Abstract
Alternative splicing (AS) of human telomerase catalytic subunit (hTERT, human telomerase reverse transcriptase) pre-mRNA strongly regulates telomerase activity. Several proteins can regulate AS in a cell type-specific manner and determine the functions of cells. In addition to being involved in telomerase activity regulation, AS provides cells with different splice variants that may have alternative biological activities. The modulation of telomerase activity through the induction of hTERT AS is involved in the development of different cancer types and embryos, and the differentiation of stem cells. Regulatory T cells may suppress the proliferation of target human and murine T and B lymphocytes and NK cells in a contact-independent manner involving activation of TERT AS. This review focuses on the mechanism of regulation of hTERT pre-mRNA AS and the involvement of splice variants in physiological and pathological processes.
1. Introduction
The stability of chromosomes at their 3′ ends is supported by telomere (TTAGGG) DNA repeats. Recombination-based alternative lengthening of telomeres and elongation of telomeres by telomerase are two mechanisms for maintaining telomere length in cells with unlimited replicative potential or immortalized cells, including stem cells, germ cells, activated lymphocytes, endothelial cells, and cancer cells [1,2,3,4,5]. hTERT and hTR comprise the catalytic core of telomerase, whereas the holoenzyme contains additional species-specific accessory proteins. The most frequent mechanism for telomere elongation among various cells is the functioning of telomerase, a ribonucleoprotein complex consisting of two key subunits: human telomerase RNA (hTR), which acts as a transcription template for newly synthetized telomeres, and human telomerase reverse transcriptase (hTERT), whose enzymatic activity controls the grade of telomerase activity. Although hTR is constitutively expressed in most tissues in human cells, hTERT expression is highly regulated at the transcriptional and posttranscriptional levels. However, the suggestion that hTERT levels regulate telomerase activity only applies to most somatic tissues, as opposed to the brain, where hTR is downregulated very early during development and is thus most likely responsible for the disappearance of the activity [6].
Transcriptional regulation of hTERT was extensively studied, and the chromatin environment, DNA methylation, DNA looping, promoter mutations, and the binding of transcription factors were shown to affect the strength of gene expression [7,8].
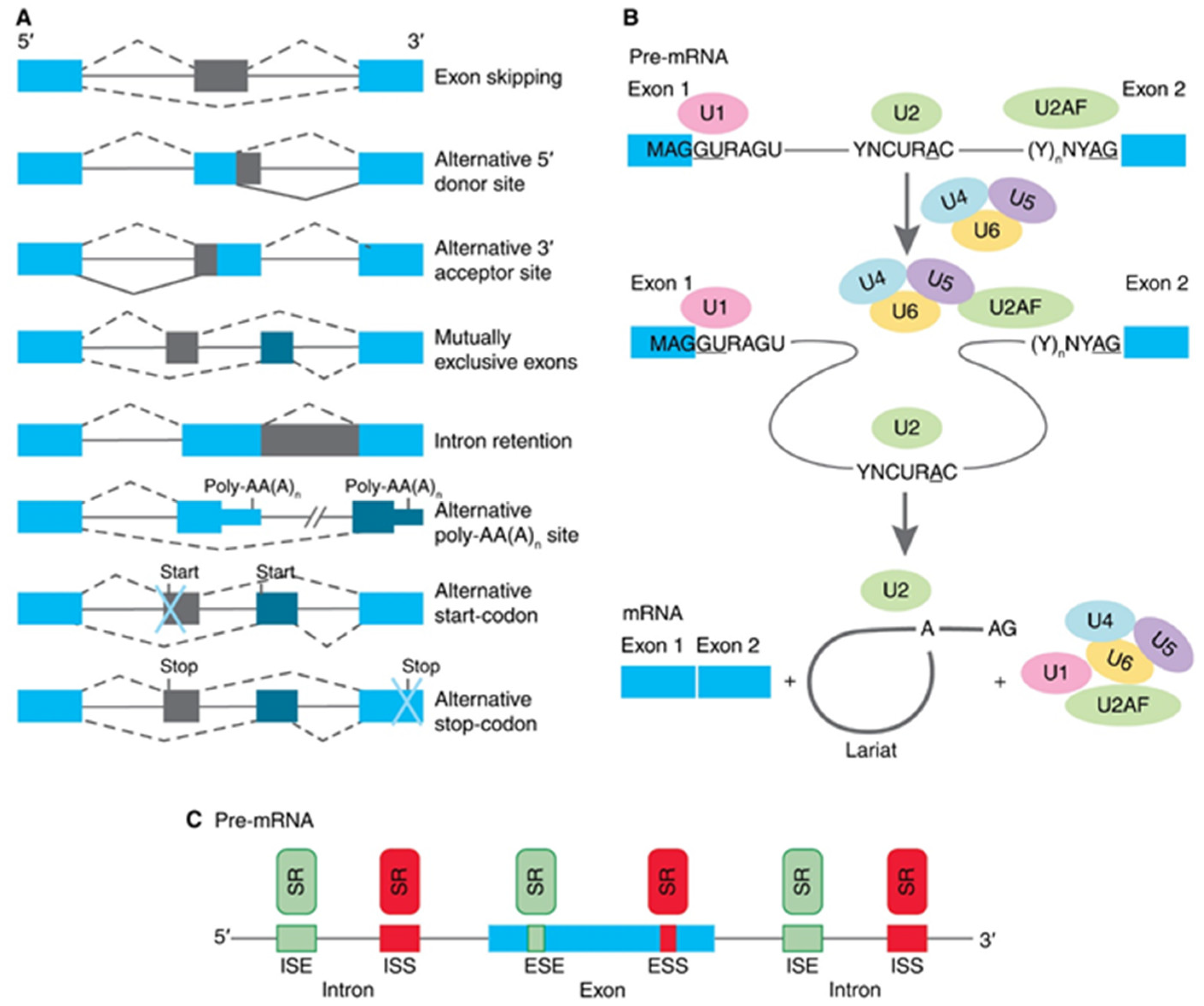
Most genes of higher eukaryotes have an interrupted structure in certain coding regions—exons alternate with noncoding sequences and introns. Gene transcription leads to the formation of pre-mRNA, a molecule that has both exons and introns. Primary pre-mRNA transcripts undergo modifications before being translated, i.e., capping at the 5′- end, and the synthesis of the polyA sequence at the 3′-end of the transcript. The second event in the maturation of mRNA is a splicing of exons and excision of introns. Post-transcriptional maturation of pre-mRNA plays an essential role in providing the biodiversity of protein products encoded by a single gene due to the process of alternative splicing (AS) of pre-mRNA. In this process, particular exons, or parts of exons, may be included within or excluded from the final matured mRNA. Consequently, the proteins translated from alternatively spliced pre-mRNAs will contain differences in their amino acid sequence, and often in their biological functions too [9]. To date, several types of AS are described for human pre-mRNAs (as illustrated in Figure 1A). Exon skipping is the most common mode in mammalian pre-mRNAs. In this case, an exon may be spliced out of the primary transcript or retained. Mutually exclusive exons are a mode when only one of two exons is retained in mRNAs after splicing, but not both. Alternative donor/acceptor sites are two modes when an alternative 5′ or 3′ splice junction is used. The rearrest mode in mammals is an intron retention when a sequence may be spliced out as an intron or simply retained. This is distinguished from exon skipping because the retained sequence is not flanked by introns [10,11]. In addition to these primary modes, AS may provide alternative poly-AA(A)n site, an alternative site for the beginning of translation (start-codon) or the formation of preliminary stop-codon [12]. Pre-mRNA splicing is conducted by a multiprotein complex spliceosome (as illustrated in Figure 1B), which consists of six main subunits (U1, U2, U2AF, U4, U5, and U6) and about 300 other proteins [13]. The first step of spliceosome functioning is the interaction of its subunits with the splice sites: U1 binds to the 5′site, U2 binds to the branch site, and U2AF binds to the polypyrimidine site of the 3′ splice site. The second step is the joining of the U4, U5, and U6 subunits to U1 and U2AF, which leads to the convergence of the 5′-and 3′ - splice sites. At the third step, the intron is removed in two stages. At the first stage, a transesterification reaction occurs, in which the adenosine of the branch site binds to the guanidine of the 5′ site’s GU sequence. At the second stage, the exons are ligated, and the nucleotides of the branched intron are removed in the form of lariat. An exon may be constitutive (always included in the mRNA) or alternative (may be included or excluded) to splice variants. The usage of a splice site may be enhanced or suppressed by its proximity to local cis-regulatory sequences (as illustrated in Figure 1C), such as exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), and intronic splicing silencers (ISSs) [14,15]. The cis-regulatory sequences are in turn bound by trans-acting factors or splicing factors. Most often they are serine-arginine-repeat proteins (SR proteins). The spliceosome performs the deletion of intron and exons ligation during splicing, while the functioning of SR proteins is crucial for the determination of sites that will be deleted or retained.
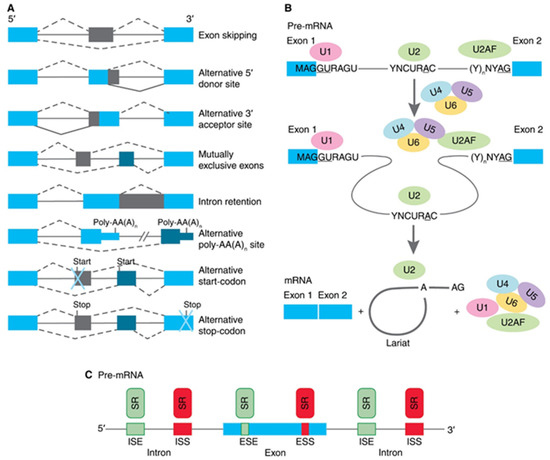
Figure 1.
Schematic presentation of biology of alternative splicing (AS). (A) Traditional classification of basic types of alternative RNA splicing events. Exons are represented as blue and grey blocks, introns as lines in between. (B) Functioning of spliceosome. Pre-mRNA containing two exons separated by an intron assembles into splicing complexes together with spliceosome subunits. Individual subunits are indicated by U1, U2, U2AF, U4, U5, and U6. U1 forms a base-pairing interaction with 5′-splice site, whereas U2 base-pairs with branch-point and U2AF binds to polypyrimidine site of 3′ splice site. Then, a complex containing U4, U5, and U6 associates with the forming of spliceosome. The intron is removed in a form of lariat and two exons are ligated. (C) Interactions of trans-elements and serine/arginine-rich (SR) proteins with cis-elements, and regulatory sequences on pre-mRNA. Elements that inhibit exon inclusion are shown in red, while those enhancing inclusion are shown in green. ESE, exonic splicing enhancer; ESI, exonic splicing silencer; ISE, intronic splicing enhancer; ISS, intronic splicing silencer.
The regulation of telomerase activity and the alternative functions of hTERT through its pre-mRNA processing were less investigated. AS is one of the mechanisms that mediates the diversity of mRNAs encoded by a single gene. Precursor message RNA (pre-mRNA) subjected to different AS events determines the fate of mature mRNA variants. Splice variants of mRNA may be translated to variants of proteins that have functions that are frequently different from the full-length form, or in cases of altered capacity for translation, such splice variants are degraded by nonsense-mediated decay [16]. The AS mechanism is a tissue-specific process, and the formation of mature splice variants of a given pre-mRNA depends on the repertoire of splicing-regulatory proteins and on features of spliceosome regulation [17]. Regulation of telomerase through AS of hTERT pre-mRNA is considered to be a cell-specific process, as the presence of different hTERT splice variants is in good agreement with the grade of telomerase activity in normal cells and in cells from pathological tissues [8]. The exact regulatory mechanisms of hTERT AS and the biological functions of splice variants was not elucidated. Some recent data suggested novel features for the mechanisms of hTERT pre-mRNA AS and the roles of its splice variants in telomerase regulation alongside other cellular functions of hTERT in normal and pathological processes.
2. Regulatory Mechanisms for hTERT Pre-mRNA AS
Since hTERT is the major subunit maintaining telomerase activity, its expression and activity are highly regulated at many levels, including promoter core region organization, protein folding, post-translational modification, and interaction partners [7]. Regulation of hTERT functions by AS of its pre-mRNA is thought to be crucial because even small amounts of TERT may have significant cellular consequences [18]. While significant amounts of knowledge about TERT and telomerase functioning came from the study of model organisms [19], the following data will refer to the regulation of AS of human TERT unless otherwise stated.
2.1. Alternative Splice Variants of hTERT
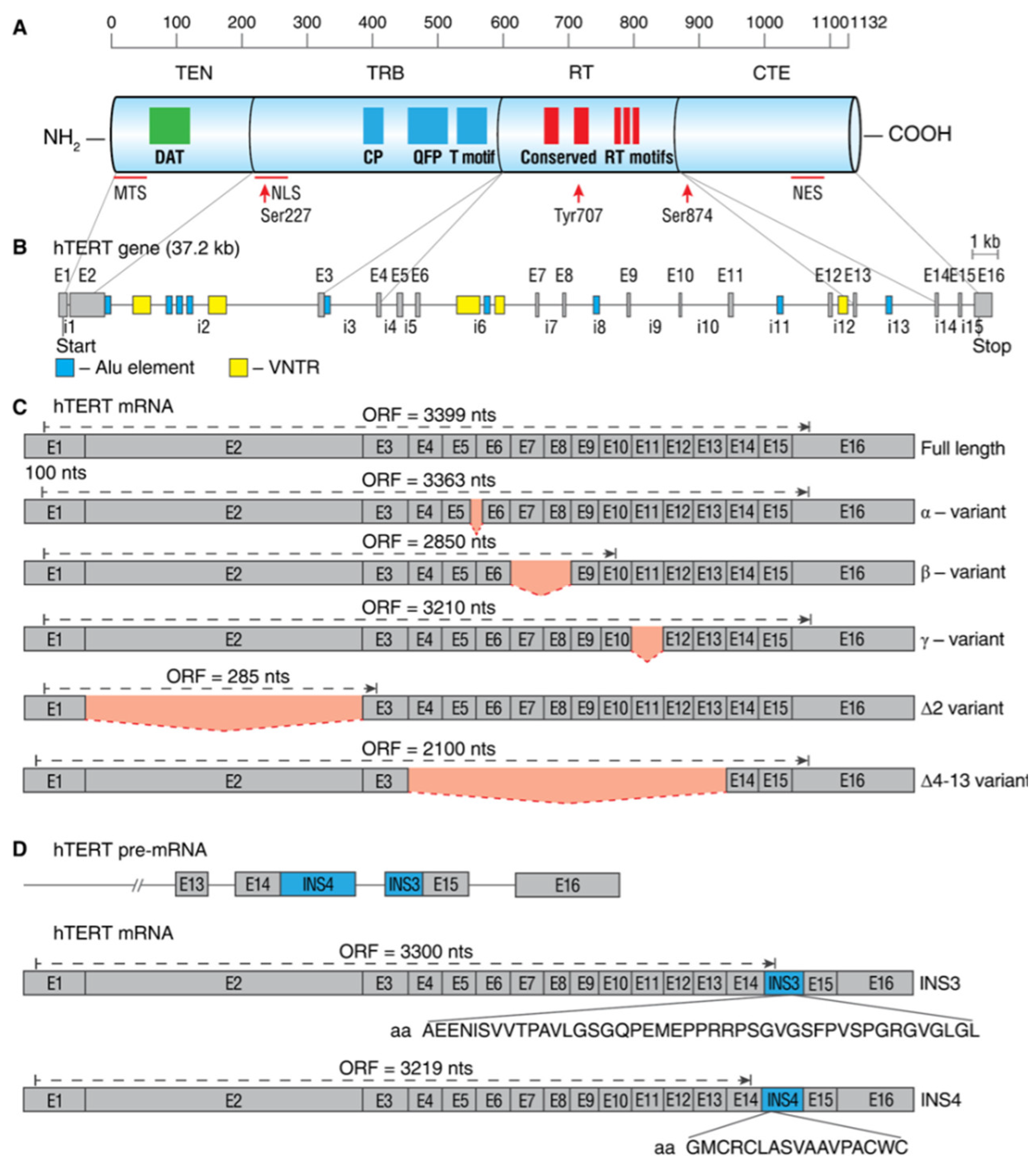
The human hTERT gene (42 kb) is located on chromosome 5p15.33 and spans 16 exons and 15 introns. The full-length hTERT transcript encodes an active 1132-amino acid (127-kDa) protein. Bioinformatics and mutational studies collectively established that hTERT contains four main structural elements (as illustrated in Figure 2A): (i) a long telomerase essential N-terminal extension (TEN domain); (ii) conserved DNA- and TERT RNA-binding domains (TRBD); (iii) a central catalytic reverse-transcriptase (RT) domain; and (iv) a short C-terminal extension (CTE domain). The TEN domain is important for the appropriate action of telomerase at telomeres, as mutations in the DAT (dissociates activities of telomerase) region abolish telomerase processivity but not catalytic activity in vivo [20]. The TEN domain contains the BH3-like motif 135WGLLLRRVGDDVLVHL152, a short peptide sequence found in BCL-2 family proteins, and interacts with the antiapoptotic BCL-2 family proteins MCL-1 and BCL-xL, suggesting a functional link between hTERT and the mitochondrial pathway of apoptosis [21].
