Circulating microRNAs Related to Bone Metabolism in HIV-Associated Bone Loss
Abstract
1. Introduction
2. Patients and Methods
2.1. Patient Recruitment
2.2. Samples Collection and Storage
2.3. RNA Isolation/RT qPCR
MiRNA Primer Assays
2.4. DXA Measurements
2.5. Biochemical Assays
2.6. Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Study Population
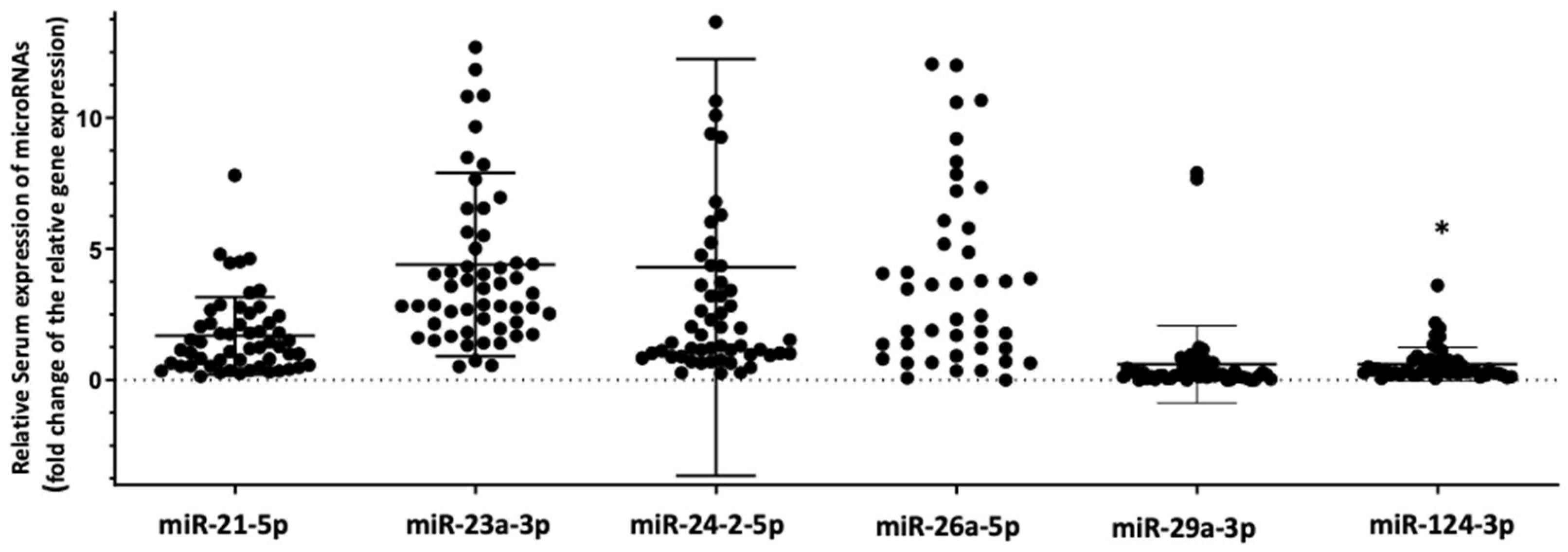
3.2. Differential Expression of the Selected Panel of miRNAs Linked to Bone Metabolism in HIV+ Individuals with Osteoporosis Compared to HIV+ Individuals with Normal Bone Mass
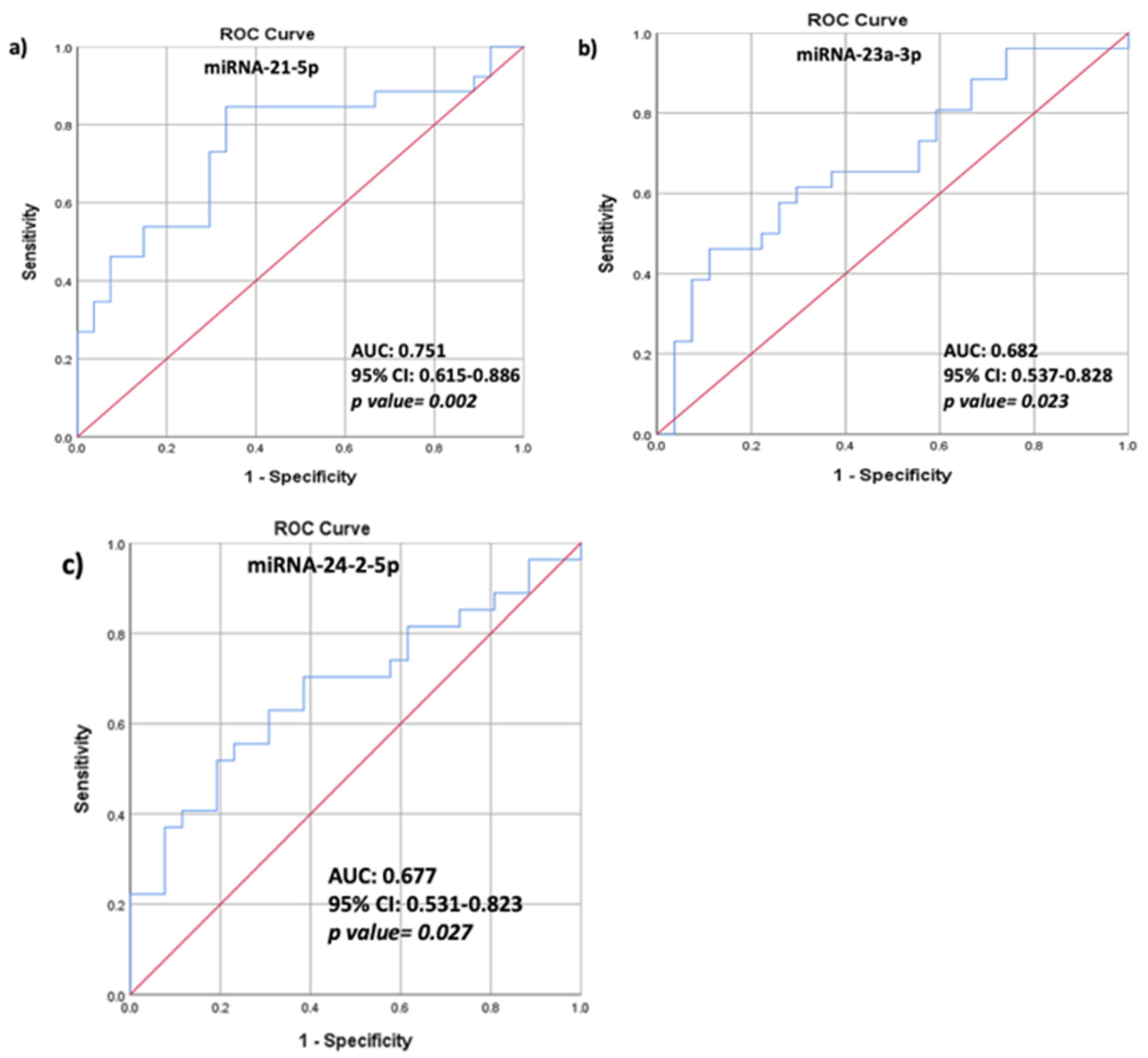
3.3. Correlations between HAART, BMD Values, TBS Values, Biochemical Characteristics, and Relative Serum miRNA Expression
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shaiykova, A.; Pasquet, A.; Goujard, C.; Lion, G.; Durand, E.; Bayan, T.; Lachatre, M.; Choisy, P.; Ajana, F.; Bourdic, K.; et al. Reduced bone mineral density among HIV-infected, virologically controlled young men: Prevalence and associated factors. AIDS 2018, 32, 2689–2696. [Google Scholar] [CrossRef] [PubMed]
- Gonciulea, A.; Wang, R.; Althoff, K.N.; Palella, F.J.; Lake, J.; Kingsley, L.A.; Brown, T.T. An increased rate of fracture occurs a decade earlier in HIV+ compared with HIV− men. AIDS 2017, 31, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.B.; Gerstoft, J.; Kronborg, G.; Larsen, C.S.; Pedersen, C.; Pedersen, G.; Obel, N. Incidence of low and high-energy fractures in persons with and without HIV infection: A Danish population-based cohort study. AIDS 2012, 26, 285–293. [Google Scholar] [CrossRef]
- Pramukti, I.; Lindayani, L.; Chen, Y.C.; Yeh, C.Y.; Tai, T.W.; Fetzer, S.; Ko, N.Y. Bone fracture among people living with HIV: A systematic review and meta-regression of prevalence, incidence, and risk factors. PLoS ONE 2020, 15, e0233501. [Google Scholar] [CrossRef]
- Compston, J. HIV infection and osteoporosis. BoneKey Rep. 2015, 4, 636. [Google Scholar] [CrossRef] [PubMed]
- Compston, J. HIV infection and bone disease. J. Intern. Med. 2016, 280, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Vikulina, T.; Fan, X.; Yamaguchi, M.; Roser-Page, S.; Zayzafoon, M.; Guidot, D.M.; Ofotokun, I.; Weitzmann, M.N. Alterations in the immuno-skeletal interface drive bone destruction in HIV-1 transgenic rats. Proc. Natl. Acad. Sci. USA 2010, 107, 13848–13853. [Google Scholar] [CrossRef] [PubMed]
