MicroRNA Expression in Extracellular Vesicles from Nasal Lavage Fluid in Chronic Rhinosinusitis
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Subjects
2.2. NLF Collection
2.3. Isolation of EVs from the NLF
2.4. EV Characterization
2.5. MicroRNA Isolation from EVs
2.6. Nanostring nCounter System MiRNA Assay
2.7. Prediction of Target Genes and Associated Pathways
2.8. Statistical Analysis
3. Results
3.1. Clinical Characteristics of the Study Participants
3.2. Phenotypic Characterization of NLF-EVs
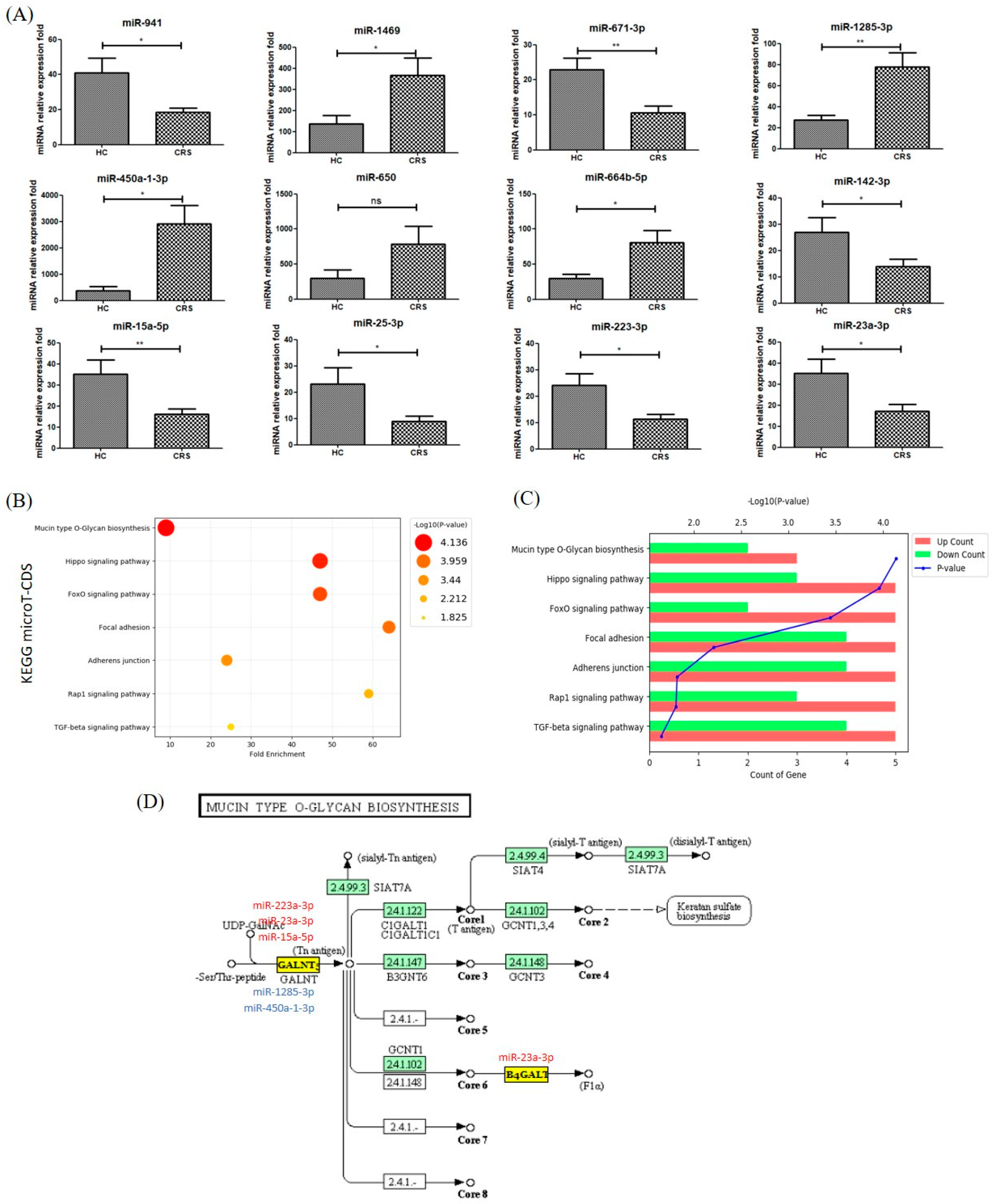
3.3. MicroRNAs Were Differentially Expressed in NLF-EVs of CRS Patients
3.4. Gene Ontology and Pathway Classification of In Silico Analysis of MiRNA Target Genes in CRS vs. Healthy NLF-EVs
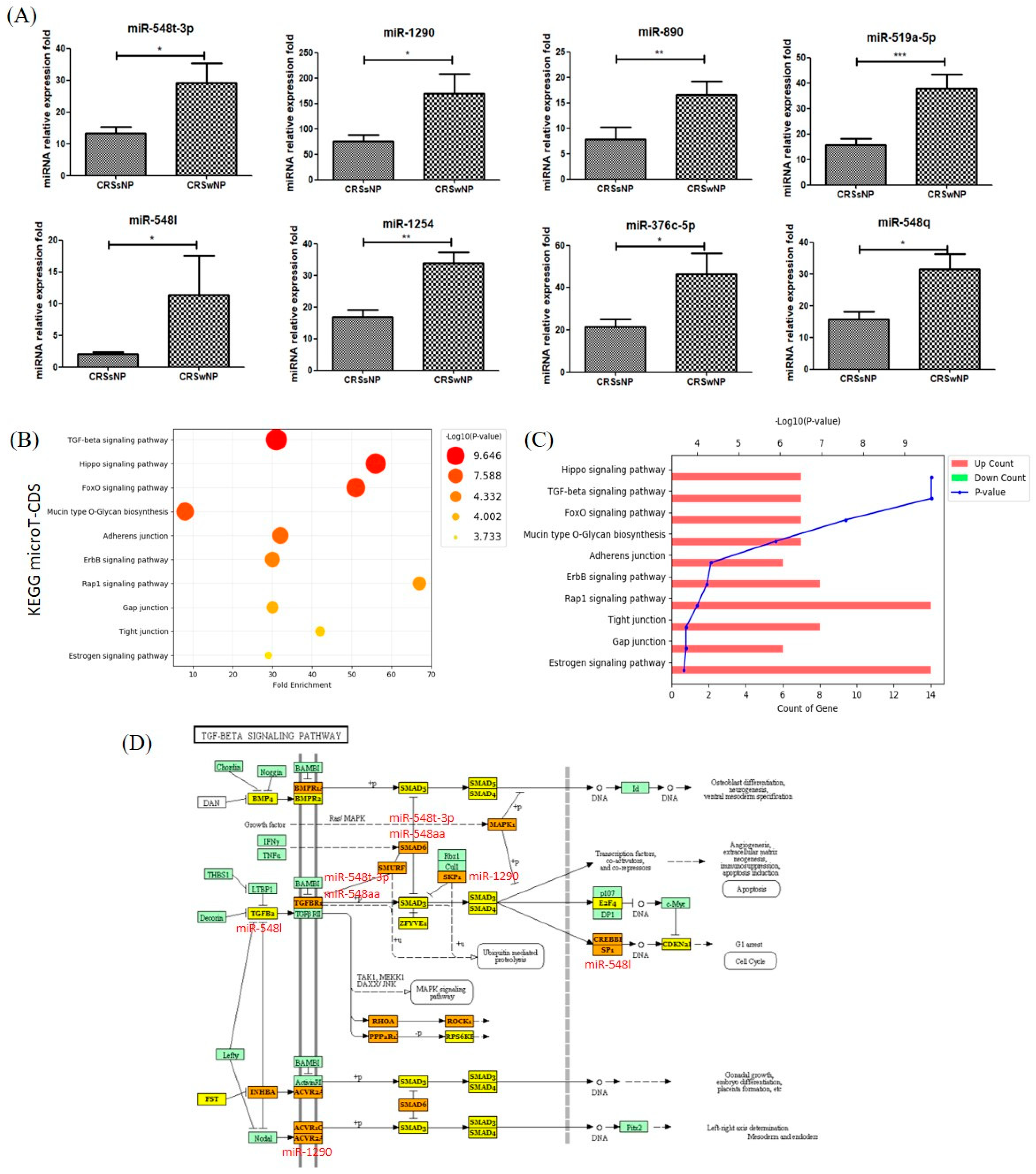
3.5. Gene Ontology and Pathway Classification of in Silico Analysis of MiRNA Target Genes in CRSsNP vs. CRSwNP NLF-EVs
3.6. Functional Validation of Representative MiRNA Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRS | Chronic rhinosinusitis |
| EVs | Extracellular vesicles |
| miRNA | MicroRNA |
| NLF | Nasal lavage fluid |
| JESREC | Japanese Epidemiological Survey of Refractory Eosinophilic Chronic Rhinosinusitis |
| KEGG | Kyoto Encyclopedia Gene and Genome database |
| GalNAc-Ts | N-acetylgalactosanimyltransferases |
| GalNAc | N-acetylgalactosamine |
| TGF-β | Transforming growth factor beta |
| MET | MET proto-oncogene |
| EGFR | Epidermal growth factor receptor |
| ACTB | Beta-actin |
| PARP1 | Poly(ADP-ribose) polymerase 1 |
| TGFBR | Transforming growth factor beta receptor |
| SMAD | Small mothers against decapentaplegic |
| MAPK | Mitogen-activated protein kinase |
References
- Wu, G.; Yang, G.; Zhang, R.; Xu, G.; Zhang, L.; Wen, W.; Lu, J.; Liu, J.; Yu, Y. Altered microRNA Expression Profiles of Extracellular Vesicles in Nasal Mucus From Patients With Allergic Rhinitis. Allergy Asthma Immunol. Res. 2015, 7, 449–457. [Google Scholar] [CrossRef]