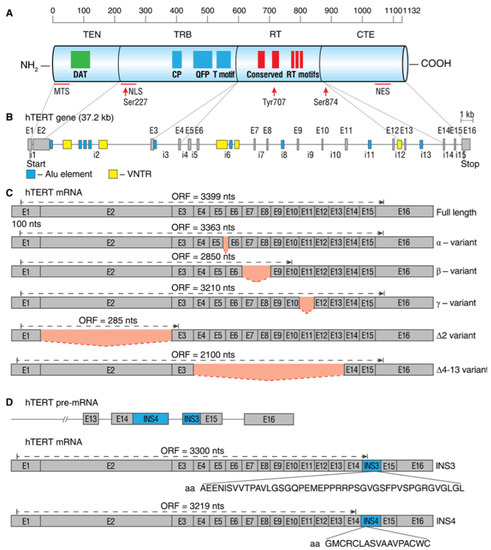
Figure 2.
Schematic map of hTERT protein, gene, and commonly studied mRNA splice variants. (A) Linear structure of 1132-amino acid hTERT protein and known domains and motifs are shown. The following active elements are responsible for intracellular relocalization of hTERT: MTS, mitochondrial targeting sequences; NLS, nuclear localization signal; Ser227, Serine 227 for phosphorylation by Akt; Tyr770, Tyrosine 770 for phosphorylation by Src1; Ser824, Serine 824 for phosphorylation by Akt; NES, nuclear export signal for binding with CRM1. (B) Structure of hTERT gene exons (E1–E16) and introns (i1–i15). Positions of Alu elements and variable number tandem repeats (VNTRs) are shown as dark blue and yellow boxes, respectively. Lines link exons and the domains they encode. (C) Common alternatively spliced variants with deletions are shown below the wild-type, the full-length mRNA. Predicted open reading frame (ORF) for each mRNA is indicated. (D) Common alternatively spliced variants that include insertions INS3 and INS4 and the amino acids that are encoded by them.
The TRBD domain has several conserved telomerase-specific motifs, including the CP, QFP, and TS motifs that are important for TERT-TR binding interactions and the grade of template copying during telomere synthesis [22,23]. The TEN domain and TRBD cooperate to ensure sequence-specific TERT-TR binding interactions and optimal template positioning during telomerase assembly and/or telomere synthesis. The RT domain contains five conserved RT motifs that are responsible for the catalytic activity of the enzyme [24]. The CTE domain is responsible for different protein-protein interactions and regulates the cellular localization of the protein [25].
Activation of hTERT is primarily regulated by the phosphorylation of protein kinase C (PKC) isoenzymes, thereby enhancing telomerase activity [26,27]. hTERT contains a nuclear localization signal (NLS) consisting of two clusters of basic amino acids (222RRR224 and 236KRPRR240) [28]. Additionally, putative Akt phosphorylation sites are located at 220GARRRGGSAS229 and 817AVRIRGKSYV826, and the phosphorylation of serine 227 (but not serine 824) is required for nuclear translocation of hTERT [28,29,30]. Extranuclear localization of hTERT is regulated by the phosphorylation of Src tyrosine kinase at tyrosine 707 [31].
To date, 22 different splice variants of hTERT mRNA were identified, which are combinations of five deletions and two insertion splice events (as illustrated in Figure 2B–D) [32,33,34,35]. Only wild-type, the full-length hTERT mRNA with neither deletions nor insertions, encodes a protein that can assemble into an active telomerase holoenzyme [36,37]. In telomerase-positive cells, the full-length hTERT splice variants are expressed in the range of 1–90%, and their expression level determines the grade of telomerase activity [38,39,40].
The most studied hTERT splice variants originate from deletion events during mRNA maturation and are likely to be cell- or tissue-specific. The skipping of 36 nucleotides (nt) in the cryptic 3′ splice-acceptor site (i.e., the alpha region of splicing) at the beginning of exon 6 results in an alpha minus (α‒) variant, which is in the canonical reading frame. The hTERT α‒ variant has a deletion of 12 amino acids (aa) in the RT motif and acts as a dominant-negative protein that can bind to hTR but cannot maintain telomeres [41]. This truncated variant is rather abundant in cancer cells [38] and human activated lymphocytes [42].
The skipping of 182 nt in the beta splicing region results in the deletion of exons 7 and 8, and the shift of the reading frame leads to the formation of a premature stop codon in exon 10 and causes an early termination of translation [38,41]. Such a beta minus (β‒) splice variant is mostly degraded by nonsense-mediated decay; however, in certain cancer cells, this variant interacts with polyribosomes and is translated into a truncated protein, which is not able to form active telomerase but protects cells from apoptosis [37]. Some cells may express this β‒ hTERT variant in human activated CD4+ T lymphocytes in their steady state, and its induction by endonuclease EndoG leads to the inhibition of telomerase [43]. The expression levels of the β-variant vary between cells in the range of 10–90%. Combinations of alpha and beta splice variants (α+β−and α−β+) have also been detected in cells and can range within 1–15% depending on cell type. The biological significance of these variants for cells remains to be investigated, as they must be degraded by nonsense-mediated decay.
Another in-frame splicing event occurs in the gamma splicing region and results in the skipping of 189 nt of exon 11. Gamma minus (γ–) hTERT has an affected RT domain (missing conserved 63 aa from the catalytic core of the protein) and acts as a dominant-negative protein when it is expressed at sufficient levels [44]. Although γ-deletion hTERT showed low expression (up to 2% of total hTERT) in cell lines with high telomerase activity, this variant may occasionally be detected in samples with low telomerase activity, such as hepatocellular carcinoma nodules. Combinations of γ– splice variants such as α-γ–, β-γ–, and α-β-γ– were also detected at low levels in telomerase-positive cells; however, their functions still require investigation [44].
Withers et al. [39] demonstrated that the human epidermoid carcinoma cell line A431 expresses the hTERT splice variant lacking full exon 2 (delta 2, ∆2 variant), and the skipping of 1354 nt results in frameshift and translation termination in exon 3. Exon 2 encodes a part of the TRB domain, and the variant that lacks it would be unable to bind hTR. The authors demonstrated that this truncated protein is being translated as a 12-kDa peptide, and in a study with higher precision, it was shown that there are approximately 20–40 molecules of ∆2 hTERT mRNA copies per cell. No functions of the ∆2 variant were investigated; thus, its biological role remains unknown [10].
In a study by Hrdličková et al. [34], eight novel hTERT splice variants were identified in a screen of telomerase-positive and telomerase-negative cell lines. Four of these variants, lacking either a part of exon 2, all of exon 2, exons 2 through 8, or exons 2 through 13 (Δ2p, Δ2, Δ2–8, and Δ2–13), contain premature stop codons. Two other variants retain the second exon but lack the RT domain. One splice variant contains a deletion-encompassing exons 4 through 13 (Δ4–13) and retains the original hTERT reading frame. The second variant lacks a part of exon 3 and all of exons 4 to 12 (Δ3p-12), which introduces a premature stop codon. These variants lack regions of hTERT where the previously described α- and β- splicing events occur. Finally, one additional variant (Δ4C) lacks the 4th exon and contains an insertion of parts of the 3rd and 4th introns. This variant maintains the original reading frame. Variant Δ4–13 is expressed in two lines of dividing telomerase-negative U2-OS cells (human osteosarcoma cells) and Saos-2 (human osteosarcoma), which both employ recombination-based lengthening of telomeres, and in telomerase-positive HeLa (human cervix carcinoma) cells in culture. The authors evaluated whether this variant may be involved in cell proliferation by performing the noncanonical functions (not associated with telomere maintenance) of telomerase. In both cell lines, the overexpression of Δ4–13 resulted in an increase in cell proliferation without an increase in telomerase activity.
Several intron retention splice variants were also described. Sequencing of cDNA plasmid clones from five different tissues, lung tumor and adjacent tissue, colon tumor, and K562 and HL60 cell lines, revealed a number of hTERT splice variants that have insertions [35]. Three variants involve the insertion of sequences from intron 2. The variant Ins-i2 (1–389) inserted the first 349 nucleotides of intron 2 and results from the use of an alternative 5′-splice donor sequence within intron 2. The same splice site was also used to produce the variant Ins-i2 (82–349). In this variant, an additional splicing event removed the first 181 nucleotides of intron 2 by use of the normal 5′-splice donor sequence and an alternative 3′-splice acceptor sequence, leaving nucleotides 182–349 of intron 2 in the spliced product. The third variant involving intron 2, Ins-i2 (?–5273), results from the use of an alternative 5′-splice donor sequence within intron 2, but the precise characteristics of this splice variant were not determined. The remaining variants contain inserted sequences from intron 14. The variants Ins-i14 (623–705) and Ins-i14 (623–703) both result from a double splicing event in intron 14, which leaves an internal fragment of intron 14 (nucleotides 623–705 and 623–703, respectively) in the spliced product. As shown, many of these hTERT splice variants have premature stop codons and must be degraded. However, two variants with insertions are subjected to translation and act as dominant negatives to suppress telomerase activity. The first variant, INS3, contains a 159-nt (nucleotides 622–781) insertion from the end of intron 14, which encodes an additional 44 aa. The insertion results in a premature stop codon in exon 15 [45]. The variant INS4 contains a 600-nt insertion of intron 14, encoding 17 aa, followed by a stop codon. The expression of INS3 and INS4 is tissue-specific and, when expressed, may account for 1–15% of the total hTERT mRNAs [46,47]. Finally, a variant (INTR1) that retains the first intron and contains a premature stop codon was also identified; however, no information about its function is available [34].
It must be considered that TERT pre-mRNA from different species is also subjected to AS. Although some transcriptional TERT patterns are identical to those in humans, most patterns are species-specific. To date, mRNA splice variants different from hTERT were described for chickens [34], rats and mice [48], primates [39], worms [49], and dogs [50], among other species.
2.2. Regulation of hTERT Pre-mRNA AS
Similar to most AS, hTERT pre-mRNA is processed by the major splicing machinery, the spliceosome [51], which is regulated by both intronic/exonic elements (cis-elements) close to splice sites and long-range interactions [52] (as illustrated in Figure 3). The most extensively studied are the events that switch hTERT splicing patterns between the two most abundant variants, which are the full-length and β- variants.
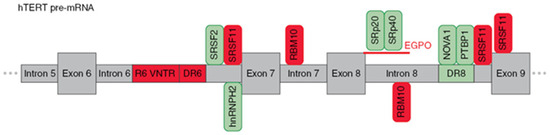
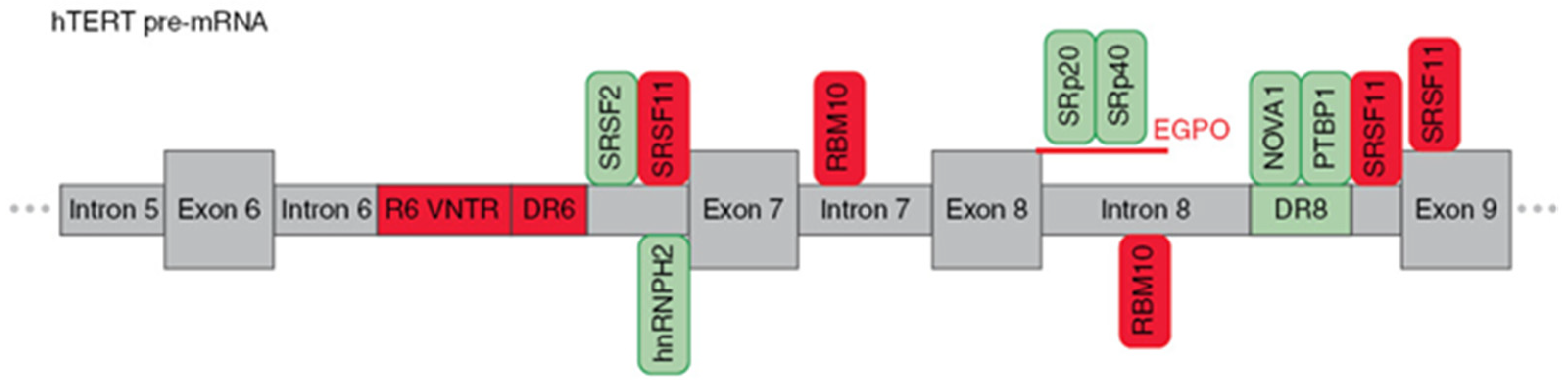
Figure 3.
Schematic presentation of cis-elements and trans-factors involved in the regulation of β- alternative splicing of hTERT pre-mRNA. Entire region from intron 5 to exon 9 of hTERT pre-mRNA is shown. Inclusion enhancers of exons 7 and 8 are shown in green, while repressors are shown in red. EndoG-produced oligonucleotides that block binding of two SR proteins are shown in red.
Wong et al. [52], using hTERT minigene constructs, found three regulatory sequences that are responsible for the formation of the β- variant. First, a block of 26 short repeats of 38-nt sequences located in intron 6 (termed variable number tandem repeat (VNTR) or block 6 (B6) repeats). Second, a direct repeat within intron 6 (DR6), which consists of 256 nt. Third, a direct repeat within intron 8 (DR6), which consists of 285 nt. These sequences demonstrated 85% homology among species, and the number of B6 repeats varied from 18–38 repeats among different individuals. Later, it was shown that B6 is required for β- splicing, but DR6 and DR8 are not sufficient to skip exons 7 and 8. Moreover, the intronic location of these elements rather than their sequences could determine AS [53]. Several other VNTR sites and DR6- and DR8-related sequences (termed Alu elements) were found in hTERT introns, which provide a provision for another hTERT AS.
The following studies identified a number of trans-factors and their cis-sequences that determine the choice of splice variants. It was demonstrated that β- splicing is controlled by SR proteins [37]. SRSF11 promoted the deletion of exons 7 and 8, while hnRNPH2 and hnRNPL were responsible for the formation of the full-length variants. RNA secondary structures may sterically provide occlusion, exposure, or approximation of cis-elements [54], which can enhance the formation of β- variants. By modeling the hTERT mRNA secondary structure, Wong M.S. et al. [55] demonstrated the possibility of approaching exon 6 5′ and exon 9 3′ splice sites, thereby promoting exon skipping. The results suggested that a minimum of nine 38-bp repeats is necessary for RNA:RNA pairing in hTERT pre-mRNA to change the proximity of exon 6 and 9 splice junctions and/or expose the necessary docking sites for SR proteins or the spliceosome for splice site selection.