- Titanji, K.; Vunnava, A.; Sheth, A.N.; Delille, C.; Lennox, J.L.; Sanford, S.E.; Foster, A.; Knezevic, A.; Easley, K.A.; Weitzmann, M.N.; et al. Dysregulated B cell expression of RANKL and OPG correlates with loss of bone mineral density in HIV infection. PLoS Pathog 2014, 10, e1004497. [Google Scholar] [CrossRef]
- De Menezes, E.G.; Machado, A.A.; Barbosa, F., Jr.; de Paula, F.J.; Navarro, A.M. Bone metabolism dysfunction mediated by the increase of proinflammatory cytokines in chronic HIV infection. J. Bone Miner. Metab. 2017, 35, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Raynaud-Messina, B.; Bracq, L.; Dupont, M.; Souriant, S.; Usmani, S.M.; Proag, A.; Pingris, K.; Soldan, V.; Thibault, C.; Capilla, F.; et al. Bone degradation machinery of osteoclasts: An HIV-1 target that contributes to bone loss. Proc. Natl. Acad. Sci. USA 2018, 115, E2556–E2565. [Google Scholar] [CrossRef] [PubMed]
- Makras, P.; Boubouchairopoulou, N.; Katsarolis, I.; Athanasakis, K. Cost-effective osteoporosis treatment thresholds for people living with HIV infection in Greece. J. Musculoskelet. Neuronal. Interact. 2017, 17, 292–298. [Google Scholar]
- Makras, P.; Athanasakis, K.; Boubouchairopoulou, N.; Rizou, S.; Anastasilakis, A.D.; Kyriopoulos, J.; Lyritis, G.P. Cost-effective osteoporosis treatment thresholds in Greece. Osteoporos Int. 2015, 26, 1949–1957. [Google Scholar] [CrossRef] [PubMed]
- Yavropoulou, M.P.; Yovos, J.G. The “dark matter” of DNA and the regulation of bone metabolism: The role of non-coding RNAs. J. Musculoskelet Neuronal Interact. 2018, 18, 18–31. [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Gantier, M.P.; McCoy, C.E.; Rusinova, I.; Saulep, D.; Wang, D.; Xu, D.; Irving, A.T.; Behlke, M.A.; Hertzog, P.J.; Mackay, F.; et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011, 39, 5692–5703. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Haleyurgirisetty, M.; Lee, S.; Hewlett, I.; Devadas, K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine 2019, 43, 307–316. [Google Scholar] [CrossRef]
- Zhan, J.; Qin, S.; Lu, L.; Hu, X.; Zhou, J.; Sun, Y.; Yang, J.; Liu, Y.; Wang, Z.; Tan, N.; et al. miR-34a is a common link in both HIV- and antiretroviral therapy-induced vascular aging. Aging (Albany NY) 2016, 8, 3298–3310. [Google Scholar] [CrossRef]
- Cheng, K.; Rai, P.; Plagov, A.; Lan, X.; Mathieson, P.W.; Saleem, M.A.; Husain, M.; Malhotra, A.; Singhal, P.C. Rapamycin-induced modulation of miRNA expression is associated with amelioration of HIV-associated nephropathy (HIVAN). Exp. Cell Res. 2013, 319, 2073–2080. [Google Scholar] [CrossRef][Green Version]
