Context-Specific Osteogenic Potential of Mesenchymal Stem Cells
Abstract
1. Introduction
2. Results
2.1. MSCs of Adipose Tissue are More Sensitive to Notch-Dependent Induction of Osteogenic Differentiation in Comparison to VICs
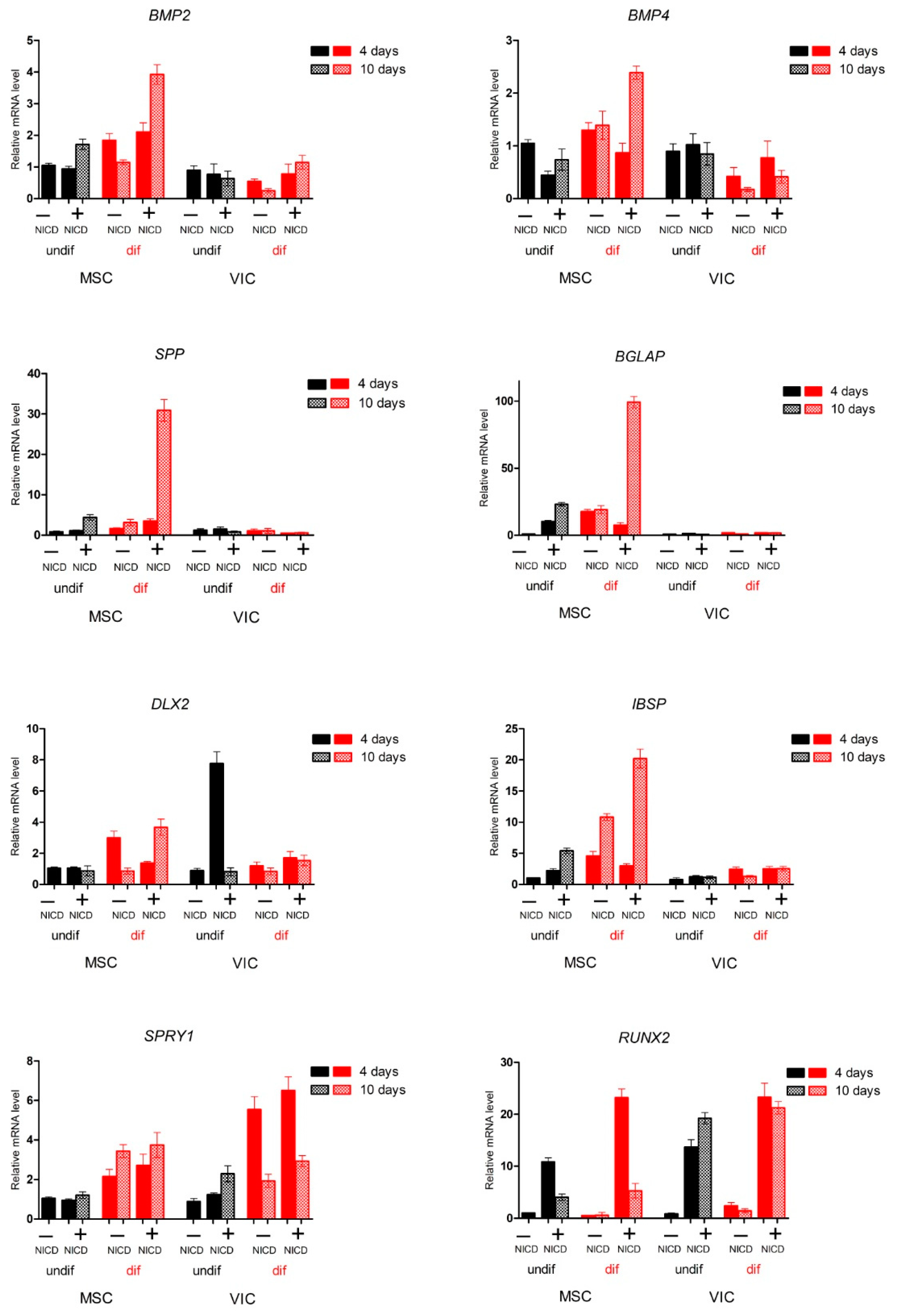
2.2. Proosteogenic Gene Expression Is Different between MSCs and VICs at Osteogenic Differentiation
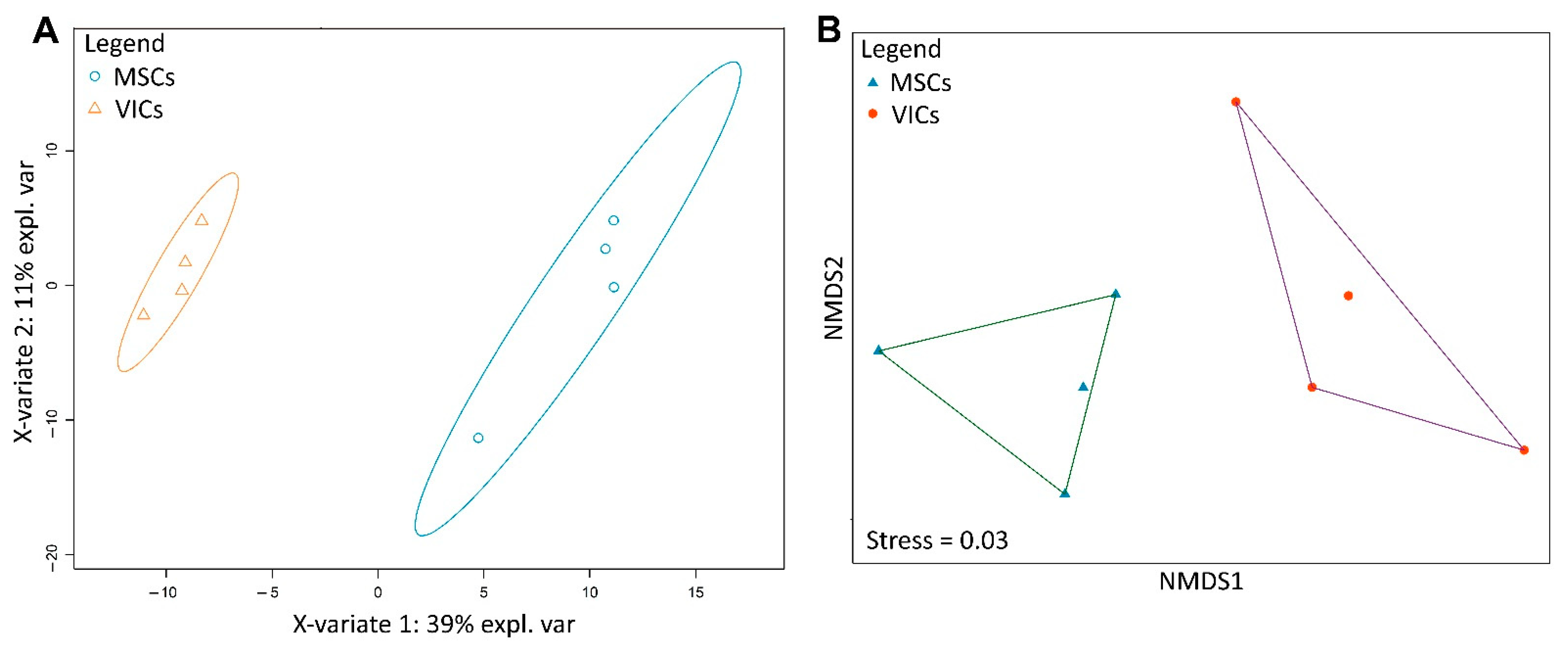
2.3. Metabolic Profiles of MSCs and VICs Differ after Osteogenic Differentiation
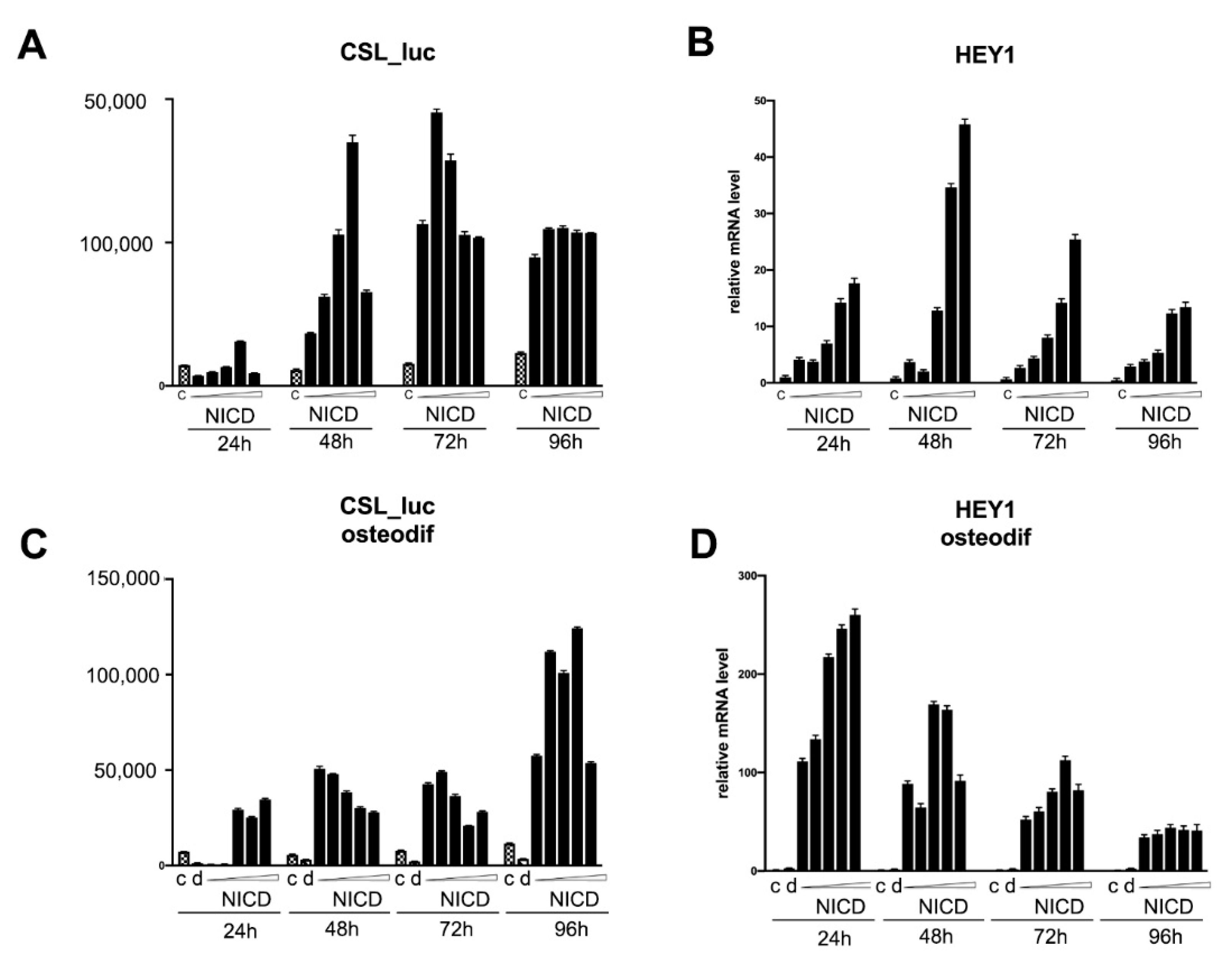
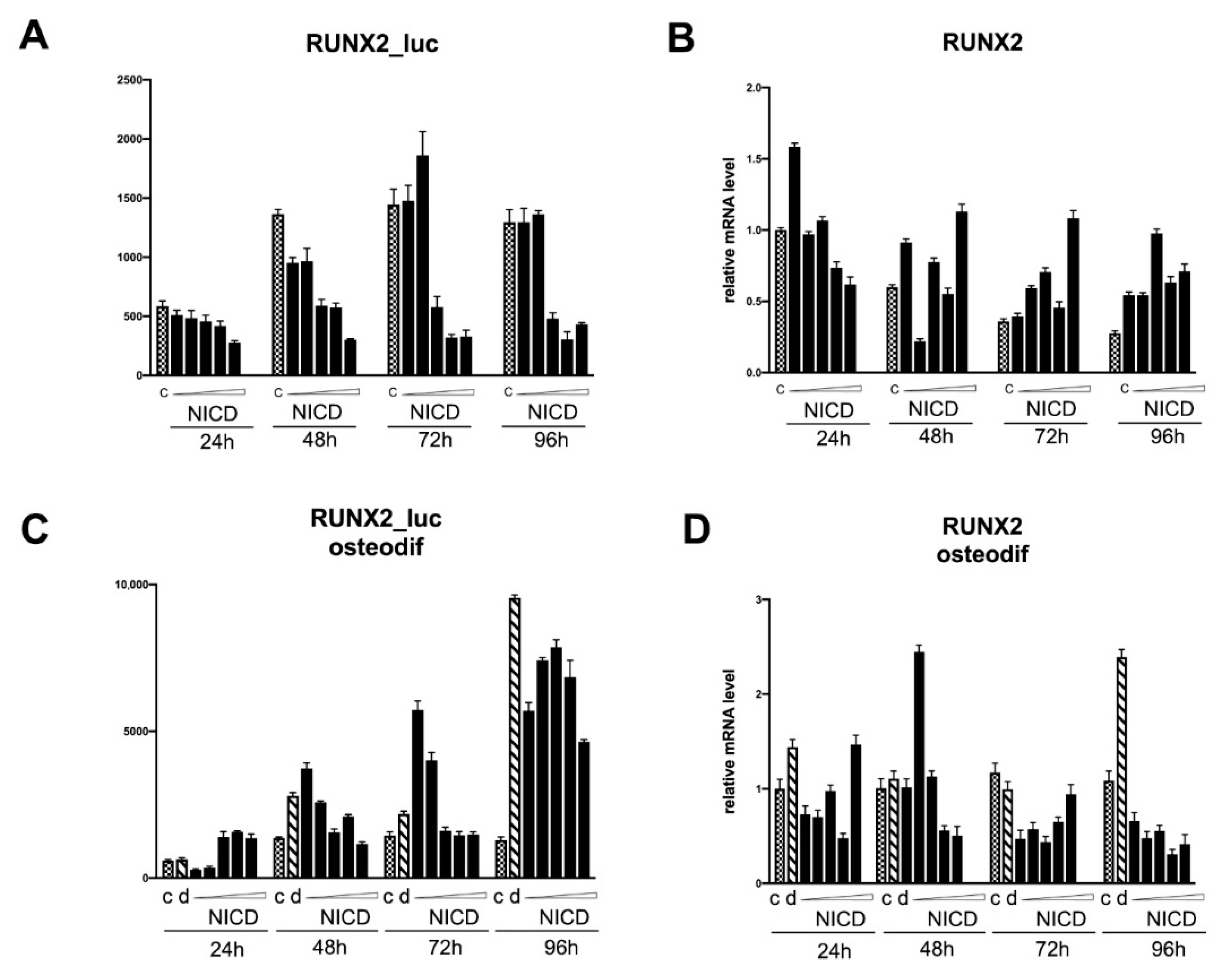
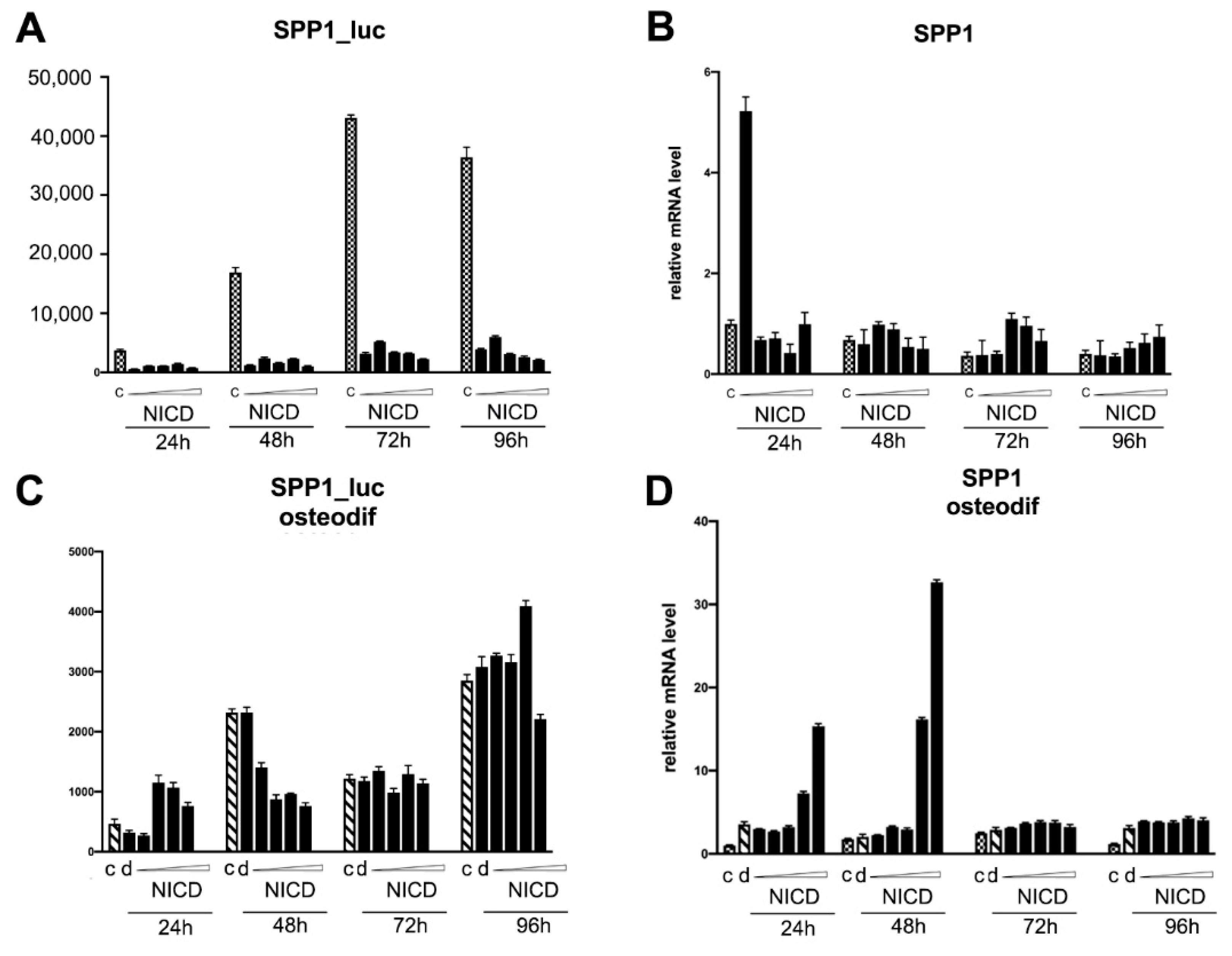
2.4. Assessment of the Transcriptional Activity of the Promoters of the Proosteogenic Markers RUNX2 and SPP1 Depending on the Level of Notch Activation
3. Discussion
4. Materials and Methods
4.1. Isolation, In Vitro Culture, and Differentiation of Primary Cell Lines
4.2. Genetic Constructs and Lentivirus Production
4.3. qPCR Analysis
4.4. Promoter Activity Assay
4.5. Metabolomic Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mastrolia, I.; Foppiani, E.M.; Murgia, A.; Candini, O.; Samarelli, A.V.; Grisendi, G.; Veronesi, E.; Horwitz, E.M.; Dominici, M. Challenges in Clinical Development of Mesenchymal Stromal/Stem Cells: Concise Review. STEM CELLS Transl. Med. 2019, 8, 1135–1148. [Google Scholar] [CrossRef]
