- Riman, T.; Dickman, P.W.; Nilsson, S.; Correia, N.; Nordlinder, H.; Magnusson, C.M.; Persson, I.R. Risk factors for epithelial borderline ovarian tumors: Results of a Swedish case–control study. Gynecol. Oncol. 2001, 83, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Mayr, D.; Hirschmann, A.; Löhrs, U.; Diebold, J. KRAS and BRAF mutations in ovarian tumors: A comprehensive study of invasive carcinomas, borderline tumors and extraovarian implants. Gynecol. Oncol. 2006, 103, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-L.; Kurman, R.J.; Dehari, R.; Wang, T.-L.; Shih, I.-M. Mutations of BRAF and KRAS precede the development of ovarian serous borderline tumors. Cancer Res. 2004, 64, 6915–6918. [Google Scholar] [CrossRef]
- Anglesio, M.S.; Arnold, J.M.; George, J.; Tinker, A.V.; Tothill, R.; Waddell, N.; Simms, L.; Locandro, B.; Fereday, S.; Traficante, N. Mutation of ERBB2 provides a novel alternative mechanism for the ubiquitous activation of RAS-MAPK in ovarian serous low malignant potential tumors. Mol. Cancer Res. 2008, 6, 1678–1690. [Google Scholar] [CrossRef] [PubMed]
- El-Balat, A.; Schmeil, I.; Gasimli, K.; Sänger, N.; Karn, T.; Ahr, A.; Becker, S.; Arsenic, R.; Holtrich, U.; Engels, K. Claudin-1 is linked to presence of implants and micropapillary pattern in serous borderline epithelial tumours of the ovary. J. Clin. Pathol. 2018, 71, 1060–1064. [Google Scholar] [CrossRef]
- Malpica, A.; Wong, K.-K. The molecular pathology of ovarian serous borderline tumors. Ann. Oncol. 2016, 27 (Suppl. S1), i16–i19. [Google Scholar] [CrossRef] [PubMed]
- Ozretić, P.; Trnski, D.; Musani, V.; Maurac, I.; Kalafatić, D.; Orešković, S.; Levanat, S.; Sabol, M. Non-canonical Hedgehog signaling activation in ovarian borderline tumors and ovarian carcinomas. Int. J. Oncol. 2017, 51, 1869–1877. [Google Scholar] [CrossRef]
- Hirst, J.; Crow, J.; Godwin, A. Ovarian cancer genetics: Subtypes and risk factors. In Ovarian Cancer—From Pathogenesis to Treatment; Devaja, O., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Verhaak, R.G.; Tamayo, P.; Yang, J.-Y.; Hubbard, D.; Zhang, H.; Creighton, C.J.; Fereday, S.; Lawrence, M.; Carter, S.L.; Mermel, C.H. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J. Clin. Investig. 2012, 123, 517–525. [Google Scholar] [CrossRef]
- Ducie, J.; Dao, F.; Considine, M.; Olvera, N.; Shaw, P.A.; Kurman, R.J.; Shih, I.-M.; Soslow, R.A.; Cope, L.; Levine, D.A. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Network, C.G.A.R. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609. [Google Scholar] [CrossRef]
- Cooke, S.L.; Ng, C.K.; Melnyk, N.; Garcia, M.J.; Hardcastle, T.; Temple, J.; Langdon, S.; Huntsman, D.; Brenton, J.D. Genomic analysis of genetic heterogeneity and evolution in high-grade serous ovarian carcinoma. Oncogene 2010, 29, 4905–4913. [Google Scholar] [CrossRef]
- Javadi, S.; Ganeshan, D.M.; Qayyum, A.; Iyer, R.B.; Bhosale, P. Ovarian cancer, the revised FIGO staging system, and the role of imaging. Am. J. Roentgenol. 2016, 206, 1351–1360. [Google Scholar] [CrossRef]