- Cheng, K.; Rai, P.; Plagov, A.; Lan, X.; Subrati, A.; Husain, M.; Malhotra, A.; Singhal, P.C. MicroRNAs in HIV-associated nephropathy (HIVAN). Exp. Mol. Pathol. 2013, 94, 65–72. [Google Scholar] [CrossRef][Green Version]
- Van Wijnen, A.J.; van de Peppel, J.; van Leeuwen, J.P.; Lian, J.B.; Stein, G.S.; Westendorf, J.J.; Oursler, M.J.; Im, H.J.; Taipaleenmaki, H.; Hesse, E.; et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr. Osteoporos Rep. 2013, 11, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, C.; Karpinski, K.; Haug, A.T.; Vester, H.; Schmitt, A.; Bauer, J.S.; van Griensven, M. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 2014, 29, 1718–1728. [Google Scholar] [CrossRef]
- Yavropoulou, M.P.; Anastasilakis, A.D.; Makras, P.; Tsalikakis, D.G.; Grammatiki, M.; Yovos, J.G. Expression of microRNAs that regulate bone turnover in the serum of postmenopausal women with low bone mass and vertebral fractures. Eur. J. Endocrinol. 2017, 176, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Anastasilakis, A.D.; Makras, P.; Pikilidou, M.; Tournis, S.; Makris, K.; Bisbinas, I.; Tsave, O.; Yovos, J.G.; Yavropoulou, M.P. Changes of circulating MicroRNAs in response to treatment with teriparatide or denosumab in postmenopausal osteoporosis. J. Clin. Endocrinol. Metab. 2018, 103, 1206–1213. [Google Scholar] [CrossRef]
- Yavropoulou, M.P.; Anastasilakis, A.D.; Makras, P.; Papatheodorou, A.; Rauner, M.; Hofbauer, L.C.; Tsourdi, E. Serum profile of microRNAs linked to bone metabolism during sequential treatment for postmenopausal osteoporosis. J. Clin. Endocrinol. Metab. 2020, 105. [Google Scholar] [CrossRef] [PubMed]
- Yavropoulou, M.P.; Vaios, V.; Makras, P.; Georgianos, P.; Batas, A.; Tsalikakis, D.; Tzallas, A.; Ntritsos, G.; Roumeliotis, S.; Eleftheriadis, T.; et al. Expression of circulating MicroRNAs linked to bone metabolism in chronic kidney disease-mineral and bone disorder. Biomedicines 2020, 8, 601. [Google Scholar] [CrossRef]
- Sugatani, T.; Vacher, J.; Hruska, K.A. A microRNA expression signature of osteoclastogenesis. Blood 2011, 117, 3648–3657. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Z.; Wang, J.; Ji, X.; Yao, Z.; Wang, X. miR21 promotes osteoclastogenesis through activation of PI3K/Akt signaling by targeting Pten in RAW264.7 cells. Mol. Med. Rep. 2020, 21, 1125–1132. [Google Scholar] [CrossRef]