- Demer, L.L.; Tintut, Y. Interactive and Multifactorial Mechanisms of Calcific Vascular and Valvular Disease. Trends Endocrinol. Metab. 2019, 30, 646–657. [Google Scholar] [CrossRef]
- Yao, Y.; Jumabay, M.; Ly, A.; Radparvar, M.; Cubberly, M.R.; Boström, K.I. A Role for the Endothelium in Vascular Calcification. Circ. Res. 2013, 113, 495–504. [Google Scholar] [CrossRef]
- Xiao, L.; Dudley, A.C. Fine-tuning vascular fate during endothelial-mesenchymal transition. J. Pathol. 2017, 241, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Nantavisai, S.; Pisitkun, T.; Osathanon, T.; Pavasant, P.; Kalpravidh, C.; Dhitavat, S.; Makjaroen, J.; Sawangmake, C. Systems biology analysis of osteogenic differentiation behavior by canine mesenchymal stem cells derived from bone marrow and dental pulp. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rutkovskiy, A.; Malashicheva, A.; Sullivan, G.; Bogdanova, M.; Kostareva, A.; Stensløkken, K.-O.; Fiane, A.; Vaage, J. Valve Interstitial Cells: The Key to Understanding the Pathophysiology of Heart Valve Calcification. J. Am. Hear. Assoc. 2017, 6, e006339. [Google Scholar] [CrossRef]
- Canalis, E. Notch in skeletal physiology and disease. Osteoporos. Int. 2018, 29, 2611–2621. [Google Scholar] [CrossRef] [PubMed]
