1. Introduction
Recently discovered NEET proteins are iron–sulfur (Fe–S) cluster-containing proteins that are present in all the kingdoms of life. Members of this family are characterized by at least one highly conserved 39-amino-acid motif called CDGSH Iron–Sulfur Domain (CISD) harboring a redox-active 2Fe–2S cluster coordinated by an unusual 3Cys–1His quartet instead of the usual 4Cys. In mammals, three NEET proteins have been identified: mitoNEET (also known as CISD1), CISD2 (NAF-1, ERIS, Noxp70 or Miner1) and CISD3 (Miner2 or MiNT) [
1,
2]. Even if their specific functions are still debated, they are clearly involved in various processes including ageing, autophagy, apoptosis and the regulation of calcium, iron metabolism, Fe–S cluster and reactive oxygen species (ROS) homeostasis. The molecular mechanisms underlying the involvement of NEET proteins in these multiple processes remain poorly understood [
2].
Several studies reveal that various types of cancers are accompanied by overexpression of NEET proteins [
3] and the CISD2 expression level has been proposed as an independent prognostic marker for survival in cancer patients including pancreatic [
4] and gastric cancers [
5]. More generally, the overexpression of NEET proteins promotes the proliferation of cancer cells, supports tumor growth and metastasis. Conversely, their depletion leads to decreased cancer cell proliferation in breast cancer [
6] and neuroblastomas [
7]. Recessive mutations in the
CISD2 gene coding for CISD2 are the causative factor for Wolfram syndrome 2 (or WFS2), a rare autosomal recessive neurodegenerative disorder leading to severe neurological disabilities and a drastic decrease in life span [
8].
MitoNEET and CISD2 are dimers that assemble one cluster per protomer whereas CISD3 is monomeric and assembles two clusters. While the latter resides inside the mitochondrial matrix, mitoNEET and CISD2 are respectively bound to the mitochondrial and endoplasmic reticulum (ER) membranes by a specific N-terminal anchor with the remaining parts of the protein, including the Fe–S cluster domain, laying into the cytosol [
2]. Their cytosolic domains share 65% identity and 79% similarity (
Figure 1) with a similar unique folding that exhibits two distinct domains: the Fe–S cluster binding domain and a β-cap domain [
2].
One of the most outstanding biochemical properties of the NEET proteins is their ability to transfer their cluster to acceptor apoproteins (cluster transfer reaction) [
9,
10]. By combining in vitro and in cellulo approaches, our recent data demonstrated the involvement of the human mitoNEET in a novel Fe–S trafficking pathway to quickly rebuild a cluster in IRP-1/cytosolic aconitase following an oxidative insult [
11]. Remarkably, the mitoNEET cluster is extremely stable when reduced, even at acidic pH, and can hardly be lost or transferred [
12]. However, it can be reversibly oxidized to [2Fe–2S]
2+ by hydrogen peroxide (H
2O
2) and be further reduced by biological thiols suggesting a redox sensory function of mitoNEET [
13,
14]. Only an oxidized mitoNEET cluster can be transferred in vitro to physiological cluster acceptors such as the cytosolic aconitase and to model cluster acceptors such as
Escherichia coli ferredoxin (FDX) [
12]. Finally, we found that the rate of mitoNEET cluster transfer is highly pH dependent because a much faster reaction occurs at slightly acidic pHs [
15]. All these results led us to propose that mitoNEET was acting as a redox switch protein with a pH-dependent cluster transfer activity controlled by the redox state of its Fe–S cluster [
15,
16].
CISD2 has been much less characterized than mitoNEET at the biochemical level and the transfer of CISD2 cluster to acceptor proteins was only shown in vitro using hyperthermophilic ferredoxin [
10] and human CIAPIN1 [
17]. No CISD2 cluster recipient has been formally identified in cellulo to date. Since human NEET proteins are playing important roles in multiple essential biological processes and in various human diseases, there is an urgent need for a better knowledge, at both biochemical and cellular levels, of each NEET member. Combining cellular and biochemical approaches, the present study focused on human CISD2 and aimed to better delineate what distinguishes it from its closest homolog mitoNEET at the molecular level. We found out that mitoNEET and CISD2 show different expression profiles in mouse tissues. Moreover, CISD2 is highly stable in cells and little sensitive to iron chelator treatments, known otherwise to lead to rapid mitoNEET decay. In vitro, the decreased stability of oxidized CISD2 and mitoNEET clusters in the presence of oxygen (aerobiosis) are similar. In contrast, at acidic pHs, the CISD2 Fe–S cluster is much more stable and is transferred more slowly than the mitoNEET cluster. As a consequence, CISD2 is a poor cluster donor compared to mitoNEET. This work reveals unexpected major differences between human CISD2 and mitoNEET, two structurally close NEET proteins.