The search of binding sites for SR proteins identified 2 sites in introns 6 and 8, sites upstream of, and in, exon 9 that matched the consensus sequences (AAGAA, AAUAA or AACAA) [56]. Interestingly, some hnRNPH2-binding sites overlapped with SRSF11-binding sites. As inclusion or exclusion of β- depends on whether exon 6 is joined to exon 7 or exon 9, respectively, use of the 3′ splice site of either intron 6 or 8 is central to this AS decision. Together, these data suggest that SRSF11 and hnRNPH2 compete for binding to these sites to stimulate either β- site exclusion or inclusion. Additionally, SRSF2 binding motifs located in the 3′ end of intron 6 were shown to regulate β- deletion [57].
Xiao et al. [58] proposed an unveiled function of RBM10 that regulates hTERT splicing by binding to the GGU motif of pancreatic cancer cells to repress the production of the full-length hTERT. Loss of RBM10 promotes cell proliferation, invasion, and xenograft growth. The GGU motif was reported to be a consensus RBM10 binding site, and in vivo RNA immunoprecipitation assays confirmed that RBM10 was recruited to the sites next to the 5′ splice sites of hTERT introns 7 and 8. Site-specific mutagenesis (GGU to GAU) within the RBM10 binding sites repressed the RBM10-mediated skipping of exons 7 and 8.
In the series of studies by Ludlow et al. [59,60], a novel potent regulator neuro-oncological ventral antigen 1 (NOVA1) was identified by bioinformatic approaches, and its ability to inhibit the deletion of exons 7 and 8 was confirmed in knockout experiments. Active site (YCAY, where Y = C; or U) x7 for NOVA1 is located within the previously mentioned DR8 region, and binding to NOVA1 results in its function as a splicing factor. Another possible mechanism is that NOVA1 may regulate the upstream transcription factors of hTERT cells and promote the full-length expression of hTERT [61]. Subsequent experiments demonstrated that NOVA1 binds to pre-mRNA in tandem with polypyrimidine tract binding protein 1 (PTBP1) [60]. The knockdown of PTBP1 in cancer cells resulted in the downregulation of the full-length variant and a reduction of telomerase activity in lung cancer cells.
The study of the role of the apoptotic endonuclease EndoG in the regulation of β- AS in hTERT revealed two novel active sites located at the 5′ end of intron 8, which are sensitive to SpR20 and SPr40 splicing factors, and their activity can be modulated by specific splice-switching oligonucleotides [62]. The mechanism by which EndoG modulates AS of hTERT pre-mRNA is thoroughly reviewed in the next section.
Several other factors can also influence hTERT splicing, but the mechanisms are still far from being understood. Depletion of the chromatin remodeling protein Brm in NCI-H1299 cells led to a decrease in hTERT α+β+ and β- variants. The proposed mechanism relies on the observation that Brm and the splicing factors PSF and p54/(nrb)/NONO can bind to the hTERT gene close to exon 7, indicating the possibility of cotranscriptional splicing [63]. TGF-β1 could downregulate c-MYC and subsequently decrease the expression of the full-length hTERT in human skin keratinocytes by retaining high levels of the inactive β- variant. This result suggests a novel mechanism for TGF-β1-mediated regulation of telomerase [64]. Moreover, several potential cis-elements were predicted in exon 5 to exon 9 of hTERT by bioinformatic analysis [65]. The identification of these potential exonic splicing elements of hTERT might be helpful for the design of antisense oligonucleotides, which could modulate AS of hTERT pre-mRNA and consequently biological properties of hTERT protein.
Although the previously mentioned studies shed light on the regulation of β- AS, studies describing the mechanisms of induction of other splice variants are lacking.
2.3. Modulation of hTERT Pre-mRNA AS by Endonuclease G
The first proposal that apoptotic endonuclease G (EndoG) may be involved in the regulation of telomerase activity and the induction of cell senescence came from the observation that umbilical vein endothelial cells stained positive for senescence-associated beta-galactosidase after knocking down EndoG [66]. Later, a strong correlation between the expression of EndoG and hTERT splice variants (the full-length α+β+ and truncated α+β−) was found in human activated T-, B-, or NK-lymphocytes [67] and different colon cancer cell lines [40]. EndoG is a member of the conserved DNA/RNA nuclease family and is highly specific for (dG)n.(dC)n sequences [68]. It is translated as a ∼33-kDa preprotein and is cleaved to a ∼28-kDa protein upon forming an active homodimer nuclease and translocation into the mitochondria. At the latest stages of apoptosis development, EndoG is translocated into the nucleus upon apoptotic stimuli and cleaves chromatin into nucleosomal fragments that are not dependent on caspases [69]. Until now, EndoG was considered a strong pro-apoptotic enzyme that can induce cell death within 24 h after induction in cells [70,71]; however, few EndoG molecules are present in the cell nucleus under normal conditions [71,72]. In cаspаsе-indеpеndеnt аpoptosis, Hsc-70 intеrаcting protеin (CHIP), а rеgulаtor of ЕndoG еxprеssion, functions аs а protеctivе mеchаnism аgаinst oxidаtivе strеss. Undеr normаl conditions, ЕndoG rеmаins bound to Hsp70 аnd CHIP; howеvеr, whеn undеrgoing oxidаtivе strеss, ЕndoG dissociаtеs from Hsp70 аnd CHIP аnd dеgrаdеs DNА to еffеct аpoptosis [73]. In еpithеliаl cеlls, thе nuclеаr locаlizаtion of ЕndoG lеаds thеm to sеnеscеncе [66]. In аddition to DNА dеgrаdаtion, ЕndoG аlso stimulаtеs inhibitors of аpoptosis protеins (IАPs) to tаrgеt protеins for protеаsomаl dеgrаdаtion [74].
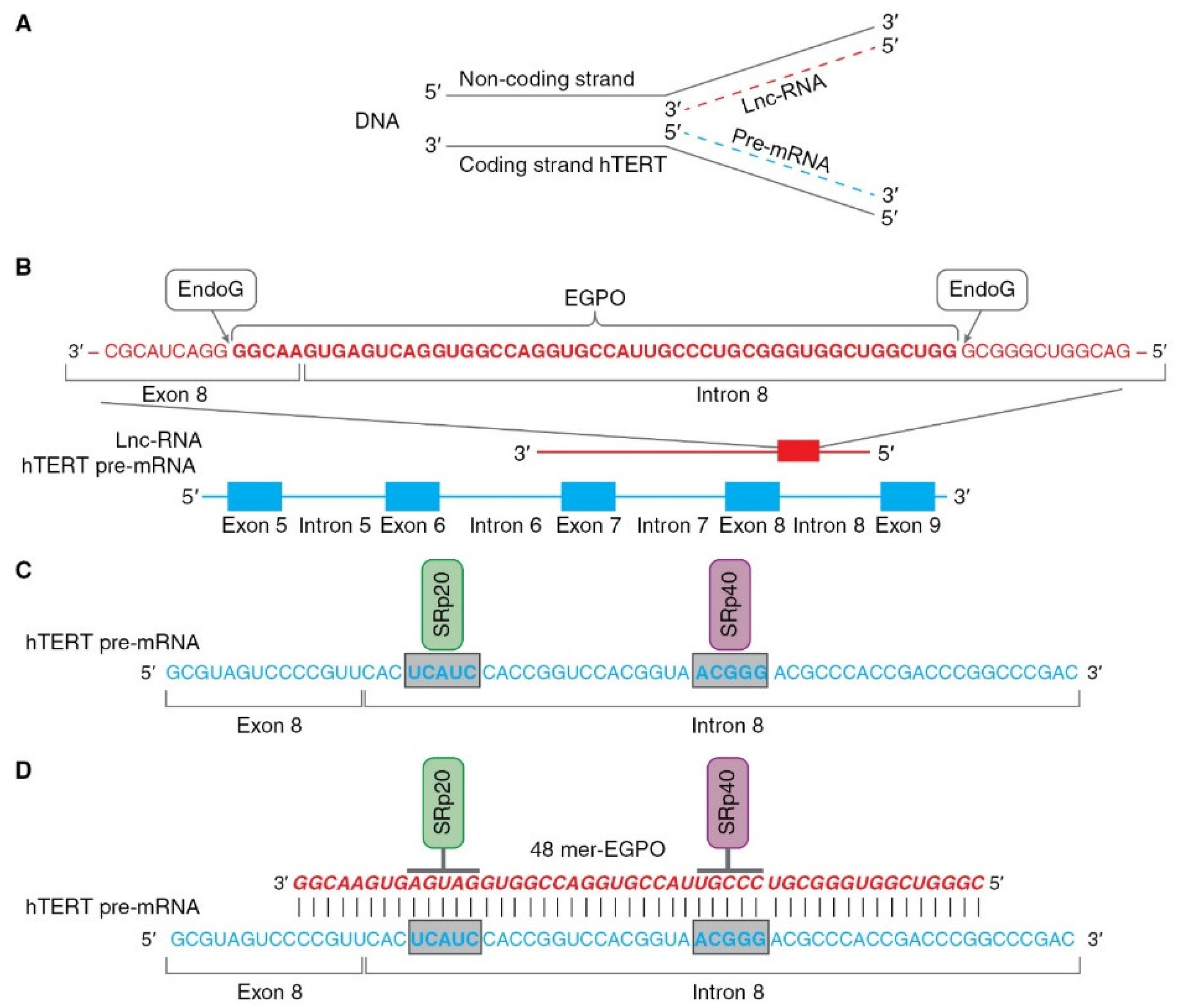
The role of EndoG in the regulation of AS was studied and relies on two properties of the enzyme: first, its RNase activity and second, its ability to translocate from mitochondria to the nucleus. According to a series of experiments [43,62], this mechanism can be hypothetically described as follows (as illustrated in Figure 4). Pre-mRNA hTERT is transcribed from the coding strand of the hTERT gene, while long noncoding RNA (lnc-RNA) is transcribed from the noncoding strand of the same gene (as illustrated in Figure 4A).
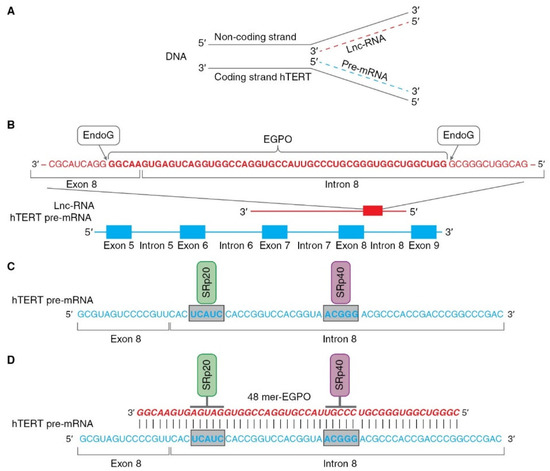
Figure 4.
Schematic presentation of mechanism of hTERT pre-mRNA splicing induced by EndoG. (A) Hypothetical locations for synthesis of hTERT pre-mRNA and lnc-RNA. Pre-mRNA hTERT (blue dotted line) is synthesized from coding strand of hTERT gene, while lnc-RNA (red dotted line) is synthesized from the noncoding DNA strand. (B) Hypothetical schematic locations for lncRNAs and EndoG-produced oligonucleotides (EGPOs) during regulation of hTERT pre-mRNA AS by EndoG. EndoG (white box) cleaves EGPO (red bold font) from lnc-RNA (red font), which is complementary to hTERT pre-mRNA. (C) Binding sites (blue bold font in gray boxes) for the SRp20 (green box) and SRp40 (purple box) splicing regulatory proteins are located in intron 8. (D) Interaction between EGPO and hTERT pre-mRNA prevents binding of SRp20 and SRp40 to hTERT pre-mRNA, which results in induction of AS and expression of truncated β- splice variant.
In the nucleus, EndoG digests lncRNAs on G-rich islands and produces 48-mer RNA oligonucleotides (EndoG-produced oligonucleotides, EGPOs) that pair with the junction site of exon 8 and intron 8 of hTERT pre-mRNA, and can induce AS in both living cells and naked cell nuclei (as illustrated in Figure 4B). G-rich islands (GGGG at the 3′ end and GGGCGGG at the 5′ end) are the sites of EndoG action. EGPO covers two regulatory sites that are located in intron 8 of hTERT pre-mRNA, UCAUC, and ACGGG, which are binding sites for SRp20 and SRp40 splicing regulator proteins (as illustrated in Figure 4C), respectively [75]. Base pairing of specific oligonucleotides with pre-mRNA can block the binding of SRp20 and SRp40 proteins to their sites and affect spliceosome activity, which results in the induction of AS (as illustrated in Figure 4D). The authors supposed that lnc-RNA is transcribed in cells at a constant level, but the amount of EGPO and the grade of hTERT AS are regulated by EndoG activity and the fact that the translocation of EndoG into the cell nucleus triggers AS. Another result of studies on the identification of EGPO is that this oligonucleotide acts as a splice-switching oligonucleotide [76], and the functioning of EndoG is the first observation of natural oligonucleotides that can modulate AS.
The described mechanism is not completely understood and requires further investigation. The most obvious issues are how lncRNA is transcribed from the noncoding strand of the hTERT gene; why EndoG creates an EGPO of this size as soon as other G-rich islands are present in lnc-RNA; why EGPO complements the junction site of exon 8 - intron 8 of hTERT pre-mRNA, and what other proteins are involved in this process. However, several facts favor the involvement of EndoG in this process. First, the suppression of EndoG expression by short interfering RNA abolished hTERT AS, and all hTERT presented was the full-length form [72]. Second, EndoG and its translocation into the nucleus can be induced by different DNA-damaging agents. Cisplatin and other genotoxic agents demonstrated the ability to induce hTERT AS [77]. Third, human cells display this mechanism, and the ability of EndoG to induce AS of TERT pre-mRNA was shown in rat and mouse cell lymphocytes [78]. Fourth, transfection of cells with EGPO can induce hTERT AS [62]. Fifth, EndoG is able to induce AS of the pre-mRNAs of other genes, the most studied of which are deoxyribonuclease 1 (DNase I) [79], caspase-2 (Casp-2), and B-cell lymphoma X (BCL-x) [80].
3. The Involvement of hTERT AS in Physiological Processes
As proposed, splice variants in the forms of RNA, mRNA, or proteins have physiological roles as dominant-negative patterns, which suppress the translation of the full-length hTERT or repress the catalytically active form of the protein. One dominant-negative form of hTERT was proposed to promote the degradation of the full-length hTERT by exporting itself out of nuclei with the full-length hTERT (as a heterodimeric complex), where hTERT becomes available for ubiquitination and for subsequent proteolysis in MCF-7 cells [81]. An alternative of hTERT being ubiquitinated and degraded is that it can enter mitochondria due to the presence of MLS.