- Youngstrom, D.W.; Hankenson, K.D. Contextual Regulation of Skeletal Physiology by Notch Signaling. Curr. Osteoporos. Rep. 2019, 17, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Sciaudone, M.; Gazzerro, E.; Priest, L.; Delany, A.M.; Canalis, E. Notch 1 Impairs Osteoblastic Cell Differentiation. Endocrinology 2003, 144, 5631–5639. [Google Scholar] [CrossRef]
- Nofziger, D.; Miyamoto, A.; Lyons, K.; Weinmaster, G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 1999, 126, 1689–1702. [Google Scholar] [CrossRef]
- Deregowski, V.; Gazzerro, E.; Priest, L.; Rydziel, S.; Canalis, E. Notch 1 Overexpression Inhibits Osteoblastogenesis by Suppressing Wnt/β-Catenin but Not Bone Morphogenetic Protein Signaling. J. Biol. Chem. 2006, 281, 6203–6210. [Google Scholar] [CrossRef] [PubMed]
- Urbanek, K.; Lesiak, M.; Krakowian, D.; Koryciak-Komarska, H.; Likus, W.; Czekaj, P.; Kusz, D.; Sieroń, A.L. Notch signaling pathway and gene expression profiles during early in vitro differentiation of liver-derived mesenchymal stromal cells to osteoblasts. Lab. Investig. 2017, 97, 1225–1234. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bagheri, L.; Pellati, A.; Rizzo, P.; Aquila, G.; Massari, L.; De Mattei, M.; Ongaro, A. Notch pathway is active during osteogenic differentiation of human bone marrow mesenchymal stem cells induced by pulsed electromagnetic fields. J. Tissue Eng. Regen. Med. 2017, 12, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Ongaro, A.; Pellati, A.; Bagheri, L.; Rizzo, P.; Caliceti, C.; Massari, L.; De Mattei, M. Characterization of Notch Signaling During Osteogenic Differentiation in Human Osteosarcoma Cell Line MG63. J. Cell. Physiol. 