- Zeppernick, F.; Meinhold-Heerlein, I. The new FIGO staging system for ovarian, fallopian tube, and primary peritoneal cancer. Arch. Gynecol. Obstet. 2014, 290, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Yemelyanova, A.; Vang, R.; Kshirsagar, M.; Lu, D.; Marks, M.A.; Shih, I.M.; Kurman, R.J. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: An immunohistochemical and nucleotide sequencing analysis. Mod. Pathol. 2011, 24, 1248–1253. [Google Scholar] [CrossRef]
- Mota, A.; Triviño, J.C.; Rojo-Sebastian, A.; Martínez-Ramírez, Á.; Chiva, L.; González-Martín, A.; Garcia, J.F.; Garcia-Sanz, P.; Moreno-Bueno, G. Intra-tumor heterogeneity in TP53 null high grade serous ovarian carcinoma progression. BMC Cancer 2015, 15, 1–11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McAlpine, J.N.; Porter, H.; Köbel, M.; Nelson, B.H.; Prentice, L.M.; Kalloger, S.E.; Senz, J.; Milne, K.; Ding, J.; Shah, S.P. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod. Pathol. 2012, 25, 740–750. [Google Scholar] [CrossRef]
- Brachova, P.; Thiel, K.W.; Leslie, K.K. The consequence of oncomorphic TP53 mutations in ovarian cancer. Int. J. Mol. Sci. 2013, 14, 19257–19275. [Google Scholar] [CrossRef]
- Moschetta, M.; George, A.; Kaye, S.; Banerjee, S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann. Oncol. 2016, 27, 1449–1455. [Google Scholar] [CrossRef]
- Pal, T.; Permuth-Wey, J.; Betts, J.A.; Krischer, J.P.; Fiorica, J.; Arango, H.; LaPolla, J.; Hoffman, M.; Martino, M.A.; Wakeley, K. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2005, 104, 2807–2816. [Google Scholar]
- Bai, H.; Cao, D.; Yang, J.; Li, M.; Zhang, Z.; Shen, K. Genetic and epigenetic heterogeneity of epithelial ovarian cancer and the clinical implications for molecular targeted therapy. J. Cell. Mol. Med. 2016, 20, 581–593. [Google Scholar] [CrossRef]
- Tew, W.P.; Lacchetti, C.; Ellis, A.; Maxian, K.; Banerjee, S.; Bookman, M.; Jones, M.B.; Lee, J.-M.; Lheureux, S.; Liu, J.F. PARP inhibitors in the management of ovarian cancer: ASCO guideline. Obstet. Gynecol. Surv. 2020, 75, 739–741. [Google Scholar] [CrossRef]
- Franzese, E.; Centonze, S.; Diana, A.; Carlino, F.; Guerrera, L.P.; Di Napoli, M.; De Vita, F.; Pignata, S.; Ciardiello, F.; Orditura, M. PARP inhibitors in ovarian cancer. Cancer Treat. Rev. 2019, 73, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gadducci, A.; Guerrieri, M.E. PARP inhibitors in epithelial ovarian cancer: State of art and perspectives of clinical research. Anticancer. Res. 2016, 36, 2055–2064. [Google Scholar]
- Gomez, M.K.; Illuzzi, G.; Colomer, C.; Churchman, M.; Hollis, R.L.; O’Connor, M.J.; Gourley, C.; Leo, E.; Melton, D.W. Identifying and Overcoming Mechanisms of PARP Inhibitor Resistance in Homologous Recombination Repair-Deficient and Repair-Proficient High Grade Serous Ovarian Cancer Cells. Cancers 2020, 12, 1503. [Google Scholar] [CrossRef]
- Xie, H.; Wang, W.; Xia, B.; Jin, W.; Lou, G. Therapeutic applications of PARP inhibitors in ovarian cancer. Biomed. Pharmacother. 2020, 127, 110204. [Google Scholar] [CrossRef] [PubMed]
- Takaya, H.; Nakai, H.; Takamatsu, S.; Mandai, M.; Matsumura, N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Kadife, E.; Raza, A.; Short, M.; Jubinsky, P.T.; Kannourakis, G. Ovarian cancer, cancer stem cells and current treatment strategies: A potential role of magmas in the current treatment methods. Cells 2020, 9, 719. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP inhibitors: Clinical relevance, mechanisms of action and tumor resistance. Front. Cell Dev. Biol. 2020, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, C.; Rouzier, R.; Geyl, C.; Cortez, A.; Castela, M.; Lis, R.; Daraï, E.; Touboul, C. Predictive markers of chemoresistance in advanced stages epithelial ovarian carcinoma. Gynecol. Oncol. 2015, 136, 112–120. [Google Scholar] [CrossRef]