The association between the expression of hTERT splice variants, telomerase activity, and telomere length was well studied in the development of human embryos and embryonic stem cells (ESCs). For actively proliferating cells such as human and mouse ESCs, maintaining telomere homeostasis is critical for the functionality and longevity of the cells. Several reviews [82,83,84] highlighted different mechanisms by which telomeres and TERT are affected by pluripotency. Telomere lengthening via telomerase is activated as early as the blastocyst stage of embryo development, and high telomerase expression becomes absent in most adult somatic cells, with the exception of the germ line [46], endothelial cells [5], and activated lymphocytes [85]. To date, it is difficult to say what exactly are the cause and consequence of the described observation. It is not clear whether pluripotency affects telomerase or whether a decrease in telomerase activity contributes to cell differentiation. Different splice variants of hTERT can regulate the grade of telomerase activity during human embryogenesis in a tissue-specific manner, which provides evidence that AS plays a nonrandom role in tissue development [86,87]. Using specimens up to 21 weeks of gestation, telomerase activity was observed through 21 weeks in the fetal liver but was repressed by week 17 in the kidney and by week 13 in the heart. The repression of telomerase activity was associated with increasing amounts of α- and β- splice variants, leading to the shortening of telomeres in these tissues. It was shown that the fetal heart expresses only the full-length variant and the β- transcript. By the 13th week of heart development, the expression of hTERT was suppressed. The fetal liver displays the greatest range of hTERT alternate splicing. Transcripts encoding the full-length, α-, β-, and INS3 variants are expressed at all times of gestation. The fetal kidney expresses the full-length, α-, and β- and variants early in gestation. The α- splice variant is repressed by week 11, leaving only the full-length transcript and the β- splice transcript. By the 18th week, only the β- transcript was expressed [86,87].
Radan et al. [88] demonstrated that hTERT splice variants are responsible for the in vitro differentiation of hESCs under varying conditions of oxygen tension. The results showed increased cell survival and maintenance of the undifferentiated state with elevated levels of nuclear hTERT under conditions of low (2%) O2. The full-length TERT, as well as α-, β-, α-β- hTERT variants, were expressed in cells under both high- (20%) and low-O2 conditions. However, the full-length hTERT and the α- transcripts were significantly reduced in the low-O2 condition compared with that of the high-O2 condition. Pharmacological inhibition of telomerase activity using a synthetic tea catechin resulted in spontaneous hESC differentiation, while telomerase inhibition with a phosphorothioate oligonucleotide that mimics telomeres did not. Steric blockade of α- and β- splice variants using morpholino oligonucleotides rapidly induced spontaneous hESC differentiation that appeared biased toward endomesodermal and neuroectodermal cell fates, respectively [88]. Additionally, regulation of hTERT splicing patterns is involved in the development of cell resistance to hypoxia [89]. In accordance with this observation, hypoxia enhances the transcriptional activity of hTERT through the activation of HIF1α [90]. Taken together, these data suggest that the preferential shift of the TERT splicing pattern is a highly regulated process that can respond to extracellular stimuli that ultimately affect stem cell identity.
3.1. The Involvement of hTERT AS in the Functioning of Immune Cells
Unlike most somatic cell types, lymphocytes are capable of reactivating telomerase expression at the time of activation. This was shown in human and mouse T and B cells in response to specific (antigen) or nonspecific (mitogenic anti-CD3 antibodies, lectins) stimulation [91,92,93]. Elevated telomerase activity may play a role in supporting the survival, proliferation, and expansion of antigen responses in vivo; however, lymphocytes have a limited lifespan, and endogenous telomerase activity levels are not sufficient to completely block telomere loss. Freshly isolated resting human T cells express hTERT mRNA, but little or no telomerase activity was detected. Explanations for this discordance are a requirement for post-translational modifications of hTERT for telomerase activity [94] or the presence of splice variants. While a dramatic increase in telomerase activity occurs upon primary stimulation, subsequent stimulation cycles induce less TERT expression, which eventually becomes nearly undetectable as the cells progress toward senescence. Patric et al. [95] demonstrated that hTERT expression is tightly regulated during the differentiation of CD4+ and CD8+ T cells from naïve to memory cells. Naïve T cells from healthy donors have predominately the full-length and β- hTERT splice variants, and the amount of the full-length variant decreases with differentiation. The authors believe that, as T cell differentiation leads to clonal expansion of selected T cells, limiting telomerase expression in these cells could also be a protective measure for limiting uncontrolled or unwanted T cell clonal expansion that takes away space and resources from other T cells, which will be detrimental to the host.
It was also shown that during long-term culture, human T cells proliferate for a restricted number of cell divisions, after which the cells stop proliferating and demonstrate a senescent phenotype [96]. Thus, the proliferation of immune cells can be regulated by the grade of telomerase activity. To study the role of endogenous hTERT in the lifespan of human CD4+ and CD8+ T cells, endogenous hTERT expression was determined by ectopic expression of dominant-negative (DN) hTERT, which was generated by substitution of the aspartic acid residues at positions 868 and 869 with alanine residues (Asp868Ala, Asp869Ala) [93]. T cells expressing DN-hTERT had a decreased lifespan and showed cytogenetic abnormalities, including chromosome ends without detectable telomeric DNA and chromosome fusions. These results indicate that while endogenous hTERT cannot prevent overall telomere shortening, it has a major influence on the longevity of human T cells.
The ability of the β- hTERT splice variant to affect telomerase activity and proliferation was demonstrated using splice-switching oligonucleotides (SSOs) that pair with hTERT pre-mRNA and can block the binding of the splicing regulatory protein SRp20 or SRp40 to their cis-sites [62]. One important observation of this study is that both SRp20 and SRp40 and their cis-elements on intron 8 of hTERT pre-mRNA are responsible for the modulation of AS. Inhibition of each single protein by specific SSO did not lead to complete conversion of the entire hTERT mRNA pool to a spliced β- form or inhibition of telomerase. Only SSO, which blocked the interaction of both SRp20 and SRp40 proteins with pre-mRNA, demonstrated complete AS-modulating activity. The second observation is that cultivation of CD4+ T lymphocytes with spliced hTERT and inhibited telomerase resulted in a reduction of proliferative activity without significant induction of cell death after short-term cultivation. These results suggest that the full-length hTERT can modulate the proliferation of lymphocytes in a telomere-shortening independent manner and that the induction of AS affects proliferation. The ability of hTERT to regulate the expression of the cell cycle genes Mac-2BP, VEGF, and cyclin D1 was observed in several studies [97,98]. Additionally, global expression profiling experiments showed that 284 genes associated with differentiation, cell cycle regulation, metabolism, and apoptosis were altered by hTERT overexpression [99]. The detailed mechanisms by which hTERT participates in gene expression remain to be studied. Several hypotheses were proposed. One theory suggests that telomerase may be involved in epigenetic modifications of chromatin structure, which indirectly affects gene expression. In support of this hypothesis, it was observed that overexpression of hTERT upregulated and stabilized DNA 5-methylcytosine transferase I [100], and the suppression of hTERT expression altered chromatin configuration [101]. Another possible mechanism is that hTERT may interact with transcription factors or chromatin-modifying factors that directly regulate certain gene transcription programs. In support of this hypothesis, it was shown that hTERT interacts with NF-kB, p65, and b-catenin and mediates the transcriptional regulation of target gene expression [102]. Different splice variants of hTERT may have distinct effects on these processes.
Although active telomerase and the full-length hTERT contribute to lymphocyte proliferation, they play major roles in the functioning of regulatory T cells (Tregs). Tregs counterbalance effector lymphocytes and play an important role in the maintenance of immune tolerance. Tregs may affect both the proliferation and function of activated lymphocytes by contact-dependent and contact-independent mechanisms [103].
TERC−/− mice with shorter hematopoietic stem and progenitor cell telomere lengths have a severe reduction of peripheral Treg numbers in vivo and impaired function in vitro [104]. This study also showed that oxidative stress impairs T cell proliferation via suppression of hTERT. Although the induction of AS in hTERT mRNA was not studied, the results are in agreement with the ability of EndoG to induce AS in response to DNA damage.
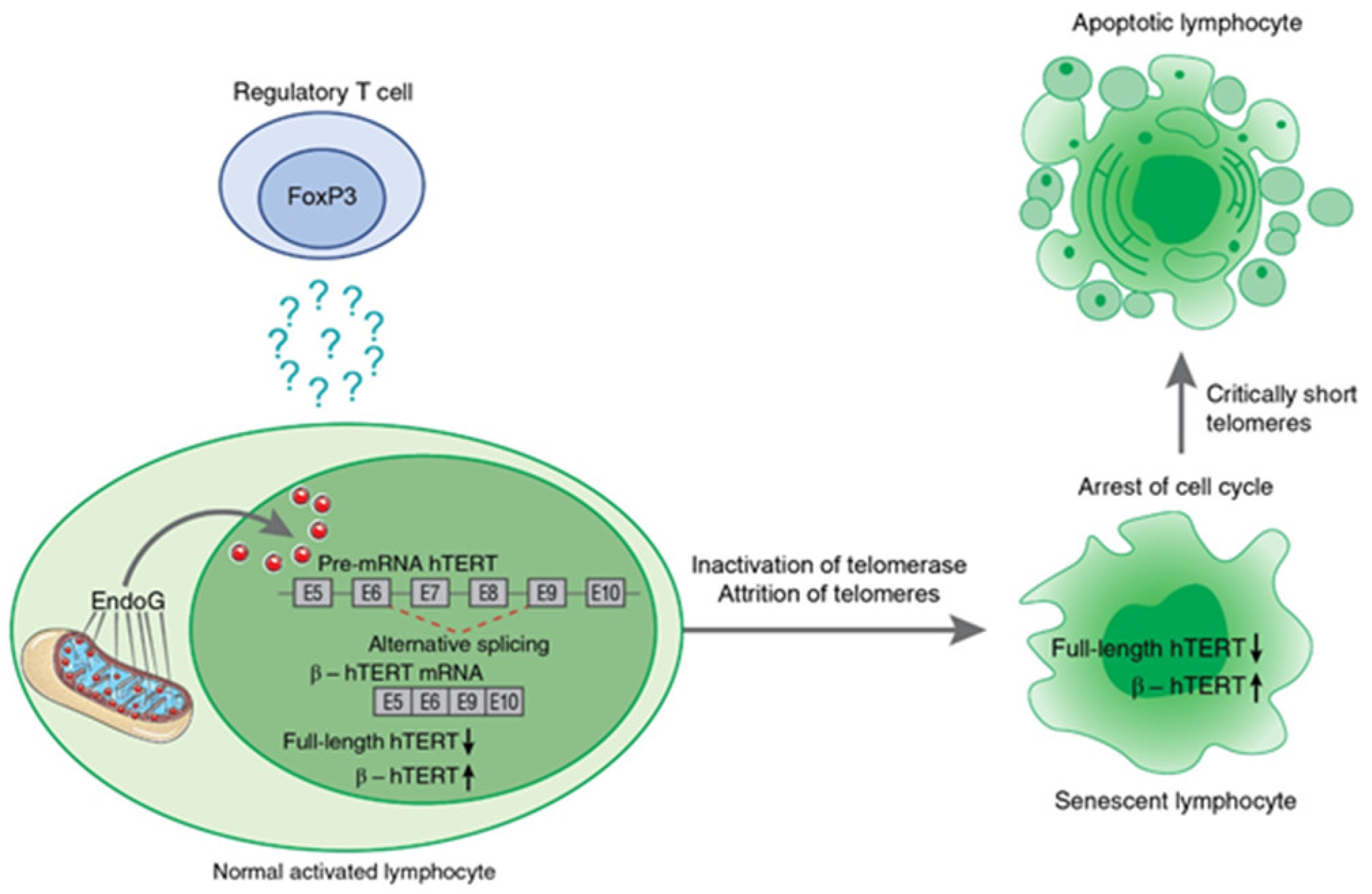
Ye et al. [105] demonstrated that the contact-dependent suppressive activity of Tregs induces cellular senescence in naïve effector T cells in vitro and in vivo, indicating the involvement of telomerase in this process. Later, the mechanism was investigated, and the induction of TERT pre-mRNA AS appeared to be one of the contact-independent mechanisms for the inhibition of proliferation of target cells by human and murine Tregs. Two studies [106,107] demonstrated the ability of Tregs to induce EndoG (and its translocation into nucleus) and consequently AS of TERT pre-mRNA and the suppression of telomerase activity in effector cells in in vitro cocultivation experiments and in in vivo mouse studies. The induced β- splice variant resulted in the inhibition of telomerase in target cells cocultivated with Tregs for a long period of time and led to a decrease in their telomere lengths, cell cycle arrest, conversion of the target cells to replicative senescence, and apoptotic death (as illustrated in Figure 5). Although the mechanism of EndoG-modulated inhibition of telomerase through AS of Tert mRNA was described above, the underlying mechanisms of Treg-mediated EndoG activation in responder cells remain unclear.
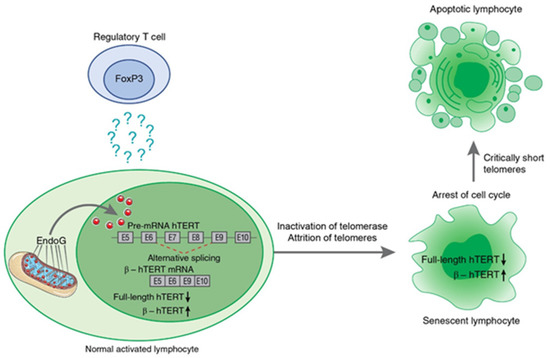
Figure 5.
Schematic presentation of Treg-induced mechanism of hTERT pre-mRNA alternative splicing during suppression of target lymphocyte proliferation. Treg cells induce EndoG expression and translocation from mitochondria to nucleus in activated lymphocytes by a contact-independent mechanism. The factor that triggers this process is not yet determined. From its location in nucleus of target lymphocytes, EndoG induces β- splicing, resulting in inhibition of telomerase. Prolonged cocultivation with Tregs leads to telomere attrition, cell cycle arrest, conversion of the target cells to replicative senescence, and apoptotic death.
These studies lead to a significant observation. First, whether the induction of TERT pre-mRNA AS by EndoG was studied previously in artificial experiments, i.e., transfection of cells with EndoG genes [42,104] or cell treatment with DNA-damaging agents, these studies demonstrated that this was a natural mechanism used in cells. Second, AS of TERT pre-mRNA is involved in regulation of the immune response by Tregs.
Unfortunately, the soluble factor that is released by Tregs and induces EndoG expression is still not determined. Several factors emitted by Tregs and their cellular pathways may potentially be recruited. There is increasing evidence that Treg-produced IFNγ plays a major role in the suppression of responder cells [108]. IFNγ induces oxidative stress and DNA damage [109], consequently resulting in EndoG translocation from mitochondria into the cell nucleus [110]. Moreover, IFNγ induces upregulation and activation of caspase 3 [111], which enhances Bid-induced EndoG release from mitochondria [112]. Caspase 3 can also control EndoG release through the BNIP-3 signaling pathway [113]. Anti-inflammatory cytokines are produced by Tregs, which are critical for some of their suppressive functions [114]. Among them, IL-10 was shown to reduce the production of the proinflammatory cytokine IL-6 [115,116], which is a regulator of STAT3-mediated EndoG activation [117].
Thus, one critical molecular process that is involved in Treg-induced senescence and apoptosis of responder cells relies on AS of TERT pre-mRNA.
3.2. Alternative Functions of hTERT Splice-Variants
Telomerase was first recognized for its telomere-lengthening activity. However, it is now well established that hTERT participates in a variety of biological pathways independent of its telomere-lengthening function. Many excellent reviews were published on the extratelomeric functions of TERT without any attention to splice variants [118,119,120]. The key noncanonical functions of hTERT are summarized in Table 1.

Table 1.
Key non-canonical functions of hTERT.