2016, 231, 2652–2663. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Ke, Y.; Gao, S. Intermittent activation of notch signaling promotes bone formation. Am. J. Transl. Res. 2017, 9, 2933–2944. [Google Scholar]
- Semenova, D.; Bogdanova, M.; Kostina, A.; Golovkin, A.; Kostareva, A.; Malashicheva, A. Dose-dependent mechanism of Notch action in promoting osteogenic differentiation of mesenchymal stem cells. Cell Tissue Res. 2019, 379, 169–179. [Google Scholar] [CrossRef]
- Kostina, A.; Shishkova, A.; Ignatieva, E.; Irtyuga, O.; Bogdanova, M.; Levchuk, K.; Golovkin, A.; Zhiduleva, E.; Uspenskiy, V.; Moiseeva, O.; et al. Different Notch signaling in cells from calcified bicuspid and tricuspid aortic valves. J. Mol. Cell. Cardiol. 2018, 114, 211–219. [Google Scholar] [CrossRef]
- Kostina, A.; Semenova, D.; Kostina, D.; Uspensky, V.; Kostareva, A.; Malashicheva, A. Human aortic endothelial cells have osteogenic Notch-dependent properties in co-culture with aortic smooth muscle cells. Biochem. Biophys. Res. Commun. 2019, 514, 462–468. [Google Scholar] [CrossRef]
- Aquila, G.; Kostina, A.; Sega, F.V.D.; Shlyakhto, E.; Kostareva, A.; Marracino, L.; Ferrari, R.; Rizzo, P.; Malaschicheva, A. The Notch pathway: a novel therapeutic target for cardiovascular diseases? Expert Opin. Ther. Targets 2019, 23, 695–710. [Google Scholar] [CrossRef]