- Lässer, C.; O’Neil, S.E.; Ekerljung, L.; Ekström, K.; Sjöstrand, M.; Lötvall, J. RNA-containing Exosomes in Human Nasal Secretions. Am. J. Rhinol. Allergy 2011, 25, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Nocera, A.L.; Miyake, M.M.; Seifert, P.; Han, X.; Bleier, B.S. Exosomes mediate interepithelial transfer of functional P-glycoprotein in chronic rhinosinusitis with nasal polyps. Laryngoscope 2017, 127, E295–E300. [Google Scholar] [CrossRef] [PubMed]
- Xuan, L.; Luan, G.; Wang, Y.; Lan, F.; Zhang, X.; Hao, Y.; Zheng, M.; Wang, X.; Zhang, L. MicroRNAs regulating mucin type O-glycan biosynthesis and transforming growth factor β signaling pathways in nasal mucosa of patients with chronic rhinosinusitis with nasal polyps in Northern China. Int. Forum Allergy Rhinol. 2019, 9, 106–113. [Google Scholar] [CrossRef]
- Tokunaga, T.; Sakashita, M.; Haruna, T.; Asaka, D.; Takeno, S.; Ikeda, H.; Nakayama, T.; Seki, N.; Ito, S.; Murata, J.; et al. Novel scoring system and algorithm for classifying chronic rhinosinusitis: The JESREC Study. Allergy 2015, 70, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Hur, J.Y.; Lee, J.S.; Kim, I.A.; Kim, H.J.; Kim, W.S.; Lee, K.Y. Extracellular vesicle-based EGFR genotyping in bronchoalveolar lavage fluid from treatment-naive non-small cell lung cancer patients. Transl. Lung Cancer Res. 2019, 8, 1051–1060. [Google Scholar] [CrossRef]
- Hashimoto, K.; Goto, S.; Kawano, S.; Aoki-Kinoshita, K.F.; Ueda, N.; Hamajima, M.; Kawasaki, T.; Kanehisa, M. KEGG as a glycome informatics resource. Glycobiology 2006, 16, 63R–70R. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein. Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.A.; Nymon, A.B.; Ringelberg, C.S.; Lesseur, C.; Hazlett, H.F.; Howard, L.; Marsit, C.J.; Ashare, A. Pulmonary microRNA profiling: Implications in upper lobe predominant lung disease. Clin. Epigenetics 2017, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fan, L.; Yu, H.; Zhang, J.; He, Y.; Feng, D.; Wang, F.; Li, X.; Liu, Q.; Li, Y.; et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology 2019, 70, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Vartholomatos, G.; Tzavaras, T.; Hatziapostolou, M.; Fackelmayer, F.O.; Sandaltzopoulos, R.; Polytarchou, C.; Kolettas, E. Senescence-associated microRNAs target cell cycle regulatory genes in normal human lung fibroblasts. Exp. Gerontol. 2017, 96, 110–122. [Google Scholar] [CrossRef]
- Fesen, K.; Silveyra, P.; Fuentes, N.; Nicoleau, M.; Rivera, L.; Kitch, D.; Graff, G.R.; Siddaiah, R. The role of microRNAs in chronic pseudomonas lung infection in Cystic fibrosis. Respir. Med. 2019, 151, 133–138. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, D.; Moon, H.-G.; Lee, H.; A Haspel, J.; Hu, K.; Xie, L.; Jin, Y. MicroRNA-15a/16 Regulates Apoptosis of Lung Epithelial Cells After Oxidative Stress. Mol. Med. 2016, 22, 233–243. [Google Scholar] [CrossRef]
- Wang, S.; Min, J.; Yu, Y.; Yin, L.; Wang, Q.; Shen, H.; Yang, J.; Zhang, P.; Xiao, J.; Wang, Z. Differentially expressed miRNAs in circulating exosomes between atrial fibrillation and sinus rhythm. J. Thorac. Dis. 2019, 11, 4337–4348. [Google Scholar] [CrossRef]
- Gomez, J.L.; Chen, A.; Diaz, M.P.; Zirn, N.; Gupta, A.; Britto, C.; Sauler, M.; Yan, X.; Stewart, E.; Santerian, K.; et al. A Network of Sputum MicroRNAs Is Associated with Neutrophilic Airway Inflammation in Asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 51–64. [Google Scholar] [CrossRef]
- Boxberger, N.; Hecker, M.; Zettl, U.K. Dysregulation of Inflammasome Priming and Activation by MicroRNAs in Human Immune-Mediated Diseases. J. Immunol. 2019, 202, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Inchley, C.S.; Sonerud, T.; O Fjærli, H.; Nakstad, B. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect. Dis. 2015, 15, 150. [Google Scholar] [CrossRef]