- Hu, C.H.; Sui, B.D.; Du, F.Y.; Shuai, Y.; Zheng, C.X.; Zhao, P.; Yu, X.R.; Jin, Y. miR-21 deficiency inhibits osteoclast function and prevents bone loss in mice. Sci. Rep. 2017, 7, 43191. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.Q.; Gordon, J.A.; Beloti, M.M.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA 2010, 107, 19879–19884. [Google Scholar] [CrossRef]
- Dai, Y.; Zheng, C.; Li, H. Inhibition of miR-23a-3p promotes osteoblast proliferation and differentiation. J. Cell Biochem. 2019. [Google Scholar] [CrossRef]
- Zhao, W.; Wu, C.; Dong, Y.; Ma, Y.; Jin, Y.; Ji, Y. MicroRNA-24 Regulates osteogenic differentiation via targeting T-Cell factor-1. Int. J. Mol. Sci. 2015, 16, 11699–11712. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, L.; Hu, J.; Zhang, L.; Liao, L.; Liu, S.; Wu, D.; Yang, P.; Shen, L.; Chen, J.; et al. MiR-26a Rescues bone regeneration deficiency of mesenchymal stem cells derived from osteoporotic mice. Mol. Ther. 2015, 23, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Kapinas, K.; Kessler, C.B.; Delany, A.M. miR-29 suppression of osteonectin in osteoblasts: Regulation during differentiation and by canonical Wnt signaling. J. Cell Biochem. 2009, 108, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Mi, W.; Shi, Q.; Chen, X.; Wu, T.; Huang, H. miR-33a-5p modulates TNF-alpha-inhibited osteogenic differentiation by targeting SATB2 expression in hBMSCs. FEBS Lett. 2016, 590, 396–407. [Google Scholar] [CrossRef]
- Aslani, S.; Abhari, A.; Sakhinia, E.; Sanajou, D.; Rajabi, H.; Rahimzadeh, S. Interplay between microRNAs and Wnt, transforming growth factor-beta, and bone morphogenic protein signaling pathways promote osteoblastic differentiation of mesenchymal stem cells. J. Cell Physiol. 2019, 234, 8082–8093. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, H.J.; Park, C.K.; Kim, Y.G.; Lee, H.J.; Kim, J.Y.; Kim, H.H. MicroRNA-124 regulates osteoclast differentiation. Bone 2013, 56, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, Y.; Shiozaki, Y.; Sugimoto, Y.; Takigawa, T.; Tanaka, M.; Matsukawa, A.; Ozaki, T. miRNA-133a-5p Inhibits the Expression of Osteoblast Differentiation-Associated Markers by Targeting the 3′ UTR of RUNX2. DNA Cell Biol. 2018, 37, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Y.; Zhang, Y.; Ma, L.; Lin, L.; Meng, J.; Jiang, L.; Wang, L.; Zhou, P.; Zhang, Y. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharm. 2017, 89, 1178–1186. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, 169–173. [Google Scholar] [CrossRef]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional microRNA targets in protein coding sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jang, I.; Jun, Y.; Yoon, S.; Ko, M.; Kwon, Y.; Choi, I.; Chang, H.; Ryu, D.; Lee, B.; et al. MiRGator v3.0: A microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013, 41, 252–257. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [PubMed]
- Yavropoulou, M.P.; Vaios, V.; Pikilidou, M.; Chryssogonidis, I.; Sachinidou, M.; Tournis, S.; Makris, K.; Kotsa, K.; Daniilidis, M.; Haritanti, A.; et al. Bone quality assessment as measured by trabecular bone score in patients with end-stage renal disease on dialysis. J. Clin. Densitom. 2017, 20, 490–497. [Google Scholar] [CrossRef]
- Swaminathan, S.; Murray, D.D.; Kelleher, A.D. The role of microRNAs in HIV-1 pathogenesis and therapy. AIDS 2012, 26, 1325–1334. [Google Scholar] [CrossRef]
- Reynoso, R.; Laufer, N.; Hackl, M.; Skalicky, S.; Monteforte, R.; Turk, G.; Carobene, M.; Quarleri, J.; Cahn, P.; Werner, R.; et al. MicroRNAs differentially present in the plasma of HIV elite controllers reduce HIV infection in vitro. Sci. Rep. 2014, 4, 5915. [Google Scholar] [CrossRef]
- Wei, F.; Yang, S.; Guo, Q.; Zhang, X.; Ren, D.; Lv, T.; Xu, X. MicroRNA-21 regulates osteogenic differentiation of periodontal ligament stem cells by targeting Smad5. Sci. Rep. 2017, 7, 16608. [Google Scholar] [CrossRef] [PubMed]
- Erlandson, K.M.; O’Riordan, M.; Labbato, D.; McComsey, G.A. Relationships between inflammation, immune activation, and bone health among HIV-infected adults on stable antiretroviral therapy. J. Acquir. Immune. Defic. Syndr. 2014, 65, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Rey, D.; Treger, M.; Sibilia, J.; Priester, M.; Bernard-Henry, C.; Cheneau, C.; Javier, R.M. Bone mineral density changes after 2 years of ARV treatment, compared to naive HIV-1-infected patients not on HAART. Infect. Dis. 2015, 47, 88–95. [Google Scholar] [CrossRef]
- Serrano, S.; Marinoso, M.L.; Soriano, J.C.; Rubies-Prat, J.; Aubia, J.; Coll, J.; Bosch, J.; Del Rio, L.; Vila, J.; Goday, A.; et al. Bone remodelling in human immunodeficiency virus-1-infected patients. A histomorphometric study. Bone 1995, 16, 185–191. [Google Scholar] [CrossRef]

| Gene Symbol miScript Primer Assay (Catalog # Qiagen) | Predicted Target-Genes | miRNA Sequence | Predicted Mechanism of Action |
|---|---|---|---|