- Roy, L.; Cowden Dahl, K.D. Can stemness and chemoresistance be therapeutically targeted via signaling pathways in ovarian cancer? Cancers 2018, 10, 241. [Google Scholar] [CrossRef]
- Davidson, B.; Trope, C.G.; Reich, R. Epithelial–mesenchymal transition in ovarian carcinoma. Front. Oncol. 2012, 2, 33. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Zarrabi, A.; Hushmandi, K.; Kalantari, M.; Mohammadinejad, R.; Javaheri, T.; Sethi, G. Association of the epithelial–mesenchymal transition (EMT) with cisplatin resistance. Int. J. Mol. Sci. 2020, 21, 4002. [Google Scholar] [CrossRef]
- Iwatsuki, M.; Mimori, K.; Yokobori, T.; Ishi, H.; Beppu, T.; Nakamori, S.; Baba, H.; Mori, M. Epithelial–mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010, 101, 293–299. [Google Scholar] [CrossRef]
- Haslehurst, A.M.; Koti, M.; Dharsee, M.; Nuin, P.; Evans, K.; Geraci, J.; Childs, T.; Chen, J.; Li, J.; Weberpals, J. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 2012, 12, 1–10. [Google Scholar] [CrossRef]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-mesenchymal transition in cancer: A historical overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Voulgari, A.; Pintzas, A. Epithelial–mesenchymal transition in cancer metastasis: Mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim. Biophys. Acta (BBA) Rev. Cancer 2009, 1796, 75–90. [Google Scholar] [CrossRef]
- Rohnalter, V.; Roth, K.; Finkernagel, F.; Adhikary, T.; Obert, J.; Dorzweiler, K.; Bensberg, M.; Müller-Brüsselbach, S.; Müller, R. A multi-stage process including transient polyploidization and EMT precedes the emergence of chemoresistent ovarian carcinoma cells with a dedifferentiated and pro-inflammatory secretory phenotype. Oncotarget 2015, 6, 40005. [Google Scholar] [CrossRef]
- Loret, N.; Denys, H.; Tummers, P.; Berx, G. The role of epithelial-to-mesenchymal plasticity in ovarian cancer progression and therapy resistance. Cancers 2019, 11, 838. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-M.; Chuang, C.-M.; Wang, M.-L.; Yang, Y.-P.; Chuang, J.-H.; Yang, M.-J.; Yen, M.-S.; Chiou, S.-H.; Chang, C.-C. Gene set− based integrative analysis revealing two distinct functional regulation patterns in four common subtypes of epithelial ovarian cancer. Int. J. Mol. Sci. 2016, 17, 1272. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-M.; Chuang, C.-M.; Wang, M.-L.; Yang, M.-J.; Chang, C.-C.; Yen, M.-S.; Chiou, S.-H. Gene set-based functionome analysis of pathogenesis in epithelial ovarian serous carcinoma and the molecular features in different FIGO stages. Int. J. Mol. Sci. 2016, 17, 886. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-M.; Wang, M.-L.; Lu, K.-H.; Yang, Y.-P.; Juang, C.-M.; Wang, P.-H.; Hsu, R.-J.; Yu, M.-H.; Chang, C.-C. Integrating the dysregulated inflammasome-based molecular functionome in the malignant transformation of endometriosis-associated ovarian carcinoma. Oncotarget 2018, 9, 3704. [Google Scholar] [CrossRef]