- Falo-Sanjuan, J.; Bray, S.J. Decoding the Notch signal. Dev. Growth Differ. 2020, 62, 4–14. [Google Scholar] [CrossRef]
- Perepelina, K.; Klauzen, P.; Kostareva, A.; Malashicheva, A. Tissue-Specific Influence of Lamin A Mutations on Notch Signaling and Osteogenic Phenotype of Primary Human Mesenchymal Cells. Cells 2019, 8, 266. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Schulze, K.L.; Bellen, H.J. Introduction to Notch Signaling. Adv. Struct. Saf. Stud. 2014, 1187, 1–14. [Google Scholar] [CrossRef]
- Isobe, Y.; Koyama, N.; Nakao, K.; Osawa, K.; Ikeno, M.; Yamanaka, S.; Okubo, Y.; Fujimura, K.; Bessho, K. Comparison of human mesenchymal stem cells derived from bone marrow, synovial fluid, adult dental pulp, and exfoliated deciduous tooth pulp. Int. J. Oral Maxillofac. Surg. 2016, 45, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Manokawinchoke, J.; Sumrejkanchanakij, P.; Boonprakong, L.; Pavasant, P.; Egusa, H.; Osathanon, T. NOTCH2 participates in Jagged1-induced osteogenic differentiation in human periodontal ligament cells. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hansamuit, K.; Osathanon, T.; Suwanwela, J. Effect of Jagged1 on the expression of genes in regulation of osteoblast differentiation and bone mineralization ontology in human