| hsa-miRNA-21-5p MS00009079 | SPRY1; PDCD4; FASLG | 5′UAGCUUAUCAGACUGAUGUUGA | Increases osteoclastogenesis and is up-regulated during RANKL-induced osteoclastogenesis [26,27,28]. |
| hsa-miRNA-23a-3p MS00031633 | RUNX2; SATB2 | 5′AUCACAUUGCCAGGGAUUUCC | Decreases osteoblastogenesis through inhibition of the RUNX2 gene [29,30]. |
| hsa-miRNA-24-2-5p MS00009205 | TCF-1; CALB1; SATB2 | 5′UGCCUACUGAGCUGAAACACAG | Decreases osteogenic differentiation through targeting the expression of transcription factor TCF-1 in osteoblastic cells [31]. |
| hsa-miRNA-26a-5p MS00029239 | ΤΟΒ1; IGF-1 | 5′UUCAAGUAAUCCAGGAUAGGCU | Increases bone formation through repressing TOB1 protein expression, negative regulator of BMP/SMAD signaling pathway [32]. |
| hsa-miRNA-29a-3p MS00003262 | SPARC | 5′UAGCACCAUCUGAAAUCGGUUA | Decreases osteonectin-bone matrix protein-synthesis [33]. |
| hsa-miRNA-33a-5p MS00003304 | SATB2; DKK-1; WIF1; OSTF1;β-catenin | 5′GUGCAUUGUAGUUGCAUUGCA | Decreases osteoblastogenesis through targeting SATB2 [34] and modulates Wnt signaling [35]. |
| hsa-miRNA-124-3p MS00006622 | NFATC1; NFATC2 | 5′UAAGGCACGCGGUGAAUGCC | Decreases osteoclastogenesis by suppressing the NFATc1 gene [36]. |
| hsa-miRNA-133a-3p MS00031423 | RUNX2; MEG3 | 5′UUUGGUCCCCUUCAACCAGCUG | Decreases osteoblastogenesis by targeting the RUNX2 gene [37] and modulates the expression of long non-coding RNA MEG3 [38]. |
| Cel-miRNA-39-3p MS00019789 | Spike-in control | 5′UCACCGGGUGUAAAUCAGCUUG | |
| hsa-SNORD95-11 | miScript PCR control | ||
| hsa-SNORD96A-11 | miScript PCR control | ||
| hsa-RNU6-2-1 | miScript PCR control |
| Parameters | HIV+/OP+ (Cases, n = 30) | HIV+/OP− (Controls, n = 30) | HIV− (n = 30) | p Value |
|---|---|---|---|---|
| Age (years) | 54.7 ± 8.9 | 52.6 ± 6.0 | 54.7 ± 5.4 | NS |
| BMI (kg/m2) | 27.5 ± 2.5 | 23.5 ± 3.7 | 25.5 ± 1.53 | NS |
| Smoking,n (%) | NS | |||
| Duration of HAART (yrs) | 11.1 ± 6.6 | 12.6 ± 5.4 | ΝΑ | NS |
| Duration of treatment with NRTIs (yrs) | 10.9 ± 6.2 | 11.7 ± 5.4 | ΝΑ | NS |
| Duration of treatment with TDF (yrs) | 6.6 ± 4.5 | 5.1 ± 3.2 | ΝΑ | NS |
| Duration of treatment with NNRTIs (yrs) | 2.9 ± 4.5 | 4.9 ± 5.5 | ΝΑ | NS |
| Duration of treatment with PIs (yrs) | 6.7 ± 4.8 | 5.4 ± 5.2 | ΝΑ | NS |
| Duration of treatment with INs (yrs) | 1.2 ± 2.0 | 1.2 ± 1.9 | ΝΑ | NS |
| LS T-score | −1.78 ± 1.1 | 0.67 ± 1.4 | −0.35 ± 1.13 | a |
| LS-BMD (g/m2) | 1.007 ± 0.14 | 1.305 ± 0.17 | 1.177 ± 0.13 | a |
| TBS | 1.24 ± 0.12 | 1.279 ± 0.13 | 1.291 ± 0.15 | a |
| LFN T-score | −1.81 ± 1.0 | 0.14 ± 0.85 | −0.68 ± 0.78 | a |