- Chang, C.-M.; Yang, Y.-P.; Chuang, J.-H.; Chuang, C.-M.; Lin, T.-W.; Wang, P.-H.; Yu, M.-H.; Chang, C.-C. Discovering the deregulated molecular functions involved in malignant transformation of endometriosis to endometriosis-associated ovarian carcinoma using a data-driven, function-based analysis. Int. J. Mol. Sci. 2017, 18, 2345. [Google Scholar] [CrossRef]
- Chang, C.-C.; Su, K.-M.; Lu, K.-H.; Lin, C.-K.; Wang, P.-H.; Li, H.-Y.; Wang, M.-L.; Lin, C.-K.; Yu, M.-H.; Chang, C.-M. Key immunological functions involved in the progression of epithelial ovarian serous carcinoma discovered by the gene ontology-based immunofunctionome analysis. Int. J. Mol. Sci. 2018, 19, 3311. [Google Scholar] [CrossRef] [PubMed]
- Su, K.-M.; Lin, T.-W.; Liu, L.-C.; Yang, Y.-P.; Wang, M.-L.; Tsai, P.-H.; Wang, P.-H.; Yu, M.-H.; Chang, C.-M.; Chang, C.-C. The Potential Role of Complement System in the Progression of Ovarian Clear Cell Carcinoma Inferred from the Gene Ontology-Based Immunofunctionome Analysis. Int. J. Mol. Sci. 2020, 21, 2824. [Google Scholar] [CrossRef]
- Chang, C.-M.; Li, Y.-F.; Lin, H.-C.; Lu, K.-H.; Lin, T.-W.; Liu, L.-C.; Su, K.-M.; Chang, C.-C. Dysregulated Immunological Functionome and Dysfunctional Metabolic Pathway Recognized for the Pathogenesis of Borderline Ovarian Tumors by Integrative Polygenic Analytics. Int. J. Mol. Sci. 2021, 22, 4105. [Google Scholar] [CrossRef]
- Eddy, J.A.; Hood, L.; Price, N.D.; Geman, D. Identifying tightly regulated and variably expressed networks by Differential Rank Conservation (DIRAC). PLoS Comput. Biol. 2010, 6, e1000792. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Van Diest, P.J.; van Dam, P.; Henzen-Logmans, S.C.; Berns, E.; Van der Burg, M.; Green, J.; Vergote, I. A scoring system for immunohistochemical staining: Consensus report of the task force for basic research of the EORTC-GCCG. European Organization for Research and Treatment of Cancer-Gynaecological Cancer Cooperative Group. J. Clin. Pathol. 1997, 50, 801. [Google Scholar] [CrossRef]
- Charafe-Jauffret, E.; Tarpin, C.; Bardou, V.J.; Bertucci, F.; Ginestier, C.; Braud, A.C.; Puig, B.; Geneix, J.; Hassoun, J.; Birnbaum, D. Immunophenotypic analysis of inflammatory breast cancers: Identification of an ‘inflammatory signature’. J. Pathol. 2004, 202, 265–273. [Google Scholar] [CrossRef]
- Dessimoz, C.; Škunca, N. The Gene Ontology Handbook; Springer: Basingstoke, UK, 2017. [Google Scholar]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Investig. 2009, 119, 1429–1437. [Google Scholar] [CrossRef]
- Győrffy, B.; Lánczky, A.; Szállási, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef]
- Song, G.; Chen, L.; Zhang, B.; Song, Q.; Yu, Y.; Moore, C.; Wang, T.-L.; Shih, I.-M.; Zhang, H.; Chan, D.W. Proteome-wide tyrosine phosphorylation analysis reveals dysregulated signaling pathways in ovarian tumors*[S]. Mol. Cell. Proteom. 2019, 18, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zheng, X.; Ling, R.; Gao, J.; Leung, K.-S.; Wong, M.-H.; Yang, S.; Liu, Y.; Dong, M.; Bai, H. Whole Transcriptome Analyses Identify Pairwise Gene Circuit Motif in Serous Ovarian Cancer. 2021. PREPRINT (Version 1). Available online: https://doi.org/10.21203/rs.3 (accessed on 20 July 2021).