- Velasco-Torres, Y.; Ruiz-López, V.; Pérez-Bautista, O.; Buendía-Roldan, I.; Ramírez-Venegas, A.; Pérez-Ramos, J.; Falfán-Valencia, R.; Ramos, C.; Montaño, M. miR-34a in serum is involved in mild-to-moderate COPD in women exposed to biomass smoke. BMC Pulm. Med. 2019, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tombak, A.; Ay, O.I.; Erdal, M.E.; Sungur, M.A.; Uçar, M.A.; Akdeniz, A.; Tiftik, E.N. MicroRNA Expression Analysis in Patients with Primary Myelofibrosis, Polycythemia vera and Essential Thrombocythemia. Indian J. Hematol. Blood Transfus. 2015, 31, 416–425. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deng, J.-N.; Li, Y.-Q.; Liu, Y.; Li, Q.; Hu, Y.; Xu, J.-Q.; Sun, T.-Y.; Xie, L.-X. Exosomes derived from plasma of septic patients inhibit apoptosis of T lymphocytes by down-regulating bad via hsa-miR-7-5p. Biochem. Biophys. Res. Commun. 2019, 513, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Mullany, L.E.; Herrick, J.S.; Wolff, R.K.; Stevens, J.R.; Slattery, M.L. Association of cigarette smoking and microRNA expression in rectal cancer: Insight into tumor phenotype. Cancer Epidemiol. 2016, 45, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, X.; Zhou, W.; Li, D.; Chang, A.; Wang, Y.; Wu, Y.; Zhou, Q. Plasma microRNA expression signature involving miR-548q, miR-630 and miR-940 as biomarkers for nasopharyngeal carcinoma detection. Cancer Biomarkers 2018, 23, 579–587. [Google Scholar] [CrossRef]
- Huan, L.C.; Wu, J.-C.; Chiou, B.-H.; Chen, C.-H.; Ma, N.; Chang, C.Y.; Tsen, Y.-K.; Chen, S.C. MicroRNA regulation of DNA repair gene expression in 4-aminobiphenyl-treated HepG2 cells. Toxicology 2014, 322, 69–77. [Google Scholar] [CrossRef]