| LFN BMD (g/m2) | 0.810 ± 0.08 | 1.05 ± 0.10 | 1.000 ± 0.11 | a |
| LH T-score | −1.71 ± 0.69 | 0.26 ± 0.77 | −0.61 ± 0.81 | a |
| LH BMD (g/m2) | 0.892 ± 0.16 | 1.152 ± 0.14 | 1.013 ± 0.11 | a |
| RFN T-score | −2.01 ± 0.62 | −0.48 ± 0.6 | −0.38 ± 0.93 | a |
| RFN BMD (g/m2) | 0.804 ± 0.07 | 1.059 ± 0.09 | 0.962 ± 0.09 | a |
| RH T-score | −1.77 ± 0.64 | 0.14 ± 0.65 | −1.00 ± 0.93 | a |
| RH BMD (g/m2) | 1.053 ± 0.84 | 1.119 ± 0.09 | 0.939 ± 0.12 | a |
| Serum creatinine (NR: 0.5–1.2 mg/dL) | 1.0 ± 0.2 | 0.9 ± 0.1 | 1.0 ± 0.1 | NS |
| Serum calcium # (NR:8.2–10.6 mg/dL) | 9.2 ± 0.43 | 9.3 ± 0.4 | 9.2 ± 0.23 | NS |
| Serum phosphate (NR:2.7–4.5 mg/dL) | 2.9 ± 0.59 | 2.9 ± 0.6 | 3.2 ± 0.45 | NS |
| Intact PTH (NR:10–65 pg/mL) | 36.7 ± 12.56 | 44.2 ± 10.19 | 42.7 ± 15.1 | NS |
| Serum 25-OH-vitamin D (ng/mL) | 16.45 ± 7.66 | 16.5 ± 7.1 | 23.9 ± 16.3 | NS |
| Serum P1NP (ng/mL) | 43.8 ± 12.4 | 46.9 ± 11.9 | 35.7 ± 9.3 | NS |
| Serum β-CTX (ng/L) | 320 ± 119.2 | 330 ± 95.8 | 288 ± 116.1 | NS |
| Clinical/morphometric vertebral fractures | 0 | 0 | 0 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yavropoulou, M.P.; Kolynou, A.; Makras, P.; Pikilidou, M.; Nanoudis, S.; Skoura, L.; Tsachouridou, O.; Ntritsos, G.; Tzallas, A.; Tsalikakis, D.G.; et al. Circulating microRNAs Related to Bone Metabolism in HIV-Associated Bone Loss. Biomedicines 2021, 9, 443. https://doi.org/10.3390/biomedicines9040443
Yavropoulou MP, Kolynou A, Makras P, Pikilidou M, Nanoudis S, Skoura L, Tsachouridou O, Ntritsos G, Tzallas A, Tsalikakis DG, et al. Circulating microRNAs Related to Bone Metabolism in HIV-Associated Bone Loss. Biomedicines. 2021; 9(4):443. https://doi.org/10.3390/biomedicines9040443
Chicago/Turabian StyleYavropoulou, Maria P., Artemis Kolynou, Polyzois Makras, Maria Pikilidou, Sideris Nanoudis, Lemonia Skoura, Olga Tsachouridou, Georgios Ntritsos, Alexandros Tzallas, Dimitrios G. Tsalikakis, and et al. 2021. "Circulating microRNAs Related to Bone Metabolism in HIV-Associated Bone Loss" Biomedicines 9, no. 4: 443. https://doi.org/10.3390/biomedicines9040443
APA StyleYavropoulou, M. P., Kolynou, A., Makras, P., Pikilidou, M., Nanoudis, S., Skoura, L., Tsachouridou, O., Ntritsos, G., Tzallas, A., Tsalikakis, D. G., Tsave, O., Metallidis, S., & Chatzidimitriou, D. (2021). Circulating microRNAs Related to Bone Metabolism in HIV-Associated Bone Loss. Biomedicines, 9(4), 443. https://doi.org/10.3390/biomedicines9040443