2. Materials and Methods
2.1. Protein Sequence Alignment
Pairwise sequence alignment of human mitoNEET and CISD2 was performed with the EMBOSS Water Website (
www.ebi.uk), that uses the Smith–Waterman algorithm to calculate the local alignment of two sequences. The protein comparison is presented with the “pair” output format. A space is introduced for a mismatch or a gap.
2.2. Cell Culture and Treatment
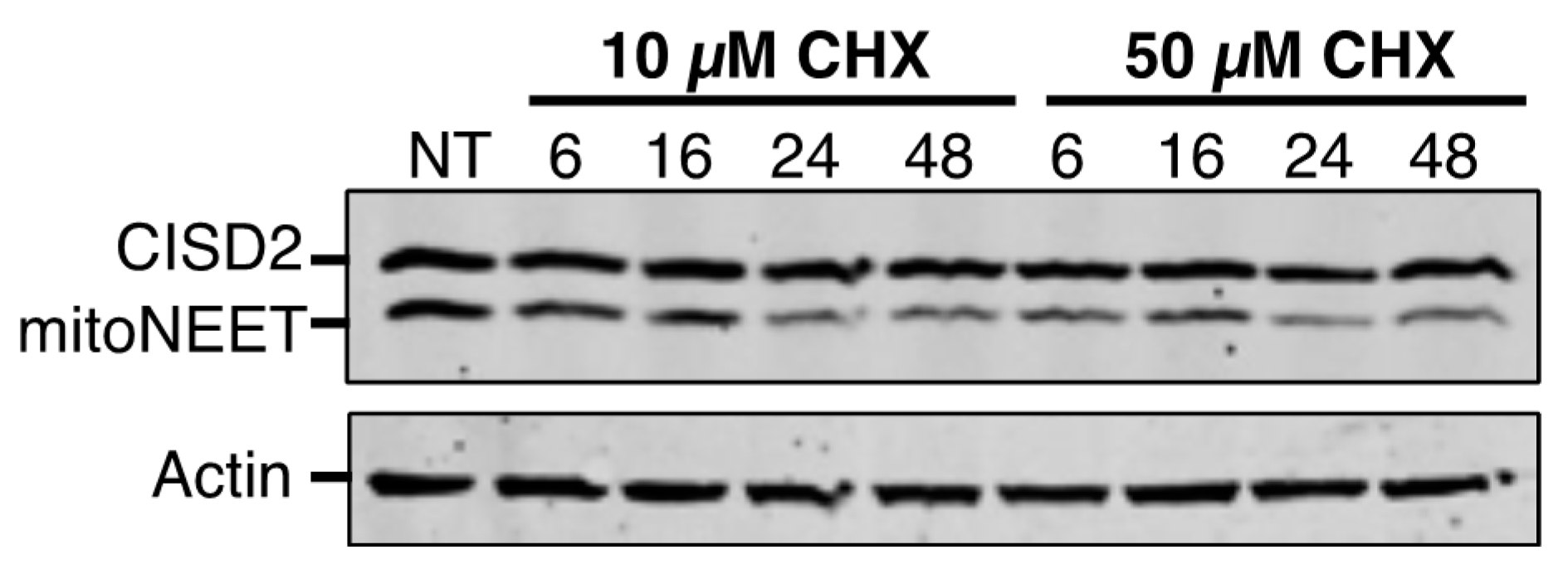
Human epithelial carcinoma (HeLa) cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Sigma–Aldrich, Saint Louis, MI, USA) containing 4.5 g/L glucose, 1 mM stable L-glutamine and supplemented with 1% penicillin–streptomycin and 10% fetal bovine serum (Lonza, Bâle, Switzerland) under 5% CO2 and humidified atmosphere. Cycloheximide (CHX, 10 and 50 µM) and desferrioxamine (DFO, 50 µM) were from Sigma–Aldrich. Salicylaldehyde isonicotinoyl hydrazone (SIH, 50 µM) was a kind gift from P. Ponka (McGill University, Montreal, QC, Canada).