- Zheng, X.-H.; Cui, C.; Ruan, H.-L.; Xue, W.-Q.; Zhang, S.-D.; Hu, Y.-Z.; Zhou, X.-X.; Jia, W.-H. Plasma microRNA profiles of nasopharyngeal carcinoma patients reveal miR-548q and miR-483-5p as potential biomarkers. Chin. J. Cancer 2014, 33, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Lambert, K.A.; Roff, A.N.; Panganiban, R.P.; Douglas, S.; Ishmael, F.T. MicroRNA-146a is induced by inflammatory stimuli in airway epithelial cells and augments the anti-inflammatory effects of glucocorticoids. PLoS ONE 2018, 13, e0205434. [Google Scholar] [CrossRef]
- Ma, Z.-X.; Tan, X.; Shen, Y.; Ke, X.; Yang, Y.-C.; He, X.-B.; Wang, Z.-H.; Dai, Y.-B.; Hong, S.-L.; Hu, G.-H. MicroRNA expression profile of mature dendritic cell in chronic rhinosinusitis. Inflamm. Res. 2015, 64, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhou, H.; Miao, Y.; Li, N.; Zhao, L.; Jia, L. MiRNA expression profiles reveal the involvement of miR-26a, miR-548l and miR-34a in hepatocellular carcinoma progression through regulation of ST3GAL5. Lab. Investig. 2017, 97, 530–542. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, S.; Cao, Y.; Yang, Y. Altered miRNAs Expression Profiles and Modulation of Immune Response Genes and Proteins During Neonatal Sepsis. J. Clin. Immunol. 2014, 34, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.W.; Schleimer, R.P.; Kern, R.C. Chronic Rhinosinusitis with Nasal Polyps. J. Allergy Clin. Immunol. Pr. 2016, 4, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Kato, A. Immunopathology of chronic rhinosinusitis. Allergol. Int. 2015, 64, 121–130. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-B.; Zhang, Z.-R.; Schluesener, H.J.; Xu, S.-Q. Role of exosomes in immune regulation. J. Cell. Mol. Med. 2006, 10, 364–375. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Görgens, A.; Bremer, M.; Ferrer-Tur, R.; Murke, F.; Tertel, T.; Horn, P.A.; Thalmann, S.; Welsh, J.A.; Probst, C.; Guerin, C.; et al. Optimisation of imaging flow cytometry for the analysis of single extracellular vesicles by using fluorescence-tagged vesicles as biological reference material. J. Extracell. Vesicles 2019, 8, 1587567. [Google Scholar] [CrossRef]