dental pulp and periodontal ligament cells. J. Oral Biol. Craniofacial Res. 2020, 10, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Osathanon, T.; Manokawinchoke, J.; Sa-Ard-Iam, N.; Mahanonda, R.; Pavasant, P.; Suwanwela, J. Jagged1 promotes mineralization in human bone-derived cells. Arch. Oral Biol. 2019, 99, 134–140. [Google Scholar] [CrossRef]
- Wagley, Y.; Chesi, A.; Acevedo, P.K.; Lu, S.; Wells, A.D.; Johnson, M.E.; Grant, S.F.A.; Hankenson, K.D. Canonical Notch signaling is required for bone morphogenetic protein-mediated human osteoblast differentiation. STEM CELLS 2020. [Google Scholar] [CrossRef] [PubMed]
- Novak, S.; Roeder, E.; Sinder, B.P.; Adams, D.J.; Siebel, C.W.; Grcevic, D.; Hankenson, K.D.; Matthews, B.G.; Kalajzic, I. Modulation of Notch1 signaling regulates bone fracture healing. J. Orthop. Res. 2020, 38, 2350–2361. [Google Scholar] [CrossRef]
- Malashicheva, A.; Bogdanova, M.; Zabirnyk, A.; Smolina, N.; Ignatieva, E.; Freilikhman, O.; Fedorov, A.; Dmitrieva, R.; Sjöberg, G.; Sejersen, T.; et al. Various lamin A/C mutations alter expression profile of mesenchymal stem cells in mutation specific manner. Mol. Genet. Metab. 2015, 115, 118–127. [Google Scholar] [CrossRef]