2.3. Preparation of Cell Extracts and Immunoblot
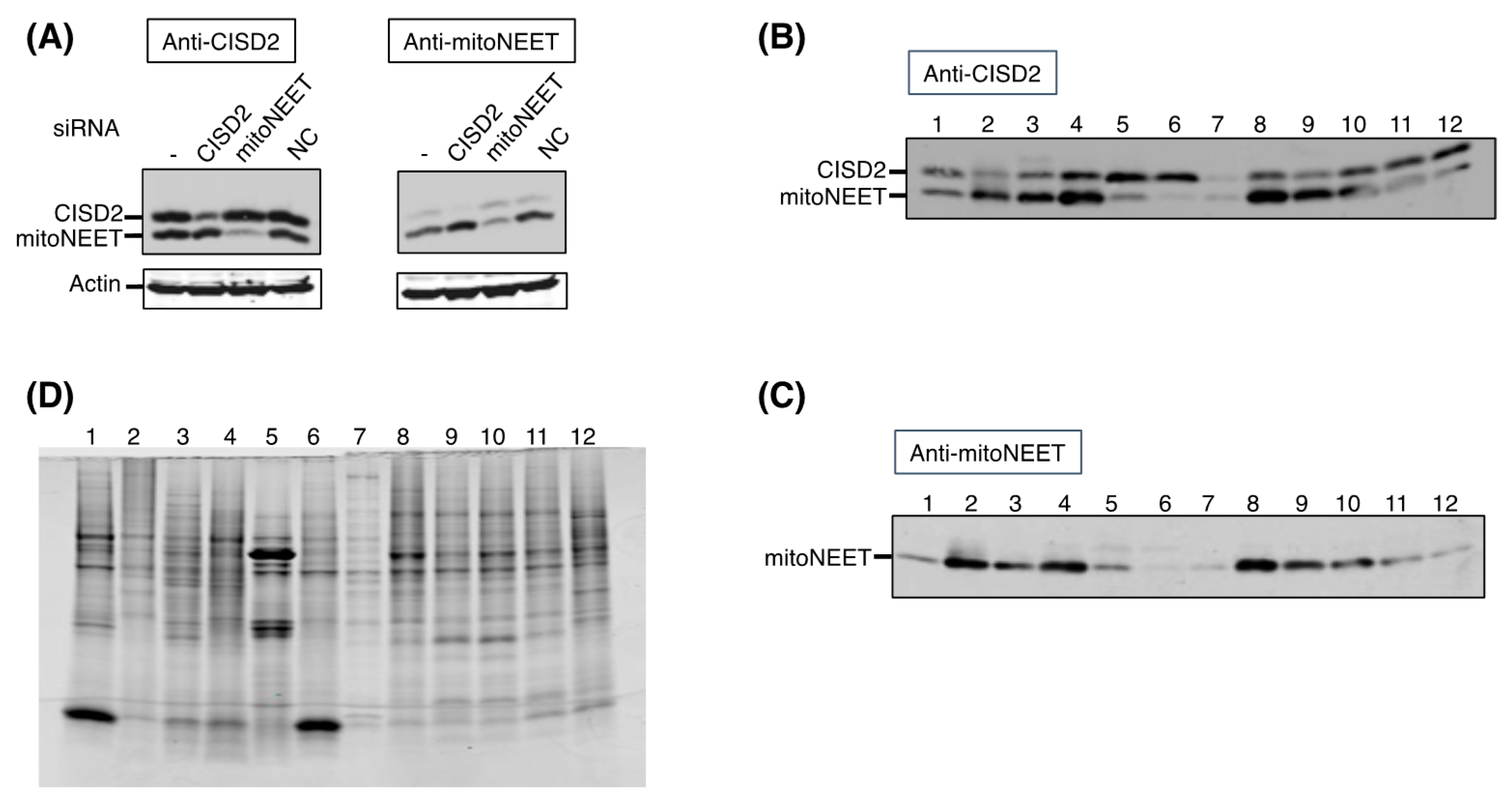
Total protein extracts from cultured HeLa cells were obtained by harvesting cells in Laemmli buffer (0.06 M Tris HCl pH 6.8, 10% glycerol, 2% SDS, protease inhibitors (Calbiochem, San Diego, CA, USA)). Protein concentrations were determined using the BCA method. Equal amounts of proteins were separated by SDS–PAGE and transferred on 0.45 µm PVDF membranes. The primary antibodies used were: anti-β-actin (Sigma–Aldrich #A5441), -CISD2 (Proteintech #13318-1-AP), -prohibitin (Novusbio #NBP2-37563), -mitoNEET (designed by Eurogentec). Secondary antibodies were anti-mouse and anti-rabbit fluorescent IRDye 800CW (LI-COR) and membranes were scanned with an Odyssey® Imaging System (LI-COR, Lincoln, NE, USA).