- Qazi, K.R.; Paredes, P.T.; Dahlberg, B.; Grunewald, J.; Eklund, A.; Gabrielsson, S. Proinflammatory exosomes in bronchoalveolar lavage fluid of patients with sarcoidosis. Thorax 2010, 65, 1016–1024. [Google Scholar] [CrossRef]
- Kesimer, M.; Gupta, R. Physical characterization and profiling of airway epithelial derived exosomes using light scattering. Methods 2015, 87, 59–63. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Zuñiga-Garcia, P.; Sanchez-Mosquera, P.; Egia, A.; Perez, A.; Loizaga, A.; Arceo, R.; Lacasa, I.; Rabade, A.; Arrieta, E.; et al. Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. J. Extracell. Vesicles 2016, 5, 29497. [Google Scholar] [CrossRef]
- Huse, D. Allergic rhinitis may worsen asthma symptoms in children; the international asthma outcomes registry. Am. J. Respir. Crit. Care Med. 1996, 153, A860. [Google Scholar]
- Bartel, S.; La Grutta, S.; Cilluffo, G.; Perconti, G.; Bongiovanni, A.; Giallongo, A.; Behrends, J.; Kruppa, J.; Hermann, S.; Chiang, D.; et al. Human airway epithelial extracellular vesicle miRNA signature is altered upon asthma development. Allergy 2019, 75, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Bao, L.; Gao, W.; Liu, S.; Ji, K.; Li, J. Differentially Expressed miRNA in Inflammatory Mucosa of Chronic Rhinosinusitis. J. Nanosci. Nanotechnol. 2015, 15, 2132–2139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-H.; Zhang, Y.-N.; Li, H.-B.; Hu, C.-Y.; Wang, N.; Cao, P.-P.; Liao, B.; Lu, X.; Cui, Y.-H.; Liu, Z. Overexpression of miR-125b, a Novel Regulator of Innate Immunity, in Eosinophilic Chronic Rhinosinusitis with Nasal Polyps. Am. J. Respir. Crit. Care Med. 2012, 185, 140–151. [Google Scholar] [CrossRef]
- Yang, H.-W.; Kim, H.-J.; Park, J.-H.; Shin, J.-M.; Lee, H.-M. Apigenin alleviates TGF-β1-induced nasal mucosa remodeling by inhibiting MAPK / NF-kB signaling pathways in chronic rhinosinusitis. PLoS ONE 2018, 13, e0201595. [Google Scholar] [CrossRef]