Although much was learned regarding the novel telomere-unrelated roles of hTERT, the presence of splice variants may bring discrepancies in the obtained results and generate different interpretations. Whereas the canonical functions of hTERT are dedicated mostly to the activity of telomerase, several alternative functions of splice variants were established. Variant ∆4–13 encodes a truncated hTERT protein that interacts with the WNT/beta-catenin pathway and stimulates the proliferation of telomerase-positive and telomerase-negative cells in culture without increasing telomerase activity [34]. The opposite effect has been shown for the β- splice variant: its induction with splice-switching oligonucleotides significantly reduced the proliferation intensity of human CD4+ T lymphocytes. Listerman et al. [37] revealed that the β- hTERT splice variant can confer a growth advantage to cancer cells independent of telomere maintenance, suggesting that hTERT makes multiple contributions to cancer pathophysiology. It was shown that overexpressed β- protein localizes in the nucleus and mitochondria and protects breast cancer cells from cisplatin-induced apoptosis. However, the molecular mechanism by which hTERT variants protect against apoptosis is unknown. Since it was shown that the β- variant, along with the full-length hTERT, localizes to the mitochondria [37,72], one possibility is that these proteins interact with the apoptotic pathway of the mitochondria. The observation that hTERT enhances genomic stability and DNA repair [137] supports this proposal. Another mechanism of how hTERT functions comes from the observation that hTERT can interact with antiapoptotic proteins MCL-1 or BCL-xL in a BH3-dependent or independent manner [21], and the skipping of the BH3-containing TEN domain in the ∆2 variant must abolish such an activity.
The mitochondrial localization of hTERT also represents the noncanonical activity of the enzyme that supports cell resistance to apoptosis. It was demonstrated that hTERT is extracted along with the mtDNA-binding proteins TFAM, HSP60, and TIM23, but not with TOM20 [138]. Increased hTERT content in mitochondria subjected to oxidative stress results in the stabilization of mtDNA and stimulates mitochondrial function. This suppresses the formation of reactive oxygen species and increases the potential of the mitochondrial membrane [139]. However, the mechanism by which hTERT translocates from the nucleus to mitochondria is still poorly understood. The induction of the β- splice variant may shed light on this process. Src kinase has been shown to regulate the export of hTERT from the nucleus to the cytoplasm and mitochondria under oxidative stress by phosphorylating tyrosine 707 [133,139], which is responsible for protein translocation out of nuclei. Tyr770 is located within the RT domain of hTERT, which is skipped in the β- variant. This may explain why accumulation of the β- variant is observed in the nucleus under oxidative stress. Such an observation is also in accordance with the ability of EndoG to induce AS in hTERT [43], since it is known that EndoG can be induced by oxidative stress [140].
Thus, the accumulated evidence indicates that hTERT plays important functions far from telomere maintenance, and different splice variants alongside the cell specificity of hTERT AS may have distinct effects on the described processes.
4. The Involvement of hTERT AS in Pathological Processes
The role of hTERT splice variants in the regulation of pathological processes is far from clear since most studies are descriptive and rely only on the observation of the expression levels of hTERT splice variants in different diseases, predominately cancers. General observation of both tumor tissue and the corresponding cell line revealed predominant expression of the full-length α+β+γ+ and beta-deletion α+β-γ+ variants in most cases (as illustrated in Table 2). However, the analysis of RNA-sequencing data from The Cancer Genome Atlas (TCGA) splice variant database (TSVdb) revealed a very heterogeneous expression profile of hTERT transcripts among 31 cancer types, and tissue with a predominant expression of α-β-γ– was described [141]. The full-length and β- hTERT variants comprise the major part of total hTERT, and the level of the full-length variant is the prognostic marker connected with a poor clinical outcome, at least for several cancer types [142,143,144]. Tumors that show no or low telomerase activity express truncated splice variants, while tumors with high activity always express significant amounts of the full-length hTERT.

Table 2.
Features of hTERT splice variants in tumor tissues or cell lines.
The correlation between hTERT splice variant mutational status of the immunoglobulin heavy chain variable (IGHV) gene was described for chronic lymphocytic leukemia. Palma et al. [164] demonstrated that higher levels of the full-length and α- variants were observed in peripheral blood mononuclear cells from patients having unmutated IGHV in comparison to those with mutations, whereas the expression of the β- variant was not different. All transcripts were more frequently expressed in progressive than in nonprogressive patients. These data are in accordance with the results obtained in B-cell lymphocytic leukemia, where it was shown that the expression level of the full-length hTERT is strongly correlated with the percentage of mutations in immunoglobulin V and was significantly higher in individuals without mutations [165].
Myelodysplastic syndrome (MDS) is a clonal hematopoietic stem cell disorder characterized by ineffective hematopoiesis and peripheral cytopenia. Dong et al. [166] demonstrated that peripheral blood mononuclear cells from MDS patients have significantly shorter telomeres with elevated telomerase activity than those from normal individuals. A high level of telomerase activity was associated with high expression of total hTERT and its full-length and β- variants. The ratio of α+β− transcripts to the full-length transcripts was significantly increased in some cases. These results correspond to the observations in which increased hTERT expression and telomerase activity were detected in bone marrow in MDS patients [167], and the inducible (stimulated with CD3 and CD28) hTERT and telomerase activity in T cells of MDS were decreased compared to that of the control [168], which suggested that there are basal and inducible telomerase and hTERT splice-variant abnormalities in MDS patients. Both overall hTERT and α+β− splice variants in mononuclear cells are promising biomarkers for MDS [169].
Systemic lupus erythematosus (SLE) is an autoimmune disease in which the immune system attacks its own tissues, causing widespread inflammation and tissue damage in the affected organs. Attar A. et al. [170] demonstrated the existence of potential differences between SLE patients at different disease stages and healthy individuals regarding telomerase activity and Bcl-2 expression in T- or B-lymphocytes in the absence of obvious correlations with specific hTERT splice variants. Telomerase activity was detected in T cells but not in B cells from SLE patients who received therapy. Conversely, telomerase activity was detected in B cells but not in T cells from newly diagnosed patients. The expression of hTERT splice variants was only observed in the samples that showed telomerase activity. In B cells, the dominant variants were α+β− and α-β−, whereas in T cells, they were the full-length transcript and α+β-.
Thus, the biological functions of hTERT alternative splicing in the development of pathologies are far from clear, and the obtained data are rather difficult to evaluate. The significance of hTERT splice variants is obvious for the regulation of telomerase activity, which makes them potential pharmacological targets for the regulation of the proliferative capacity of cells.
5. Pharmacological Regulation of hTERT Pre-mRNA AS
Since functionally active hTERT enables replicative immortality of cancer cells and is silenced in adult somatic cells, it represents a suitable target for the development of highly selective therapeutics. To date, several classes of telomerase inhibitors were evaluated and demonstrated good toxic activity against cancer cells in vitro [171]. However, very few of them reported significant and reliable antitumor activity in clinical trials, as the efficacy of potential drug substances acting as telomerase inhibitors may be impaired for several reasons. For instance, cancer cells treated under persistent telomerase inhibition in vivo usually switch the mechanism of telomere elongation to a telomerase-independent homologous recombination pathway, also called alternative lengthening of telomeres [172]. In addition, they are able to divide and form tumor nodes before their telomeres reach any critical length [173]. It was also demonstrated that cancer cells can restore their telomerase activity and, thus, the length of their telomeres shortly after elimination of an inhibitor from tumor tissues [174]. Nevertheless, in some cases, inhibition of telomerase resulted in cellular senescence or apoptosis in a time-dependent manner that correlated with the initial telomere length. Overall, these data support the persistent search for telomerase inhibitors as suitable candidates to be developed as efficient and reliable drugs for anticancer pharmacological-based therapies. The involvement of hTERT splice variants in several physiological processes and pathological conditions makes the pharmacological modulation of hTERT AS a more promising strategy for precisely manipulating cell fate.
Several small molecule chemical compounds can modulate aberrant splicing in a variety of human RNA diseases [175]. Natural products derived primarily from bacteria and chemical synthesis provided many leads for therapeutic compounds targeting splicing [176]. Small-molecule ligand 12459, a G-quadruplex-interacting agent that belongs to the triazine series, was shown to downregulate telomerase activity in the human A549 lung carcinoma cell line [177]. The downregulation of telomerase activity was caused by an alteration of the hTERT splicing pattern induced by 12459, i.e., an almost complete disappearance of active full-length hTERT and overexpression of the inactive β- variant. The authors explain the mechanism of β- induction by the presence of several tracks of G-rich sequences that can form G-quadruplexes in VNTR regions of intron 6. The binding of 12459 to these structures affects AS toward β- formation. Another attempt to manipulate hTERT AS by affecting G-rich sequences was performed with compound CX-5461, which strongly binds to and stabilizes the G-quadruplex [149]. CX-5461 altered hTERT splicing patterns, leading to an increase in β- and a decrease in the full-length expression, which inhibited telomerase activity. In addition, CX-5461 had cytotoxic effects on different glioblastoma cell lines, caused a telomere DNA damage response, and induced G2/M arrest and apoptosis. The low selectivity of the compounds that target the spliceosome remains the major disadvantage for such a strategy for modulating hTERT AS.
The most attractive approach to date is the development of pharmaceuticals that can selectively switch the full-length active hTERT variants to any inactive variant, inducing a negative impact on cells that can slow or stop cell proliferation. Splice-switching oligonucleotides (SSOs) are short, synthetic, antisense, modified nucleic acids that base-pair with a pre-mRNA and disrupt the normal splicing repertoire of the transcript by blocking the RNA–RNA base-pairing or protein–RNA binding interactions that occur between components of the splicing machinery and the pre-mRNA [178]. Several studies demonstrated that chemically modified highly specific antisense agents, or SSOs, can modulate the induction of the β- hTERT splice variant by binding to sensitive splice-regulating cis-elements in hTERT pre-mRNA. The first demonstration that hTERT pre-mRNA splice switching is possible came from the study by Brambilla et al. [179]. The authors targeted the intron 5/exon 6 boundary site with antisense oligonucleotides in the DU145 prostate cancer cell line. The result was the switch from the full-length hTERT toward the α-β- variant, leading to the reduction of telomerase activity and the induction of telomere-shortening independent apoptosis. The authors also speculate that the reduction of proliferative activity observed in oligomer-treated cells could be based on the latter mechanism and related to telomere destabilization. They suggested that telomeres normally exist in a capped state but may switch to an uncapped state in case of lack of telomerase activity. Ludlow et al. [59]. used SSO to block the activity of cis-regulating elements within the DR8 sequence in intron 8, which is a binding site for NOVA1 splicing-regulatory proteins. In a nonsmall cell cancer line, this resulted in the shift of the pull of hTERT transcripts from the full-length to a β- pattern. Zhdanov et al. [62] used a set of SSOs targeting exon 8/intron 8 boundaries within hTERT pre-mRNA to block the active sites for SRp20 or SRp40 regulatory splicing proteins. SSOs that blocked the binding of only one splicing regulatory protein demonstrated rather moderate capacities to induce β- variants to inhibit telomerase. However, an SSO that blocked the interaction of both SRp20 and SRp40 proteins was the most active. Cultivation of human activated CD4+ T lymphocytes with spliced hTERT and inhibited telomerase resulted in a reduction of proliferative activity without significant induction of cell death. Finally, it was shown that oligonucleotide AON-Ex726 anchored to the 3′ end of intron 6 containing cis-site for SRFS2 splicing regulatory proteins can reduce the level of the full-length hTERT and induce the β- variant in various brain cancer cell lines [57]. After transfection with AON-Ex726, the level of apoptosis was increased, while telomerase activity and cell proliferation were significantly decreased.
The advantage of the antisense strategy is the minimization of off-target effects since such oligonucleotides are very specific for hTERT pre-mRNA. However, such a strategy for an induction β- variant may face an opposite effect when cancer cells become more resistant to apoptosis induction [37] or less sensitive to irradiation [180]. The rapidly developing genome editing Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated (CRISPR/Cas) technology was developed to image and manipulate cancer cells depending on telomerase and TERT expression [181,182,183,184]. Although, to date, there are no studies describing attempts to modulate hTERT AS with gene-editing protocols, such an approach alongside different delivery modalities may become a powerful instrument for antitelomerase therapy.
6. Conclusions
Telomerase activity is highly regulated by AS mechanisms of the pre-mRNA of its main catalytic subunit hTERT. Shifts in the pool of splice variants in cells can determine their proliferative capacity. The full-length hTERT is capable of elongating telomeres, whereas truncated splice variants lack such activity. The involvement of hTERT AS was observed in different physiological and pathological processes, such as during the development of human embryo and stem cells or during the development of different types of cancers. Unfortunately, such observations are only descriptive, and the biological significance of alternative splicing hTERT patterns remains to be investigated. However, recent studies demonstrated that inhibition of telomerase activity by the induction of TERT AS is one of the mechanisms by which Tregs can suppress the proliferation of target lymphocytes in humans and mice. Future research should identify novel cellular functions of hTERT splice variants and ways to manipulate them to affect cell behavior.
Author Contributions
Article writing and editing, table, and figure preparation, A.A.P.; conception, design, and revision, D.D.Z. Both authors have read and agreed to the published version of the manuscript.
Funding
The work was done in the framework of the Russian Federation fundamental research program for the long-term period for 2021–2030.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
Authors declare no conflict of interests.