2.4. siRNA Transfections
HeLa cells were seeded at 3.5 × 105 cells percm2, incubated overnight and transfected with siRNA duplexes using INTERFERinTM (Polyplus Transfection, Illkirch, France) according to the manufacturer’s recommendations (Life Technologies®, Calsbad, CA, USA). For incubations with siRNA longer than three days, cells were re-transfected. The siRNA duplexes were from Life Technologies®: mitoNEET (s31650), cisd2 (s54620), negative control (4390843) and used at a final concentration of 10 nM.
2.5. Protein Expression Levels in Mouse Tissues
Samples were prepared as previously described (1). Of each tissue, 30–100 mg was sonicated in 1 mL of Buffer A (50 mM NaCl, 25 mM Tris HCl pH 7.5, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.5% Triton X-100) with cocktail protease inhibitors (Sigma–Aldrich) on ice for 15 s and then centrifuge at 14,000× g at 4 °C for 15 min. Protein content of supernatant was determined using the BCA protein assay. Ten μg of protein extracts were loaded on three 16% reducing polyacrylamide SDS–PAGE gels. Two gels were transferred on PVDF membranes for immunoblotting while the third one was stained using the SimplyBlue SafeStain kit (ThermoFisher Scientific, Waltham, MA, USA).
2.6. Purification of Holo-CISD257–135
A construct missing the 56 N-terminal amino acids of Homo sapiens (Hs) CISD2 (NP_001008389.1) was expressed using pET28b (GenScript). The open reading frame coding for CISD257–135 was cloned between NdeI and BamHI sites. To reduce protein aggregation, a C92S mutation was introduced (2). The Hs CISD257–135-C92S form will be referred to as CISD2s in this manuscript for ease of reading. The purified protein contains a thrombin cleavable N-terminal His-tag sequence (MGSSHHHHHHSSGLVPRGSHM), which was added to the N-terminal end of the protein. After thrombin cleavage, extra GSHM residues remain at the N-terminal end of CISD2s. The protein was expressed in BL21(DE3) cells cultured in LB media. When the culture reached an OD600nm of 0.6, 500 μM FeCl3 was added and the temperature was decreased to 20 °C. After 20 min incubation, protein expression was induced at 20 °C by addition of 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 24 h. Cells were harvested for purification steps performed at room temperature and aerobic conditions. Purification buffers were 0.45 µm filtered and degassed. Cell pellets from a 2 L-culture were resuspended in 100 mL of Lysis buffer (20 mM Tris HCl pH 8 500 mM NaCl, 30 mM imidazole) containing antiprotease cocktail (Sigma Fast, Sigma–Aldrich). Cells were lysed using a cell disruption system (Constant Systems Ltd., Daventry, UK). After centrifugation at 30,000 rpm for 1 h, the cleared lysate was applied on an Ni–NTA column using an Äkta FPLC (GE Healthcare, Chicago, IL, USA) and the column washed with 20 mL of Lysis buffer. Cleavage of the His-tag was performed using 100 NIH U of human thrombin (Sigma–Aldrich) in 25 mM Tris HCl pH 8, 500 mM NaCl, 2.5 mM CaCl2 for one night at 4 °C. The cleaved protein was eluted using a Lysis buffer. Colored fractions were concentrated to 5 mL using an Amicon Ultra 15 mL 3K (Sigma-Aldrich) concentrator. Then, proteins were loaded on a Superdex 75 HiLoad 16/60 size exclusion column (GE Healthcare) equilibrated with buffer A (25 mM Tris HCl pH 8, 100 