- Li, T.; Leong, M.H.; Harms, B.; Kennedy, G.; Chen, L. MicroRNA-21 as a potential colon and rectal cancer biomarker. World J. Gastroenterol. 2013, 19, 5615–5621. [Google Scholar] [CrossRef]
- Siasos, G.; Kollia, C.; Tsigkou, V.; Basdra, E.K.; Lymperi, M.; Oikonomou, E.; Kokkou, E.; Korompelis, P.; Papavassiliou, A.G. MicroRNAs: Novel diagnostic and prognostic biomarkers in atherosclerosis. Curr. Top. Med. Chem. 2013, 13, 1503–1517. [Google Scholar] [CrossRef]
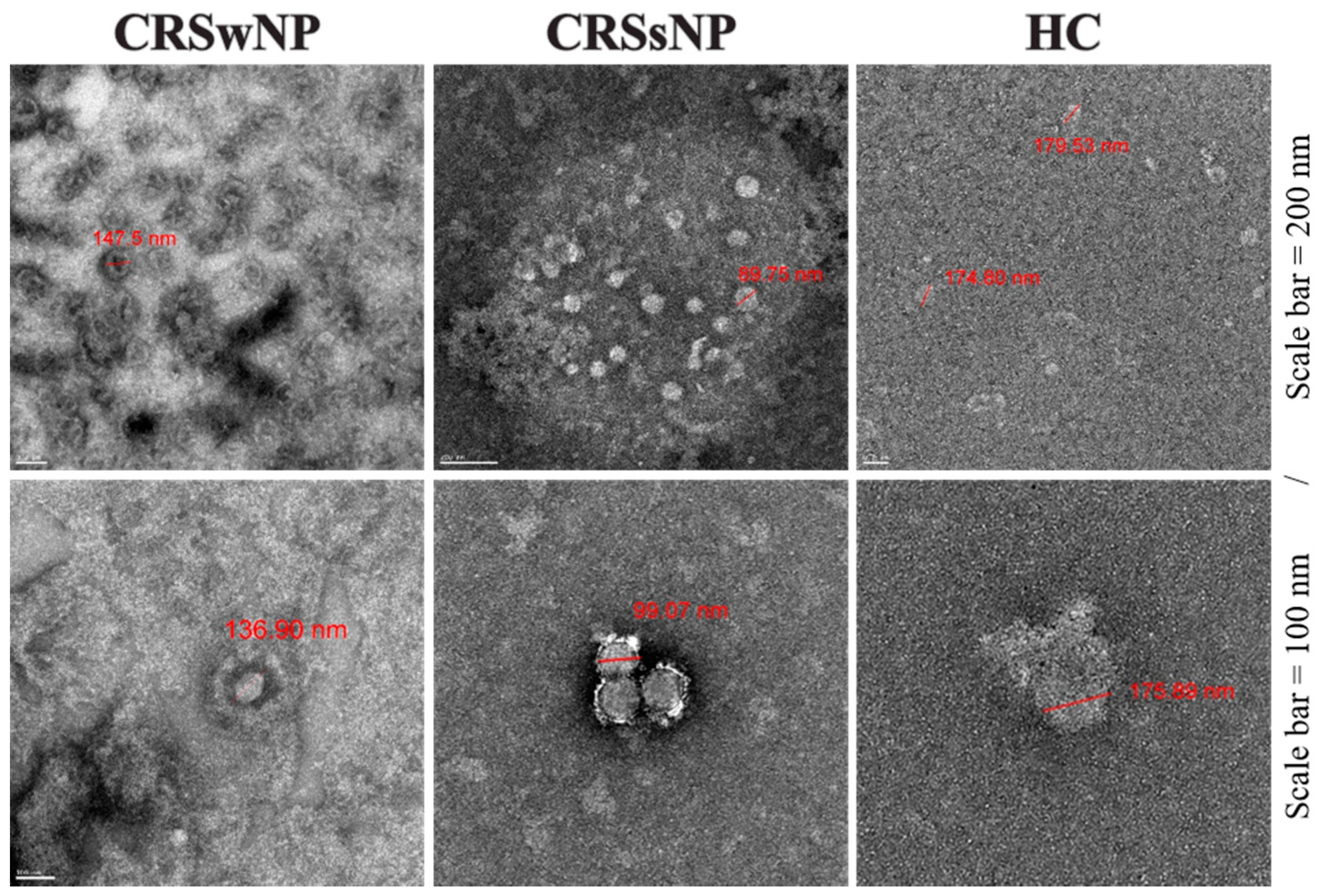

| Variables | CRSwNP 1 (n = 7) | CRSsNP 2 (n = 8) | HC 3 (n = 7) |
|---|---|---|---|
| Sex, male (%) | 3 (42.9%) | 4 (50.0%) | 6 (85.7%) |
| Age, years | 47.4 ± 23.2 | 42.0 ± 20.6 | 40.3 ± 15.2 |
| JESREC score 4 | 9.67 ± 3.27 | 7.29 ± 5.25 | NA |
| Allergy | |||
| (+) | 4 (57.1%) | 3 (37.5%) | 2 (28.6%) |
| (−) | 3 (42.9%) | 5 (62.5%) | 5 (71.4%) |
| Asthma | |||
| (+) | 7 (100%) | 7 (100%) | 7 (100%) |
| (−) | 0 (0%) | 0 (0%) | 0 (0%) |
| Subject | Median Size (IQR) (nm) | Mean Count (Cell Count/10 mm2 ± SD) |
|---|---|---|
| CRSwNP 1 | 131.56 (107.99–166.29) | 75 ± 19 |
| CRSsNP 2 | 82.32 (75.43–102.77) | 66 ± 20 |
| HC 3 | 183.93 (157.14–216.07) | 3 ± 3 |
| Total | 124.57 (93.78–157.14) | N/A |
| KEGG Pathway | MiRNA | CRS vs. HC | CRSwNP vs. CRSsNP | Target MRNA Name | Putative Disease Associations (Human Studies) |
|---|---|---|---|---|---|
| Mucin type O-Glycan biosynthesis | miR-23a-3p | Up | p > 0.05 | MET; CTNND1; LMO7; IQGAP1; EGFR; TJP1; NLK; CDH1; SMAD4; CTNNB1; CTNNA1; WASF2; CSNK2A1; FARP2; PTPRJ; ACTN4; MAPK1; TGFBR2 | Fibrosis [12], Disease Progression [13], Folic Acid Deficiency [14], Genetic Predisposition to Diseases [15] |
| miR-15a-5p | Up | p > 0.05 | ACTB; SMAD2; PVRL2; ACTG1; LMO7; IQGAP1; IGF1R; PTPRF; ACP1; CDH1; CTNNB1; CSNK2A1; RAC1; CDC42; EP300; YES1; FGFR1; MAP3K7; CREBBP; PVRL1 | Disease Progression [16] | |