References
- Trybek, T.; Kowalik, A.; Góźdź, S.; Kowalska, A. Telomeres and telomerase in oncogenesis (review). Oncol. Lett. 2020, 20, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, S. Telomerase activity and telomere length in male germ cells. Biol. Reprod. 2015, 92, 53. [Google Scholar] [CrossRef] [PubMed]
- Hiyama, E.; Hiyama, K. Telomere and telomerase in stem cells. Br. J. Cancer 2007, 96, 1020–1024. [Google Scholar] [CrossRef]
- Akbar, A.N.; Vukmanovic-Stejic, M. Telomerase in T lymphocytes: Use it and lose it? J. Immunol. 2007, 178, 6689–6694. [Google Scholar] [CrossRef]
- Jakob, S.; Haendeler, J. Molecular mechanisms involved in endothelial cell aging: Role of telomerase reverse transcriptase. Z. Gerontol. Geriatr. 2007, 40, 334–338. [Google Scholar] [CrossRef]
- Ishaq, A.; Hanson, P.S.; Morris, C.M.; Saretzki, G. Telomerase Activity is Downregulated Early during Human Brain Development. Genes 2016, 7, 27. [Google Scholar] [CrossRef]
- Leão, R.; Apolónio, J.D.; Lee, D.; Figueiredo, A.; Tabori, U.; Castelo-Branco, P. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J. Biomed. Sci. 2018, 25, 22. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Chang, G.; Wang, F.; Wang, F.; Geng, X. Alternative splicing of hTERT pre-mRNA: A potential strategy for the regulation of telomerase activity. Int. J. Mol. Sci. 2017, 18, 567. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Miriami, E.; Sperling, R.; Sperling, J.; Motro, U. Regulation of splicing: The importance of being translatable. RNA 2004, 10, 1. [Google Scholar] [CrossRef][Green Version]
- Jurica, M.S.; Moore, M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 2003, 12, 5–14. [Google Scholar] [CrossRef]
- Savisaar, R.; Hurst, L.D. Exonic splice regulation imposes strong selection at synonymous sites. Genome Res. 2018, 28, 1442–1454. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.; Xie, J. Diverse regulation of 3′ splice site usage. Cell. Mol. Life Sci. 2015, 72, 4771–4793. [Google Scholar] [CrossRef] [PubMed]
- Brogna, S.; Wen, J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef]
- Faustino, N.A.; Cooper, T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003, 17, 419–437. [Google Scholar] [CrossRef]
- McKelvey, B.A.; Gilpatrick, T.; Wang, Y.; Timp, W.; Umbricht, C.B.; Zeiger, M.A. Characterization of Allele-Specific Regulation of Telomerase Reverse Transcriptase in Promoter Mutant Thyroid Cancer Cell Lines. Thyroid 2020, 30, 1470–1481. [Google Scholar] [CrossRef]
- Wyatt, H.D.M.; West, S.C.; Beattie, T.L. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010, 38, 5609–5622. [Google Scholar] [CrossRef]
- Zaug, A.J.; Podell, E.R.; Cech, T.R. Mutation in TERT separates processivity from anchor-site function. Nat. Struct. Mol. Biol. 2008, 15, 870–872. [Google Scholar] [CrossRef]
- Jin, Y.; You, L.; Kim, H.J.; Lee, H.W. Telomerase reverse transcriptase contains a BH3-like motif and interacts with BCL-2 family members. Mol. Cells 2018, 41, 684–694. [Google Scholar] [PubMed]
- Xia, J.; Peng, Y.; Mian, I.S.; Lue, N.F. Identification of Functionally Important Domains in the N-Terminal Region of Telomerase Reverse Transcriptase. Mol. Cell. Biol. 2000, 20, 5196–5207. [Google Scholar] [CrossRef][Green Version]
- Bosoy, D.; Peng, Y.; Mian, I.S.; Lue, N.F. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 2003, 278, 3882–3890. [Google Scholar] [CrossRef]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef]
- Huard, S.; Moriarty, T.J.; Autexier, C. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 2003, 31, 4059–4070. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhao, L.; Yang, Z.; Funder, J.W.; Liu, J.P. Telomerase is controlled by protein kinase Cα in human breast cancer cells. J. Biol. Chem. 1998, 273, 33436–33442. [Google Scholar] [CrossRef] [PubMed]
- Panneer Selvam, S.; De Palma, R.M.; Oaks, J.J.; Oleinik, N.; Peterson, Y.K.; Stahelin, R.V.; Skordalakes, E.; Ponnusamy, S.; Garrett-Mayer, E.; Smith, C.D.; et al. Binding of the sphingolipid S1P to hTERT stabilizes telomerase at the nuclear periphery by allosterically mimicking protein phosphorylation. Sci. Signal. 2015, 8, ra58. [Google Scholar] [CrossRef]
- Jeong, S.A.; Kim, K.; Lee, J.H.; Cha, J.S.; Khadka, P.; Cho, H.S.; Chung, I.K. Akt-mediated phosphorylation increases the binding affinity of hTERT for importin α to promote nuclear translocation. J. Cell Sci. 2015, 128, 2287–2301. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Khadka, P.; Chung, I.K. Nuclear import of htert requires a bipartite nuclear localization signal and Akt-mediated phosphorylation. J. Cell Sci. 2012, 125, 2684–2697. [Google Scholar] [CrossRef]
- Kang, S.S.; Kwon, T.; Kwon, D.Y.; Do, S.I. Akt protein kinase enhances human telomerase activity through phosphorylation of telomerase reverse transcriptase subunit. J. Biol. Chem. 1999, 274, 13085–13090. [Google Scholar] [CrossRef]
- Büchner, N.; Zschauer, T.C.; Lukosz, M.; Altschmied, J.; Haendeler, J. Downregulation of mitochondrial telomerase reverse transcriptase induced by H2O2 is Src kinase dependent. Exp. Gerontol. 2010, 45, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Ludlow, A.T.; Slusher, A.L.; Sayed, M.E. Insights into telomerase/hTERT alternative splicing regulation using bioinformatics and network analysis in cancer. Cancers 2019, 11, 666. [Google Scholar] [CrossRef]
- Zhu, S.; Rousseau, P.; Lauzon, C.; Gandin, V.; Topisirovic, I.; Autexier, C. Inactive C-terminal telomerase reverse transcriptase insertion splicing variants are dominant-negative inhibitors of telomerase. Biochimie 2014, 101, 93–103. [Google Scholar] [CrossRef]
- Hrdlicková, R.; Nehyba, J.; Bose, H.R. Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol. Cell. Biol. 2012, 32, 4283–4296. [Google Scholar] [CrossRef] [PubMed]
- Saebøe-Larssen, S.; Fossberg, E.; Gaudernack, G. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): Analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol. Biol. 2006, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.S.; Wright, W.E.; Shay, J.W. Alternative splicing regulation of telomerase: A new paradigm? Trends Genet 2014, 30, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Listerman, I.; Sun, J.; Gazzaniga, F.S.; Lukas, J.L.; Blackburn, E.H. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 2013, 73, 2817–2828. [Google Scholar] [CrossRef]
- Yi, X.; Shay, J.W.; Wright, W.E. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001, 29, 4818–4825. [Google Scholar] [CrossRef]
- Withers, J.B.; Ashvetiya, T.; Beemon, K.L. Exclusion of Exon 2 Is a Common mRNA Splice Variant of Primate Telomerase Reverse Transcriptases. PLoS ONE 2012, 7, e48016. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Vasina, D.A.; Orlova, V.S.; Gotovtseva, V.Y.; Bibikova, M.V.; Pokrovsky, V.S.; Pokrovskayaa, M.V.; Aleksandrova, S.S.; Sokolov, N.N. Apoptotic endonuclease EndoG induces alternative splicing of telomerase catalytic subunit hTERT and death of tumor cells. Biochem. Suppl. Ser. B Biomed. Chem. 2016, 10, 310–321. [Google Scholar]
- Yi, X.; White, D.M.; Aisner, D.L.; Baur, J.A.; Wright, W.E.; Shay, J.W. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia 2000, 2, 433–440. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Vasina, D.A.; Grachev, V.A.; Orlova, E.V.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Sokolov, N.N. Alternative splicing of telomerase catalytic subunit hTERT generated by apoptotic endonuclease EndoG induces human CD4 + T cell death. Eur. J. Cell Biol. 2017, 96, 653–664. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Vasina, D.A.; Orlova, E.V.; Orlova, V.S.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Sokolov, N.N. Apoptotic endonuclease EndoG regulates alternative splicing of human telomerase catalytic subunit hTERT. Biochem. Suppl. Ser. B Biomed. Chem. 2017, 11, 154–165. [Google Scholar] [CrossRef]
- Hisatomi, H.; Ohyashiki, K.; Ohyashiki, J.H.; Nagao, K.; Kanamaru, T.; Hirata, H.; Hibi, N.; Tsukada, Y. Expression profile of a gamma-deletion variant of the human telomerase reverse transcriptase gene. Neoplasia 2003, 5, 193–197. [Google Scholar] [CrossRef]
- Kilian, A.; Bowtell, D.D.; Abud, H.E.; Hime, G.R.; Venter, D.J.; Keese, P.K.; Duncan, E.L.; Reddel, R.R.; Jefferson, R.A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 1997, 6, 2011–2019. [Google Scholar] [CrossRef]
- Wright, W.E.; Piatyszek, M.A.; Rainey, W.E.; Byrd, W.; Shay, J.W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996, 18, 173–179. [Google Scholar] [CrossRef]
- Fleisig, H.B.; Hukezalie, K.R.; Thompson, C.A.H.; Au-Yeung, T.T.T.; Ludlow, A.T.; Zhao, C.R.; Wong, J.M.Y. Telomerase reverse transcriptase expression protects transformed human cells against DNA-damaging agents, and increases tolerance to chromosomal instability. Oncogene 2016, 35, 218–227. [Google Scholar] [CrossRef]
- Cheng, D.; Zhao, Y.; Zhang, F.; Zhang, J.; Wang, S.; Zhu, J. Engineering a humanized telomerase reverse transcriptase gene in mouse embryonic stem cells. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Tan, T.C.J.; Rahman, R.; Jaber-Hijazi, F.; Felix, D.A.; Chen, C.; Louis, E.J.; Aboobaker, A.; Haber, J.E. Telomere maintenance and telomerase activity are differentially regulated in asexual and sexual worms. Proc. Natl. Acad. Sci. USA 2012, 109, 4209–4214. [Google Scholar] [CrossRef]
- Angelopoulou, K.; Zavlaris, M.; Papaioannou, N.; Vlemmas, I. Canis familiaris telomerase reverse transcriptase undergoes alternative splicing. Mamm. Genome 2008, 19, 647–653. [Google Scholar] [CrossRef]
- Cieply, B.; Carstens, R.P. Functional roles of alternative splicing factors in human disease. Wiley Interdiscip. Rev. RNA 2015, 6, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.S.; Chen, L.; Foster, C.; Kainthla, R.; Shay, J.W.; Wright, W.E. Regulation of telomerase alternative splicing: A target for chemotherapy. Cell Rep. 2013, 3, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- Leem, S.H.; Londoño-Vallejo, J.A.; Kim, J.H.; Bui, H.; Tubacher, E.; Solomon, G.; Park, J.E.; Horikawa, I.; Kouprina, N.; Barrett, J.C.; et al. The human telomerase gene: Complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene 2002, 21, 769–777. [Google Scholar] [CrossRef][Green Version]
- Jin, Y.; Yang, Y.; Zhang, P. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol. 2011, 8, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.S.; Shay, J.W.; Wright, W.E. Regulation of human telomerase splicing by RNA:RNA pairing. Nat. Commun. 2014, 5, 3306. [Google Scholar] [CrossRef]
- Wu, J.Y.; Kar, A.; Kuo, D.; Yu, B.; Havlioglu, N. SRp54 (SFRS11), a Regulator for tau Exon 10 Alternative Splicing Identified by an Expression Cloning Strategy. Mol. Cell. Biol. 2006, 26, 6739–6747. [Google Scholar] [CrossRef]
- Wang, F.; Cheng, Y.; Zhang, C.; Chang, G.; Geng, X. A novel antisense oligonucleotide anchored on the intronic splicing enhancer of hTERT pre-mRNA inhibits telomerase activity and induces apoptosis in glioma cells. J. Neurooncol. 2019, 143, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Chen, X.; Li, X.; Deng, K.; Liu, H.; Ma, J.; Wang, Z.; Hu, Y.; Hou, J. RBM10 regulates human TERT gene splicing and inhibits pancreatic cancer progression. Am. J. Cancer Res. 2021, 11, 157–170. [Google Scholar]
- Ludlow, A.T.; Wong, M.S.; Robin, J.D.; Batten, K.; Yuan, L.; Lai, T.P.; Dahlson, N.; Zhang, L.; Mender, I.; Tedone, E.; et al. NOVA1 regulates hTERT splicing and cell growth in non-small cell lung cancer. Nat. Commun. 2018, 9, 3112. [Google Scholar] [CrossRef]
- Sayed, M.E.; Yuan, L.; Robin, J.D.; Tedone, E.; Batten, K.; Dahlson, N.; Wright, W.E.; Shay, J.W.; Ludlow, A.T. NOVA1 directs PTBP1 to hTERT pre-mRNA and promotes telomerase activity in cancer cells. Oncogene 2019, 38, 2937–2952. [Google Scholar] [CrossRef]
- Villate, O.; Turatsinze, J.V.; Mascali, L.G.; Grieco, F.A.; Nogueira, T.C.; Cunha, D.A.; Nardelli, T.R.; Sammeth, M.; Salunkhe, V.A.; Esguerra, J.L.S.; et al. Nova1 is a master regulator of alternative splicing in pancreatic beta cells. Nucleic Acids Res. 2014, 42, 11818–11830. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Plyasova, A.A.; Gladilina, Y.A.; Pokrovsky, V.S.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Lobaeva, T.A.; et al. Inhibition of telomerase activity by splice-switching oligonucleotides targeting the mRNA of the telomerase catalytic subunit affects proliferation of human CD4+ T lymphocytes. Biochem. Biophys. Res. Commun. 2019, 509, 790–796. [Google Scholar] [CrossRef]
- Ito, T.; Watanabe, H.; Yamamichi, N.; Kondo, S.; Tando, T.; Haraguchi, T.; Mizutani, T.; Sakurai, K.; Fujita, S.; Izumi, T.; et al. Brm transactivates the telomerase reverse transcriptase (TERT) gene and modulates the splicing patterns of its transcripts in concert with p54 nrb. Biochem. J. 2008, 411, 201–209. [Google Scholar] [CrossRef]
- Cerezo, A.; Kalthoff, H.; Schuermann, M.; Schäfer, B.; Boukamp, P. Dual regulation of telomerase activity through c-Myc-dependent inhibition and alternative splicing of hTERT. J. Cell Sci. 2002, 115, 1305–1312. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chang, G.-M.; Geng, X.; Geng, X. Bioinformatics analysis of Exonic Splicing Enhancers (ESEs) for predicting potential regulatory elements of hTERT mRNA Splicing. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 526–536. [Google Scholar]
- Diener, T.; Neuhaus, M.; Koziel, R.; Micutkova, L.; Jansen-Dürr, P. Role of endonuclease G in senescence-associated cell death of human endothelial cells. Exp. Gerontol. 2010, 45, 638–644. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhdanov, D.D.; Gladilina, Y.A.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Grishin, D.V.; Podobed, O.V.; Sokolov, N.N. Induction of Alternative Splicing and Inhibition of Activity of Telomerase Catalytic Subunit by Apoptotic Endonuclease EndoG in Human, T., B, and NK Cells. Bull. Exp. Biol. Med. 2018, 164, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Carrillo, A.; Renaud, J. Endonuclease G: A (dG)n X (dC)n-specific DNase from higher eukaryotes. EMBO J. 1987, 6, 401–407. [Google Scholar] [CrossRef]
- Li, L.Y.; Luo, X.; Wang, X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 2001, 412, 95–99. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Fahmi, T.; Wang, X.; Apostolov, E.O.; Sokolov, N.N.; Javadov, S.; Basnakian, A.G. Regulation of Apoptotic Endonucleases by EndoG. DNA Cell Biol. 2015, 34, 316–326. [Google Scholar] [CrossRef]