- Dmitrieva, R.I.; Revittser, A.V.; Klukina, M.A.; Sviryaev, Y.; Korostovtseva, L.S.; Kostareva, A.; Zaritskey, A.Y.; Shlyakhto, E.V. Functional properties of bone marrow derived multipotent mesenchymal stromal cells are altered in heart failure patients, and could be corrected by adjustment of expansion strategies. Aging 2015, 7, 14–25. [Google Scholar] [CrossRef]
- Kostina, A.S.; Uspensky, V.E.; Irtyuga, O.B.; Ignatieva, E.V.; Freylikhman, O.; Gavriliuk, N.D.; Moiseeva, O.M.; Zhuk, S.; Tomilin, A.; Kostareva, A.; et al. Notch-dependent EMT is attenuated in patients with aortic aneurysm and bicuspid aortic valve. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2016, 1862, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2018, 15, 53–56. [Google Scholar] [CrossRef] [PubMed]
- R. Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; et al. Vegan: Community Ecology Package R Package Version 2.5-6. 2019. Available online: https://cran.r-project.org/ (accessed on 30 May 2021).
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.A.L. MixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer 5′–3′ | |
|---|---|---|
| HEY1 | forward | TGGATCACCTGAAAATGCTG |
| reverse | CGAAATCCCAAACTCCGATA | |
| GAPDH | forward | CAAGGTCATCCATGACAACTTTG |
| reverse | GTCCACCACCCTGTTGCTGTAG | |
| RUNX2 | forward | TGGATCACCTGAAAATGCTG |
| reverse | CGAAATCCCAAACTCCGATA | |
| BGLAP | forward | CCTCACACTCCTCGCCCTAT |
| reverse | CTTGGACACAAAGGCTGCAC | |
| IBSP | forward | GATTTCCAGTTCAGGGCAGTAG |
| reverse | CCATAGCCCAGTGTTGTAGC | |