| miR-223-3p | Up | p > 0.05 | PARP1; SCARB1; RASGRP1; DDIT4; GFPT1; RGS1; MECP2; MT1E; TTBK2; WDR7; FADS1; STMN1; GDI1; CHUK; SOX5; SERINC1; TMEM69; CPEB4; RNF213; ARTN; COX7A2L | Inflammation [17,18] *, Disease Progression [19,20] *, Genetic Predisposition to Disease [21], primary myelofibrosis [12,22] | |
| miR-1285-3p | Down | p > 0.05 | WBSCR17 | Inflammation [23] | |
| TGF-beta signaling pathway | miR-548q | p > 0.05 | Up | IGF1R | Disease Progression [24,25] *, Genetic Predisposition to Disease [26,27], Inflammation [28] * |
| miR-1290 | p > 0.05 | Up | CDKN2B; SKP1; ACVR2A; SP1; MAPK1; BMPR2; RPS6KB1 | Inflammation [29] | |
| miR-548l | p > 0.05 | Up | FST; SMAD2; INHBB; SKP1; ACVR2B; SMAD4; E2F5; SMAD5; ACVR2A; SP1; ACVR1C; TGFB2; BMPR1A; E2F4; PPP2R1B | Acute diseases, Acute Lung Injury [30], Inflammation [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cha, S.; Seo, E.-H.; Lee, S.H.; Kim, K.S.; Oh, C.-S.; Moon, J.-S.; Kim, J.K. MicroRNA Expression in Extracellular Vesicles from Nasal Lavage Fluid in Chronic Rhinosinusitis. Biomedicines 2021, 9, 471. https://doi.org/10.3390/biomedicines9050471
Cha S, Seo E-H, Lee SH, Kim KS, Oh C-S, Moon J-S, Kim JK. MicroRNA Expression in Extracellular Vesicles from Nasal Lavage Fluid in Chronic Rhinosinusitis. Biomedicines. 2021; 9(5):471. https://doi.org/10.3390/biomedicines9050471
Chicago/Turabian StyleCha, Seungbin, Eun-Hye Seo, Seung Hyun Lee, Kyung Soo Kim, Chung-Sik Oh, Jong-Seok Moon, and Jin Kook Kim. 2021. "MicroRNA Expression in Extracellular Vesicles from Nasal Lavage Fluid in Chronic Rhinosinusitis" Biomedicines 9, no. 5: 471. https://doi.org/10.3390/biomedicines9050471
APA StyleCha, S., Seo, E.-H., Lee, S. H., Kim, K. S., Oh, C.-S., Moon, J.-S., & Kim, J. K. (2021). MicroRNA Expression in Extracellular Vesicles from Nasal Lavage Fluid in Chronic Rhinosinusitis. Biomedicines, 9(5), 471. https://doi.org/10.3390/biomedicines9050471