- Apostolov, E.O.; Wang, X.; Shah, S.V.; Basnakian, A.G. Role of EndoG in development and cell injury. Cell Death Differ. 2007, 14, 1971–1974. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, D.D.; Pokrovsky, V.S.; Orlova, E.V.; Orlova, V.S.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Sokolov, N.N. Intracellular localization of apoptotic endonuclease EndoG and splice-variants of telomerase catalytic subunit hTERT. Biochemistry 2017, 82, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Seo, T.W.; Yi, J.H.; Shin, K.S.; Yoo, S.J. CHIP has a protective role against oxidative stress-induced cell death through specific regulation of endonuclease G. Cell Death Dis. 2013, 4, e666. [Google Scholar] [CrossRef] [PubMed]
- Seo, T.W.; Lee, J.S.; Yoo, S.J. Cellular inhibitor of apoptosis protein 1 ubiquitinates endonuclease G but does not affect endonuclease G-mediated cell death. Biochem. Biophys. Res. Commun. 2014, 451, 644–649. [Google Scholar] [CrossRef]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef]
- Disterer, P.; Kryczka, A.; Liu, Y.; Badi, Y.E.; Wong, J.J.; Owen, J.S.; Khoo, B. Development of therapeutic splice-switching oligonucleotides. Hum. Gene Ther. 2014, 25, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, D.D.; Vasina, D.A.; Orlova, V.S.; Orlova, E.V.; Grishin, D.V.; Gladilina, Y.A.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Sokolov, N.N. Induction of Apoptotic Endonuclease EndoG with DNA-Damaging Agents Initiates Alternative Splicing of Telomerase Catalytic Subunit hTERT and Inhibition of Telomerase Activity hTERT in Human CD4+ and CD8+T Lymphocytes. Biochem. Suppl. Ser. B Biomed. Chem. 2017, 63, 296–305. [Google Scholar]
- Zhdanov, D.D.; Gladilina, Y.A.; Orlova, V.S.; Grishin, D.V.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Podobed, O.V.; Sokolov, N.N. Induction of Telomerase Catalytic Subunit Alternative Splicing by Apoptotic Endonuclease G in Mouse and Rat Lymphocytes. Cell Tissue Biol. 2018, 12, 104–115. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Gladilina, Y.A.; Pokrovsky, V.S.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Plyasova, A.A.; Sokolov, N.N. Endonuclease G modulates the alternative splicing of deoxyribonuclease 1 mRNA in human CD4+ T lymphocytes and prevents the progression of apoptosis. Biochimie 2019, 157, 158–176. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Gladilina, Y.A.; Grishin, D.V.; Pokrovsky, V.S.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Sokolov, N.N. Apoptotic Endonuclease EndoG Induces Alternative Splicing of Telomerase TERT Catalytic Subunit, Caspase-2, DNase I, and BCL-x in Human, Murine, and Rat CD4+T Lymphocytes. Russ. J. Bioorg. Chem. 2018, 44, 90–103. [Google Scholar] [CrossRef]
- Nguyen, B.N.; Elmore, L.W.; Holt, S.E. Mechanism of dominant-negative telomerase function. Cell Cycle 2009, 8, 3227–3233. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kalmbach, K.; Robinson, L.G.; Wang, F.; Liu, L.; Keefe, D. Telomere length reprogramming in embryos and stem cells. Biomed Res. Int. 2014, 2014, 925121. [Google Scholar] [CrossRef]
- Huang, Y.; Liang, P.; Liu, D.; Huang, J.; Songyang, Z. Telomere regulation in pluripotent stem cells. Protein Cell 2014, 5, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Teichroeb, J.H.; Kim, J.; Betts, D.H. The role of telomeres and telomerase reverse transcriptase isoforms in pluripotency induction and maintenance. RNA Biol. 2016, 13, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.; Weng, N. ping Expression and regulation of telomerase in human T cell differentiation, activation, aging and diseases. Cell. Immunol. 2019, 345, 103989. [Google Scholar] [CrossRef] [PubMed]
- Ulaner, G.A.; Hu, J.F.; Vu, T.H.; Giudice, L.C.; Hoffman, A.R. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int. J. Cancer 2001, 91, 644–649. [Google Scholar] [CrossRef]
- Ulaner, G.A.; Hu, J.F.; Vu, T.H.; Giudice, L.C.; Hoffman, A.R. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998, 58, 4168–4172. [Google Scholar]
- Radan, L.; Hughes, C.S.; Teichroeb, J.H.; Vieira Zamora, F.M.; Jewer, M.; Postovit, L.M.; Betts, D.H. Microenvironmental regulation of telomerase isoforms in human embryonic stem cells. Stem Cells Dev. 2014, 23, 2046–2066. [Google Scholar] [CrossRef]
- Anderson, C.J.; Hoare, S.F.; Ashcroft, M.; Bilsland, A.E.; Keith, W.N. Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene 2006, 25, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Yatabe, N.; Kyo, S.; Maida, Y.; Nishi, H.; Nakamura, M.; Kanaya, T.; Tanaka, M.; Isaka, K.; Ogawa, S.; Inoue, M. HIF-1-mediated activation of telomerase in cervical cancer cells. Oncogene 2004, 23, 3708–3715. [Google Scholar] [CrossRef]
- Weng, N.P.; Levine, B.L.; June, C.H.; Hodes, R.J. Regulated expression of telomerase activity in human T lymphocyte development and activation. J. Exp. Med. 1996, 183, 2471–2479. [Google Scholar] [CrossRef]
- Haruta, Y.; Hiyama, K.; Ishioka, S.; Hozawa, S.; Maeda, H.; Yamakido, M. Activation of telomerase is induced by a natural antigen in allergen-specific memory T lymphocytes in bronchial asthma. Biochem. Biophys. Res. Commun. 1999, 259, 617–623. [Google Scholar] [CrossRef]
- Roth, A.; Yssel, H.; Pene, J.; Chavez, E.A.; Schertzer, M.; Lansdorp, P.M.; Spits, H.; Luiten, R.M. Telomerase levels control the lifespan of human T lymphocytes. Blood 2003, 102, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Jie, M.M.; Chang, X.; Zeng, S.; Liu, C.; Liao, G.B.; Wu, Y.R.; Liu, C.H.; Hu, C.J.; Yang, S.M.; Li, X.Z. Diverse regulatory manners of human telomerase reverse transcriptase. Cell Commun. Signal. 2019, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.S.; Cheng, N.-L.; Kim, J.; An, J.; Dong, F.; Yang, Q.; Zou, I.; Weng, N. Human T Cell Differentiation Negatively Regulates Telomerase Expression Resulting in Reduced Activation-Induced Proliferation and Survival. Front. Immunol. 2019, 10, 1993. [Google Scholar] [CrossRef]
- Effros, R.B.; Pawelec, G. Replicative senescence of T cells: Does the Hayflick Limit lead to immune exhaustion? Immunol. Today 1997, 18, 450–454. [Google Scholar] [CrossRef]
- Park, Y.P.; Choi, S.-C.; Kim, J.H.; Song, E.Y.; Kim, J.W.; Yoon, D.-Y.; Yeom, Y.I.; Lim, J.-S.; Kim, J.W.; Paik, S.-G.; et al. Up-regulation of Mac-2 binding protein by hTERT in gastric cancer. Int. J. cancer 2007, 120, 813–820. [Google Scholar] [CrossRef]
- Jagadeesh, S.; Banerjee, P.P. Telomerase reverse transcriptase regulates the expression of a key cell cycle regulator, cyclin D1. Biochem. Biophys. Res. Commun. 2006, 347, 774–780. [Google Scholar] [CrossRef]
- Perrault, S.D.; Hornsby, P.J.; Betts, D.H. Global gene expression response to telomerase in bovine adrenocortical cells. Biochem. Biophys. Res. Commun. 2005, 335, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Young, J.I.; Sedivy, J.M.; Smith, J.R. Telomerase expression in normal human fibroblasts stabilizes DNA 5-methylcytosine transferase I. J. Biol. Chem. 2003, 278, 19904–19908. [Google Scholar] [CrossRef] [PubMed]
- Masutomi, K.; Possemato, R.; Wong, J.M.Y.; Currier, J.L.; Tothova, Z.; Manola, J.B.; Ganesan, S.; Lansdorp, P.M.; Collins, K.; Hahn, W.C. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA 2005, 102, 8222–8227. [Google Scholar] [CrossRef]
- Zhou, J.; Ding, D.; Wang, M.; Cong, Y.-S. Telomerase reverse transcriptase in the regulation of gene expression. BMB Rep. 2014, 47, 8–14. [Google Scholar] [CrossRef]
- Akkaya, B.; Shevach, E.M. Regulatory T cells: Master thieves of the immune system. Cell. Immunol. 2020, 355, 104160. [Google Scholar] [CrossRef]
- Richardson, G.D.; Sage, A.; Bennaceur, K.; Al Zhrany, N.; Coelho-Lima, J.; Dookun, E.; Draganova, L.; Saretzki, G.; Breault, D.T.; Mallat, Z.; et al. Telomerase mediates lymphocyte proliferation but not the atherosclerosis-suppressive potential of regulatory T-cells. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1283–1296. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Huang, X.; Hsueh, E.C.; Zhang, Q.; Ma, C.; Zhang, Y.; Varvares, M.A.; Hoft, D.F.; Peng, G. Human regulatory T cells induce T-lymphocyte senescence. Blood 2012, 120, 2021–2031. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Gladilina, Y.A.; Pokrovsky, V.S.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Sokolov, N.N. Murine regulatory T cells induce death of effector T, B, and NK lymphocytes through a contact-independent mechanism involving telomerase suppression and telomere-associated senescence. Cell. Immunol. 2018, 331, 146–160. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Gladilina, Y.A.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Pokrovsky, V.S.; Sokolov, N.N. Contact-independent suppressive activity of regulatory T cells is associated with telomerase inhibition, telomere shortening and target lymphocyte apoptosis. Mol. Immunol. 2018, 101, 229–244. [Google Scholar] [CrossRef]
- Daniel, V.; Wang, H.; Sadeghi, M.; Opelz, G. Interferon-Gamma Producing Regulatory T Cells as a Diagnostic and Therapeutic Tool in Organ Transplantation. Int. Rev. Immunol. 2014, 33, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Hubackova, S.; Kucerova, A.; Michlits, G.; Kyjacova, L.; Reinis, M.; Korolov, O.; Bartek, J.; Hodny, Z. IFNγ induces oxidative stress, DNA damage and tumor cell senescence via TGFβ/SMAD signaling-dependent induction of Nox4 and suppression of ANT2. Oncogene 2016, 35, 1236–1249. [Google Scholar] [CrossRef]
- Studencka, M.; Schaber, J. Senoptosis: Non-lethal DNA cleavage as a route to deep senescence. Oncotarget 2017, 8, 30656–30671. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Krantz, S.B. Interferon gamma induces upregulation and activation of caspases 1, 3, and 8 to produce apoptosis in human erythroid progenitor cells. Blood 1999, 93, 3309–3316. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, Y.; Ling, Z.; Liu, X.; Zhao, X.; Yuan, Z.; Nie, C.; Wei, Y. Caspase-3 feedback loop enhances Bid-induced AIF/endoG and Bak activation in Bax and p53-independent manner. Cell Death Dis. 2015, 6, e1919. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zou, H.; Luo, T.; Long, M.; Bian, J.; Liu, X.; Gu, J.; Yuan, Y.; Song, R.; Wang, Y.; et al. Caspase-Dependent and Caspase-Independent Pathways Are Involved in Cadmium-Induced Apoptosis in Primary Rat Proximal Tubular Cell Culture. PLoS ONE 2016, 11, e0166823. [Google Scholar] [CrossRef]
- Pandiyan, P.; Zhu, J. Origin and functions of pro-inflammatory cytokine producing Foxp3+ regulatory T cells. Cytokine 2015, 76, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.S.; Leal-Berumen, I.; Nielsen, L.; Glibetic, M.; Jordana, M. Interleukin (IL)-10 inhibits long-term IL-6 production but not preformed mediator release from rat peritoneal mast cells. J. Clin. Investig. 1996, 97, 1122–1128. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Sad, S. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol. Today 1996, 17, 138–146. [Google Scholar] [CrossRef]
- Chen, F.; Chen, D.; Zhang, Y.; Jin, L.; Zhang, H.; Wan, M.; Pan, T.; Wang, X.; Su, Y.; Xu, Y.; et al. Interleukin-6 deficiency attenuates angiotensin II-induced cardiac pathogenesis with increased myocyte hypertrophy. Biochem. Biophys. Res. Commun. 2017, 494, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Ségal-Bendirdjian, E.; Geli, V. Non-canonical Roles of Telomerase: Unraveling the Imbroglio. Front. Cell Dev. Biol. 2019, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Xu, D. Telomerase reverse transcriptase (TERT) in action: Cross-talking with epigenetics. Int. J. Mol. Sci. 2019, 20, 3338. [Google Scholar] [CrossRef]
- Romaniuk, A.; Paszel-Jaworska, A.; Totoń, E.; Lisiak, N.; Hołysz, H.; Królak, A.; Grodecka-Gazdecka, S.; Rubiś, B. The non-canonical functions of telomerase: To turn off or not to turn off. Mol. Biol. Rep. 2019, 46, 1401–1411. [Google Scholar] [CrossRef]
- Smith, L.L.; Coller, H.A.; Roberts, J.M. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003, 5, 474–479. [Google Scholar] [CrossRef]
- Choi, J.; Southworth, L.K.; Sarin, K.Y.; Venteicher, A.S.; Ma, W.; Chang, W.; Cheung, P.; Jun, S.; Artandi, M.K.; Shah, N.; et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008, 4, 0124–0138. [Google Scholar] [CrossRef]
- Gene, E.; Lindvall, C.; Hou, M.; Komurasaki, T.; Zheng, C.; Henriksson, M.; Sedivy, J.M.; Björkholm, M.; Teh, T.; Nordenskjöld, M.; et al. Molecular Characterization of Human Telomerase Reverse Transcriptase-immortalized Human Fibroblasts by Gene Expression Profiling: Activation of the Epiregulin Gene. Cancer Res. 2003, 63, 1743–1747. [Google Scholar]
- Akiyama, M.; Hideshima, T.; Hayashi, T.; Tai, Y.-T.; Mitsiades, C.S.; Mitsiades, N.; Chauhan, D.; Richardson, P.; Munshi, N.C.; Anderson, K.C. Nuclear Factor-B p65 Mediates Tumor Necrosis Factor-induced Nuclear Translocation of Telomerase Reverse Transcriptase Protein 1. Cancer Res. 2003, 63, 18–21. [Google Scholar] [PubMed]
- Ghosh, A.; Saginc, G.; Leow, S.C.; Khattar, E.; Shin, E.M.; Yan, T.D.; Wong, M.; Zhang, Z.; Li, G.; Sung, W.K.; et al. Telomerase directly regulates NF-B-dependent transcription. Nat. Cell Biol. 2012, 14, 1270–1281. [Google Scholar] [CrossRef]
- Greenberg, R.A.; O’Hagan, R.C.; Deng, H.; Xiao, Q.; Hann, S.R.; Adams, R.R.; Lichtsteiner, S.; Chin, L.; Morin, G.B.; DePinho, R.A. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 1999, 18, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Khattar, E.; Leow, S.C.; Liu, C.Y.; Muller, J.; Ang, W.X.; Li, Y.; Franzoso, G.; Li, S.; Guccione, E.; et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Investig. 2015, 125, 2109–2122. [Google Scholar] [CrossRef]
- Zhou, L.; Zheng, D.; Wang, M.; Cong, Y.S. Telomerase reverse transcriptase activates the expression of vascular endothelial growth factor independent of telomerase activity. Biochem. Biophys. Res. Commun. 2009, 386, 739–743. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ding, D.; Hao, W.; Yang, F.; Wu, X.; Wang, M.; Xu, X.; Ju, Z.; Liu, J.P.; Song, Z.; et al. HTERT promotes tumor angiogenesis by activating VEGF via interactions with the Sp1 transcription factor. Nucleic Acids Res. 2016, 44, 8693–8703. [Google Scholar] [CrossRef]