- Lassus, H.; Sihto, H.; Leminen, A.; Nordling, S.; Joensuu, H.; Nupponen, N.; Butzow, R. Genetic alterations and protein expression of KIT and PDGFRA in serous ovarian carcinoma. Br. J. Cancer 2004, 91, 2048–2055. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, D.; Shim, J.E.; Bae, S.J.; Jung, Y.J.; Kim, S.; Lee, H.; Kim, S.H.; Jo, S.B.; Lee, J.Y. Genomic profiling of the residual disease of advanced high-grade serous ovarian cancer after neoadjuvant chemotherapy. Int. J. Cancer 2020, 146, 1851–1861. [Google Scholar] [CrossRef] [PubMed]
- Hanrahan, A.J.; Schultz, N.; Westfal, M.L.; Sakr, R.A.; Giri, D.D.; Scarperi, S.; Janikariman, M.; Olvera, N.; Stevens, E.V.; She, Q.-B. Genomic complexity and AKT dependence in serous ovarian cancer. Cancer Discov. 2012, 2, 56–67. [Google Scholar] [CrossRef]
- Mabuchi, S.; Kuroda, H.; Takahashi, R.; Sasano, T. The PI3K/AKT/mTOR pathway as a therapeutic target in ovarian cancer. Gynecol. Oncol. 2015, 137, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Wang, C.; Tong, R. ERBB2 gene expression silencing involved in ovarian cancer cell migration and invasion through mediating MAPK1/MAPK3 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5267–5280. [Google Scholar] [PubMed]
- Schaner, M.E.; Ross, D.T.; Ciaravino, G.; Sørlie, T.; Troyanskaya, O.; Diehn, M.; Wang, Y.C.; Duran, G.E.; Sikic, T.L.; Caldeira, S. Gene expression patterns in ovarian carcinomas. Mol. Biol. Cell 2003, 14, 4376–4386. [Google Scholar] [CrossRef]
- Tocci, P.; Cianfrocca, R.; Rosanò, L.; Sestito, R.; Di Castro, V.; Blandino, G.; Bagnato, A. Endothelin-1 receptor/β-arrestin1 is an actionable node that regulates YAP/TAZ signaling and chemoresistance in high-grade ovarian cancer. In Proceedings of the American Association for Cancer Research (AACR) Meeting, Washington, DC, USA, 1–5 April 2017. [Google Scholar]
- Wu, M.; Sun, Y.; Wu, J.; Liu, G. Identification of Hub Genes in High-Grade Serous Ovarian Cancer Using Weighted Gene Co-Expression Network Analysis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e922107. [Google Scholar] [CrossRef]
- Nam, E.J.; Yoon, H.; Kim, S.W.; Kim, H.; Kim, Y.T.; Kim, J.H.; Kim, J.W.; Kim, S. MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 2008, 14, 2690–2695. [Google Scholar] [CrossRef]
- Hu, Y.; Pan, J.; Shah, P.; Ao, M.; Thomas, S.N.; Liu, Y.; Chen, L.; Schnaubelt, M.; Clark, D.J.; Rodriguez, H. Integrated proteomic and glycoproteomic characterization of human high-grade serous ovarian carcinoma. Cell Rep. 2020, 33, 108276. [Google Scholar] [CrossRef]
- Ray, A.; Fornsaglio, J.; Dogan, S.; Hedau, S.; Naik, S.D.; De, A. Gynaecological cancers and leptin: A focus on the endometrium and ovary. Facts Views Vis. ObGyn 2018, 10, 5. [Google Scholar]
- Zhang, S.; Lu, Z.; Mao, W.; Ahmed, A.A.; Yang, H.; Zhou, J.; Jennings, N.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Miranda, R. CDK5 regulates paclitaxel sensitivity in ovarian cancer cells by modulating AKT activation, p21Cip1-and p27Kip1-mediated G1 cell cycle arrest and apoptosis. PLoS ONE 2015, 10, e0131833. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, V.; Provencher, D.M.; Maugard, C.M.; Le Page, C.; Ren, F.; Lussier, C.; Novak, J.; Ge, B.; Hudson, T.J.; Tonin, P.N. Discrimination between serous low malignant potential and invasive epithelial ovarian tumors using molecular profiling. Oncogene 2005, 24, 4672–4687. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, R.-C.; O’malley, B.W. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat. Rev. Cancer 2009, 9, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Wiener, J.R.; Windham, T.C.; Estrella, V.C.; Parikh, N.U.; Thall, P.F.; Deavers, M.T.; Bast, R.C., Jr.; Mills, G.B.; Gallick, G.E. Activated SRC protein tyrosine kinase is overexpressed in late-stage human ovarian cancers. Gynecol. Oncol. 2003, 88, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Lui, G.Y.; Shaw, R.; Schaub, F.X.; Stork, I.N.; Gurley, K.E.; Bridgwater, C.; Diaz, R.L.; Rosati, R.; Swan, H.A.; Ince, T.A. BET, SRC, and BCL2 family inhibitors are synergistic drug combinations with PARP inhibitors in ovarian cancer. EBioMedicine 2020, 60, 102988. [Google Scholar] [CrossRef]
- Beischlag, T.V.; Morales, J.L.; Hollingshead, B.D.; Perdew, G.H. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. ™ Eukaryot. Gene Expr. 2008, 18, 207–250. [Google Scholar] [CrossRef] [PubMed]
- Abel, J.; Haarmann-Stemmann, T. An introduction to the molecular basics of aryl hydrocarbon receptor biology. Biol. Chem. 2010, 391, 1235–1248. [Google Scholar] [CrossRef]
- Larigot, L.; Juricek, L.; Dairou, J.; Coumoul, X. AhR signaling pathways and regulatory functions. Biochim. Open 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Wang, Z.; Snyder, M.; Kenison, J.E.; Yang, K.; Lara, B.; Lydell, E.; Bennani, K.; Novikov, O.; Federico, A.; Monti, S. How the AHR Became Important in Cancer: The Role of Chronically Active AHR in Cancer Aggression. Int. J. Mol. Sci. 2021, 22, 387. [Google Scholar] [CrossRef]
- Paris, A.; Tardif, N.; Galibert, M.-D.; Corre, S. AhR and Cancer: From Gene Profiling to Targeted Therapy. Int. J. Mol. Sci. 2021, 22, 752. [Google Scholar] [CrossRef]
- Kumar, M.B.; Perdew, G.H. Nuclear receptor coactivator SRC-1 interacts with the Q-rich subdomain of the AhR and modulates its transactivation potential. Gene Expr. J. Liver Res. 1999, 8, 273–286. [Google Scholar]
- Beischlag, T.V.; Wang, S.; Rose, D.W.; Torchia, J.; Reisz-Porszasz, S.; Muhammad, K.; Nelson, W.E.; Probst, M.R.; Rosenfeld, M.G.; Hankinson, O. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell. Biol. 2002, 22, 4319. [Google Scholar] [CrossRef]
- Tomkiewicz, C.; Herry, L.; Bui, L.; Metayer, C.; Bourdeloux, M.; Barouki, R.; Coumoul, X. The aryl hydrocarbon receptor regulates focal adhesion sites through a non-genomic FAK/Src pathway. Oncogene 2013, 32, 1811–1820. [Google Scholar] [CrossRef]
- Choudhary, M.; Malek, G. The aryl hydrocarbon receptor: A mediator and potential therapeutic target for ocular and non-ocular neurodegenerative diseases. Int. J. Mol. Sci. 2020, 21, 6777. [Google Scholar] [CrossRef]
- Feng, S.; Cao, Z.; Wang, X. Role of aryl hydrocarbon receptor in cancer. Biochim. Biophys. Acta (BBA) Rev. Cancer 2013, 1836, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Mulero-Navarro, S.; Fernandez-Salguero, P.M. New trends in aryl hydrocarbon receptor biology. Front. Cell Dev. Biol. 2016, 4, 45. [Google Scholar] [CrossRef]
- Piotrowska, H.; Kucinska, M.; Murias, M. Expression of CYP1A1, CYP1B1 and MnSOD in a panel of human cancer cell lines. Mol. Cell. Biochem. 2013, 383, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Hourani, S.; Therachiyil, L.; Al-Dhfyan, A.; Agouni, A.; Zeidan, A.; Uddin, S.; Korashy, H.M. Epigenetic Regulation of Cancer Stem Cells by the Aryl Hydrocarbon Receptor Pathway. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.; Lee, S.-O.; Jin, U.-H. Role of the aryl hydrocarbon receptor in carcinogenesis and potential as a drug target. Toxicol. Sci. 2013, 135, 1–16. [Google Scholar] [CrossRef]