| DLX2 | forward | GGAGCCCCCATCCCTTATCTT |
| reverse | ATCCGCAAAGGCACCTAAACT | |
| SPRY1 | forward | GGACCCAGCCCAAGCAA |
| reverse | TTGTGCTGTGTCAGGTCCTCTT | |
| BMP2 | forward | GCCAAGCCGAGCCAACAC |
| reverse | CCCACTCGTTTCTGGTAGTTCTTC | |
| BMP4 | forward | AGCACTGGTCTTGAGTATCCTG |
| reverse | GCAGAGTTTTCACTGGTCCC | |
| SPP1 | forward | TCACCTGTGCCATACCAGTTAAA |
| reverse | TGGGTATTTGTTGTAAAGCTGCTT | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostina, A.; Lobov, A.; Semenova, D.; Kiselev, A.; Klausen, P.; Malashicheva, A. Context-Specific Osteogenic Potential of Mesenchymal Stem Cells. Biomedicines 2021, 9, 673. https://doi.org/10.3390/biomedicines9060673
Kostina A, Lobov A, Semenova D, Kiselev A, Klausen P, Malashicheva A. Context-Specific Osteogenic Potential of Mesenchymal Stem Cells. Biomedicines. 2021; 9(6):673. https://doi.org/10.3390/biomedicines9060673
Chicago/Turabian StyleKostina, Aleksandra, Arseniy Lobov, Daria Semenova, Artem Kiselev, Polina Klausen, and Anna Malashicheva. 2021. "Context-Specific Osteogenic Potential of Mesenchymal Stem Cells" Biomedicines 9, no. 6: 673. https://doi.org/10.3390/biomedicines9060673
APA StyleKostina, A., Lobov, A., Semenova, D., Kiselev, A., Klausen, P., & Malashicheva, A. (2021). Context-Specific Osteogenic Potential of Mesenchymal Stem Cells. Biomedicines, 9(6), 673. https://doi.org/10.3390/biomedicines9060673