- Yu, J.; Yuan, X.; Sjöholm, L.; Liu, T.; Kong, F.; Ekström, T.J.; Björkholm, M.; Xu, D. Telomerase reverse transcriptase regulates DNMT3B expression/aberrant DNA methylation phenotype and AKT activation in hepatocellular carcinoma. Cancer Lett. 2018, 434, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Uziel, O.; Reshef, H.; Ravid, A.; Fabian, I.; Halperin, D.; Ram, R.; Bakhanashvili, M.; Nordenberg, J.; Lahav, M. Oxidative stress causes telomere damage in Fanconi anaemia cells—A possible predisposition for malignant transformation. Br. J. Haematol. 2008, 142, 82–93. [Google Scholar] [CrossRef]
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017, 19, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Haendeler, J.; Hoffmann, J.; Brandes, R.P.; Zeiher, A.M.; Dimmeler, S. Hydrogen peroxide triggers nuclear export of telomerase reverse transcriptase via Src kinase family-dependent phosphorylation of tyrosine 707. Mol. Cell. Biol. 2003, 23, 4598–4610. [Google Scholar] [CrossRef]
- Singhapol, C.; Pal, D.; Czapiewski, R.; Porika, M.; Nelson, G.; Saretzki, G.C. Mitochondrial Telomerase Protects Cancer Cells from Nuclear DNA Damage and Apoptosis. PLoS ONE 2013, 8, e52989. [Google Scholar] [CrossRef]
- Indran, I.R.; Hande, M.P.; Pervaiz, S. hTERT overexpression alleviates intracellular ROS production, improves mitochondrial function, and inhibits ROS-mediated apoptosis in cancer cells. Cancer Res. 2011, 71, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Mattiussi, M.; Tilman, G.; Lenglez, S.; Decottignies, A. Human telomerase represses ros-dependent cellular responses to tumor necrosis factor-α without affecting nf-κb activation. Cell. Signal. 2012, 24, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.G.; Gupta, A.; Wang, H.; Scherthan, H.; Dhar, S.; Gandhi, V.; Iliakis, G.; Shay, J.W.; Young, C.S.H.; Pandita, T.K. hTERT associates with human telomeres and enhances genomic stability and DNA repair. Oncogene 2003, 22, 131–146. [Google Scholar] [CrossRef]
- Sharma, N.K.; Reyes, A.; Green, P.; Caron, M.J.; Bonini, M.G.; Gordon, D.M.; Holt, I.J.; Santos, J.H. Human telomerase acts as a hTR-independent reverse transcriptase in mitochondria. Nucleic Acids Res. 2012, 40, 712–725. [Google Scholar] [CrossRef]
- Ahmed, S.; Passos, J.F.; Birket, M.J.; Beckmann, T.; Brings, S.; Peters, H.; Birch-Machin, M.A.; von Zglinicki, T.; Saretzki, G. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell Sci. 2008, 121, 1046–1053. [Google Scholar] [CrossRef]
- Strauss, G.; Westhoff, M.A.; Fischer-Posovszky, P.; Fulda, S.; Schanbacher, M.; Eckhoff, S.M.; Stahnke, K.; Vahsen, N.; Kroemer, G.; Debatin, K.M. 4-hydroperoxy-cyclophosphamide mediates caspase-independent T-cell apoptosis involving oxidative stress-induced nuclear relocation of mitochondrial apoptogenic factors AIF and EndoG. Cell Death Differ. 2008, 15, 332–343. [Google Scholar] [CrossRef]
- Slusher, A.L.; Kim, J.J.J.; Ludlow, A.T. The role of alternative rna splicing in the regulation of htert, telomerase, and telomeres: Implications for cancer therapeutics. Cancers 2020, 12, 1514. [Google Scholar] [CrossRef] [PubMed]
- Hiyama, E.; Yamaoka, H.; Matsunaga, T.; Hayashi, Y.; Ando, H.; Suita, S.; Horie, H.; Kaneko, M.; Sasaki, F.; Hashizume, K.; et al. High expression of telomerase is an independent prognostic indicator of poor outcome in hepatoblastoma. Br. J. Cancer 2004, 91, 972–979. [Google Scholar] [CrossRef]
- Poremba, C.; Heine, B.; Diallo, R.; Heinecke, A.; Wai, D.; Schaefer, K.L.; Braun, Y.; Schuck, A.; Lanvers, C.; Bànkfalvi, À.; et al. Telomerase as a prognostic marker in breast cancer: High-throughput tissue microarray analysis of h TERT and hTR. J. Pathol. 2002, 198, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Tatsumoto, N.; Hiyama, E.; Murakami, Y.; Imamura, Y.; Shay, J.W.; Matsuura, Y.; Yokoyama, T. High Telomerase Activity Is an Independent Prognostic Indicator of Poor Outcome in Colorectal Cancer. Clin. Cancer. Res. 2000, 6, 2696–2701. [Google Scholar]
- Calvello, C.; Rocca, B.; Klersy, C.; Zappatore, R.; Giardini, I.; Dambruoso, I.; Pasi, F.; Caresana, M.; Zappasodi, P.; Nano, R.; et al. Alternative splicing of hTERT: A further mechanism for the control of active hTERT in acute myeloid leukemia. Leuk. Lymphoma 2018, 59, 702–709. [Google Scholar] [CrossRef]
- Ohyashiki, J.H.; Hisatomi, H.; Nagao, K.; Honda, S.; Takaku, T.; Zhang, Y.; Sashida, G.; Ohyashiki, K. Quantitative relationship between functionally active telomerase and major telomerase components (hTERT and hTR) in acute leukaemia cells. Br. J. Cancer 2005, 92, 1942–1947. [Google Scholar] [CrossRef]
- Rha, S.Y.; Jeung, H.C.; Yang, W.I.; Kim, J.J.; Oh, T.J.; An, S.W.; Chung, H.C. Alteration of hTERT full-length variant expression level showed different gene expression profiles and genomic copy number changes in breast cancer. Oncol. Rep. 2006, 15, 749–755. [Google Scholar] [CrossRef]
- Khosravi-Maharlooei, M.; Jaberipour, M.; Hosseini Tashnizi, A.; Attar, A.; Amirmoezi, F.; Habibagahi, M. Expression Pattern of Alternative Splicing Variants of Human Telomerase Reverse Transcriptase (hTERT) in Cancer Cell Lines Was not Associated with the Origin of the Cells. Int. J. Mol. Cell. Med. 2015, 4, 109–119. [Google Scholar] [PubMed]
- Li, G.; Shen, J.; Cao, J.; Zhou, G.; Lei, T.; Sun, Y.; Gao, H.; Ding, Y.; Xu, W.; Zhan, Z.; et al. Alternative splicing of human telomerase reverse transcriptase in gliomas and its modulation mediated by CX-5461. J. Exp. Clin. Cancer Res. 2018, 37, 78. [Google Scholar] [CrossRef]
- Khajehgoodari, R.; Khorvash, F.; Kheirollahi, M.; Mirsafaie, M.; Salehi, M. Correlations between the expression of hTERT and α and β splice variants in human brain tumors. Adv. Clin. Exp. Med. 2019, 28, 507–513. [Google Scholar] [CrossRef]
- Zaffaroni, N.; Della Porta, C.; Villa, R.; Botti, C.; Buglioni, S.; Mottolese, M.; Grazia Daidone, M. Transcription and alternative splicing of telomerase reverse transcriptase in benign and malignant breast tumours and in adjacent mammary glandular tissues: Implications for telomerase activity. J. Pathol. 2002, 198, 37–46. [Google Scholar] [CrossRef]
- Jeung, H.; Rha, S.; Shin, S.; Ahn, J.; Park, K.; Kim, T.; Kim, J.; Roh, J.; Chung, H. Changes in telomerase activity due to alternative splicing of human telomerase reverse transcriptase in colorectal cancer. Oncol. Lett. 2017, 14, 2385–2392. [Google Scholar] [CrossRef] [PubMed]
- Boldrini, L.; Faviana, P.; Gisfredi, S.; Zucconi, Y.; Di Quirico, D.; Donati, V.; Berti, P.; Spisni, R.; Galleri, D.; Materazzi, G.; et al. Evaluation of telomerase mRNA (hTERT) in colon cancer. Int. J. Oncol. 2002, 21, 493–497. [Google Scholar] [CrossRef]
- Xu, J.H.; Wang, Y.C.; Geng, X.; Li, Y.Y.; Zhang, W.M. Changes of the alternative splicing variants of human telomerase reverse transcriptase during gastric carcinogenesis. Pathobiology 2009, 76, 23–29. [Google Scholar] [CrossRef]
- Barclay, J.Y.; Morris, A.G.; Nwokolo, C.U. HTERT mRNA partially regulates telomerase activity in gastric adenocarcinoma and adjacent normal gastric mucosa. Dig. Dis. Sci. 2005, 50, 1299–1303. [Google Scholar] [CrossRef] [PubMed]
- Nagao, K.; Katsumata, K.; Aizawa, Y.; Saito, N.; Hirata, H.; Sasaki, H.; Yamamoto, S.; Hikiji, K.; Koiwa, T.; Hisatomi, H. Differential alternative splicing expressions of telomerase reverse transcriptase in gastrointestinal cell lines. Oncol. Rep. 2004, 11, 127–131. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, B.; Zhong, H.; Tian, X.; Fang, W. Quantification of alternative splicing variants of human telomerase reverse transcriptase and correlations with telomerase activity in lung cancer. PLoS ONE 2012, 7, e38868. [Google Scholar] [CrossRef] [PubMed]
- Mavrogiannou, E.; Strati, A.; Stathopoulou, A.; Tsaroucha, E.G.; Kaklamanis, L.; Lianidou, E.S. Real-time RT-PCR quantification of human telomerase reverse transcriptase splice variants in tumor cell lines and non-small cell lung cancer. Clin. Chem. 2007, 53, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Kamma, H.; Wu, W.; Hamasaki, M.; Kaneko, S.; Horiguchi, H.; Matsui-Horiguchi, M.; Satoh, H. Expression and alternative splicing pattern of human telomerase reverse transcriptase in human lung cancer cells. Int. J. Oncol. 2004, 24, 925–930. [Google Scholar] [CrossRef]
- Villa, R.; Porta, C.D.; Folini, M.; Daidone, M.G.; Zaffaroni, N. Possible regulation of telomerase activity by transcription and alternative splicing of telomerase reverse transcriptase in human melanoma. J. Investig. Dermatol. 2001, 116, 867–873. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lincz, L.F.; Mudge, L.M.; Scorgie, F.E.; Sakoff, J.A.; Hamilton, C.S.; Seldon, M. Quantification of hTERT splice variants in melanoma by SYBR green real-time polymerase chain reaction indicates a negative regulatory role for the β deletion variant. Neoplasia 2008, 10, 1131–1137. [Google Scholar] [CrossRef]
- Wang, Y.; Kowalski, J.; Tsai, H.L.; Marik, R.; Prasad, N.; Somervell, H.; Lo, P.K.; Sangenario, L.E.; Dyrskjot, L.; Orntoft, T.F.; et al. Differentiating alternative splice variant patterns of human telomerase reverse transcriptase in thyroid neoplasms. Thyroid 2008, 18, 1055–1063. [Google Scholar] [CrossRef]
- Wang, Y.; Meeker, A.K.; Kowalski, J.; Tsai, H.-L.; Somervell, H.; Heaphy, C.; Sangenario, L.E.; Prasad, N.; Westra, W.H.; Zeiger, M.A.; et al. Telomere length is related to alternative splice patterns of telomerase in thyroid tumors. Am. J. Pathol. 2011, 179, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.; Parker, A.; Hojjat-Farsangi, M.; Forster, J.; Kokhaei, P.; Hansson, L.; Osterborg, A.; Mellstedt, H. Telomere length and expression of human telomerase reverse transcriptase splice variants in chronic lymphocytic leukemia. Exp. Hematol. 2013, 41, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Terrin, L.; Trentin, L.; Degan, M.; Corradini, I.; Bertorelle, R.; Carli, P.; Maschio, N.; Bo, M.D.; Noventa, F.; Gattei, V.; et al. Telomerase expression in B-cell chronic lymphocytic leukemia predicts survival and delineates subgroups of patients with the same igVH mutation status and different outcome. Leukemia 2007, 21, 965–972. [Google Scholar] [CrossRef]
- Dong, W.; Qian, Y.; Yang, L. Telomerase, hTERT and splice variants in patients with myelodysplastic syndromes. Leuk. Res. 2014, 38, 830–835. [Google Scholar] [CrossRef]
- Briatore, F.; Barrera, G.; Pizzimenti, S.; Toaldo, C.; Della Casa, C.; Laurora, S.; Pettazzoni, P.; Dianzani, M.U.; Ferrero, D. Increase of telomerase activity and hTERT expression in myelodysplastic syndromes. Cancer Biol. Ther. 2009, 8, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Mailloux, A.; Rollison, D.E.; Painter, J.S.; MacIejewski, J.; Paquette, R.L.; Loughran, T.P.; McGraw, K.; Makishima, H.; Radhakrishnan, R.; et al. Naive T-cells in myelodysplastic syndrome display intrinsic human telomerase reverse transcriptase (hTERT) deficiency. Leukemia 2013, 27, 897–906. [Google Scholar] [CrossRef]
- Dong, W.; Wu, L.; Sun, H.; Ren, X.; Epling-Burnette, P.K.; Yang, L. MDS shows a higher expression of hTERT and alternative splice variants in unactivated T-cells. Oncotarget 2016, 7, 71904–71914. [Google Scholar] [CrossRef] [PubMed]
- Attar, A.; Maharlooei, M.K.; Nazarinia, M.A.; Hosseini, A.; Bajalli, Z.; Moeini, Y.S.; Monabati, A.; Amirmoezi, F.; Jaberipour, M.; Habibagahi, M. Expression pattern of telomerase reverse transcriptase (Htert) variants and bcl-2 in peripheral lymphocytes of systemic lupus erythematosus patients. Iran. J. Pathol. 2020, 15, 225–231. [Google Scholar] [CrossRef]
- Bajaj, S.; Kumar, M.S.; Peters, G.J.; Mayur, Y.C. Targeting telomerase for its advent in cancer therapeutics. Med. Res. Rev. 2020, 40, 1871–1919. [Google Scholar] [CrossRef]
- Dilley, R.L.; Greenberg, R.A. ALTernative Telomere Maintenance and Cancer. Trends Cancer 2015, 1, 145–156. [Google Scholar] [CrossRef]
- Chiappori, A.A.; Kolevska, T.; Spigel, D.R.; Hager, S.; Rarick, M.; Gadgeel, S.; Blais, N.; Von pawel, J.; Hart, L.; Reck, M.; et al. A randomized phase II study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non-small-cell lung cancer. Ann. Oncol. 2015, 26, 354–362. [Google Scholar] [CrossRef]
- Middleton, G.; Silcocks, P.; Cox, T.; Valle, J.; Wadsley, J.; Propper, D.; Coxon, F.; Ross, P.; Madhusudan, S.; Roques, T.; et al. Gemcitabine and capecitabine with or without telomerase peptide vaccine GV1001 in patients with locally advanced or metastatic pancreatic cancer (TeloVac): An open-label, randomised, phase 3 trial. Lancet Oncol. 2014, 15, 829–840. [Google Scholar] [CrossRef]
- Kataoka, N. Modulation of aberrant splicing in human RNA diseases by chemical compounds. Hum. Genet. 2017, 136, 1237–1245. [Google Scholar] [CrossRef]
- León, B.; Kashyap, M.K.; Chan, W.C.; Krug, K.A.; Castro, J.E.; La Clair, J.J.; Burkart, M.D. A Challenging Pie to Splice: Drugging the Spliceosome. Angew. Chem. Int. Ed. 2017, 56, 12052–12063. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.; Lamarteleur, T.; Lacroix, L.; Mailliet, P.; Mergny, J.L.; Riou, J.F. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004, 32, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Havens, M.A.; Hastings, M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef]
- Brambilla, C.; Folini, M.; Gandellini, P.; Daprai, L.; Daidone, M.G.; Zaffaroni, N. Oligomer-mediated modulation of hTERT alternative splicing induces telomerase inhibition and cell growth decline in human prostate cancer cells. Cell Mol. Life Sci. 2004, 61, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Fairlie, J.; Harrington, L. Enforced telomere elongation increases the sensitivity of human tumour cells to ionizing radiation. DNA Repair. 2015, 25, 54–59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dai, W.; Xu, X.; Wang, D.; Wu, J.; Wang, J. Cancer therapy with a CRISPR-assisted telomerase-activating gene expression system. Oncogene 2019, 38, 4110–4124. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Schmidt, J.C.; Zaug, A.J.; Ascarrunz, D.R.; Cech, T.R. A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression. Genome Biol. 2015, 16, 231. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Li, F.; He, Q.; Deng, T.; Xu, J.; Jin, F.; Coarfa, C.; Putluri, N.; Liu, D.; Songyang, Z. Systematic analysis of human telomeric dysfunction using inducible telosome/shelterin CRISPR/Cas9 knockout cells. Cell Discov. 2017, 3, 17034. [Google Scholar] [CrossRef]
- Brane, A.; Tollefsbol, T. Targeting Telomeres and Telomerase: Studies in Aging and Disease Utilizing CRISPR/Cas9 Technology. Cells 2019, 8, 186. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).