- Khorram, O.; Garthwaite, M.; Golos, T. Uterine and ovarian aryl hydrocarbon receptor (AHR) and aryl hydrocarbon receptor nuclear translocator (ARNT) mRNA expression in benign and malignant gynaecological conditions. Mol. Hum. Reprod. 2002, 8, 75–80. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsuchiya, M.; Katoh, T.; Motoyama, H.; Sasaki, H.; Tsugane, S.; Ikenoue, T. Analysis of the AhR, ARNT, and AhRR gene polymorphisms: Genetic contribution to endometriosis susceptibility and severity. Fertil. Steril. 2005, 84, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Beedanagari, S.R.; Taylor, R.T.; Bui, P.; Wang, F.; Nickerson, D.W.; Hankinson, O. Role of epigenetic mechanisms in differential regulation of the dioxin-inducible human CYP1A1 and CYP1B1 genes. Mol. Pharmacol. 2010, 78, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Salani, R.; Backes, F.J.; Fung, M.F.K.; Holschneider, C.H.; Parker, L.P.; Bristow, R.E.; Goff, B.A. Posttreatment surveillance and diagnosis of recurrence in women with gynecologic malignancies: Society of Gynecologic Oncologists recommendations. Am. J. Obstet. Gynecol. 2011, 204, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Pokhriyal, R.; Hariprasad, R.; Kumar, L.; Hariprasad, G. Chemotherapy resistance in advanced ovarian cancer patients. Biomark. Cancer 2019, 11, 1179299X19860815. [Google Scholar] [CrossRef]
- Jing, Y.; Han, Z.; Zhang, S.; Liu, Y.; Wei, L. Epithelial-Mesenchymal Transition in tumor microenvironment. Cell Biosci. 2011, 1, 1–7. [Google Scholar] [CrossRef]
- Gao, D.; Vahdat, L.T.; Wong, S.; Chang, J.C.; Mittal, V. Microenvironmental regulation of epithelial–mesenchymal transitions in cancer. Cancer Res. 2012, 72, 4883–4889. [Google Scholar] [CrossRef]
- Jung, H.-Y.; Fattet, L.; Yang, J. Molecular pathways: Linking tumor microenvironment to epithelial–mesenchymal transition in metastasis. Clin. Cancer Res. 2015, 21, 962–968. [Google Scholar] [CrossRef]
- Mladinich, M.; Ruan, D.; Chan, C.-H. Tackling cancer stem cells via inhibition of EMT transcription factors. Stem Cells Int. 2016, 2016, 1–10. [Google Scholar] [CrossRef]
- Kurrey, N.; Amit, K.; Bapat, S. Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol. Oncol. 2005, 97, 155–165. [Google Scholar] [CrossRef]
- Ganesan, R.; Mallets, E.; Gomez-Cambronero, J. The transcription factors Slug (SNAI2) and Snail (SNAI1) regulate phospholipase D (PLD) promoter in opposite ways towards cancer cell invasion. Mol. Oncol. 2016, 10, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Hwang, K.-A.; Hyun, S.-H.; Nam, K.-H.; Lee, C.-K.; Choi, K.-C. Bisphenol A and nonylphenol have the potential to stimulate the migration of ovarian cancer cells by inducing epithelial–mesenchymal transition via an estrogen receptor dependent pathway. Chem. Res. Toxicol. 2015, 28, 662–671. [Google Scholar] [CrossRef]
- Oral, D.; Erkekoglu, P.; Kocer-Gumusel, B.; Chao, M.-W. Epithelial-mesenchymal transition: A special focus on phthalates and bisphenol a. J. Environ. Pathol. Toxicol. Oncol. 2016, 35, 43–58. [Google Scholar] [CrossRef]
- Xu, Z.; Ding, W.; Deng, X. PM2. 5, fine particulate matter: A novel player in the epithelial-mesenchymal transition? Front. Physiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.-A.; Hwang, K.-A.; Choi, K.-C. Roles of dietary phytoestrogens on the regulation of epithelial-mesenchymal transition in diverse cancer metastasis. Toxins 2016, 8, 162. [Google Scholar] [CrossRef] [PubMed]
- Moretti, S.; Nucci, N.; Menicali, E.; Morelli, S.; Bini, V.; Colella, R.; Mandarano, M.; Sidoni, A.; Puxeddu, E. The aryl hydrocarbon receptor is expressed in thyroid carcinoma and appears to mediate epithelial-mesenchymal-transition. Cancers 2020, 12, 145. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).