mM NaCl). Eluted fractions were analyzed on a 16%, SDS–PAGE gel, pooled and concentrated. Finally, the proteins were loaded on a HiTrap CaptoSP ImpRes 1 mL (GE Healthcare) equilibrated with buffer B (20 mM Tris HCl pH 8) and eluted using a 40 mL linear gradient between buffer B and buffer C (20 mM Tris HCl pH 8, 1 M NaCl). Fractions were pooled, concentrated, aliquoted and stored at −80 °C. Protein concentration was measured using the Bradford assay (Bradford protein Assay, Biorad, Hercules, CA, USA) with bovine serum albumin (BSA) as standard. Protein purity was assessed to be >99% using SDS–PAGE and immunoblot using anti-His-tag (ThermoFisher #MA1-21315) and anti-CISD2 antibodies. For holo-CISD2, the optical A280nm/ A458nm ratio was near 2.4. Apo-CISD2 was obtained by loss of the cluster of holo-CISD2 at 37 °C under aerobic condition in Bis–Tris buffer at pH 6.2.
2.7. Purification of mitoNEET and Preparation of apo-FDX
Human mitoNEET44–108 was purified as previously described (3). E. coli FDX was expressed from the pET21b plasmid (a gift from Dr S. Ollagnier de Choudens, Grenoble, France) and purified as holo-FDX as described previously (1). Protein purity was assessed to be >99% using SDS–PAGE. Apo-FDX was prepared by heat cluster disassembly of purified holo-FDX in the presence of 10 mM dithiothreitol (DTT) and 10 mM EDTA followed by purification on a NAP-5 column (GE Healthcare) equilibrated with 50 mM Tris HCl pH 7, 100 mM NaCl. Protein concentrations were measured using the Bradford assay with BSA as standard.
2.8. Circular Dichroism
CD spectra were recorded with a Jasco (Lisse, France) J-810 spectropolarimeter at 25 °C using 20 µM protein solutions in quartz cuvettes with a pathway of 0.1 cm for spectra between 190 and 300 nm and 0.2 cm for spectra between 300 and 700 nm. For each spectrum, 25 accumulations were made. The spectrum of the buffer was recorded with 15 accumulations and subtracted to sample spectra, which was carried out in 10 mM Tris HCl pH 8 for holoproteins. Apoproteins were prepared by incubation overnight at 37 °C under aerobic conditions after buffer exchange to 10 mM Bis–Tris pH 6.2. CD scans from 300 to 600 nm were collected to analyze signature cluster-bound protein peaks at a scan rate of 50 nm/min. Data were processed by use of JASCO Spectra Manager II Analysis. Secondary structure composition was evaluated from the CD spectra using online Bestsel algorithm.
2.9. In Vitro CISD2 Cluster Loss and Transfer Reactions
Reaction buffers were all composed of 100 mM NaCl and 50 mM Bis–Tris (pH 5.8, 6.2 and 6.7) or Tris HCl (for pH higher than 7). For cluster transfer reaction, apo-FDX was pre-incubated with 5 mM DTT for 30 min at room temperature under anaerobic conditions to ensure cysteine reduction. Cluster loss and transfer reactions were followed by UV-visible absorption or migration of aliquots taken at defined times (1 mM DTT was added to the samples before migration) on a 16% native PAGE colored with colloidal Coomassie staining as described in (1).
UV-visible absorption spectra were recorded between 240 and 900 nm with a Cary 100 (Agilent, Santa Clara, CA, USA) spectrophotometer equipped with a temperature control apparatus set to the desired temperature. For spectra taken under anaerobic conditions including the study of the effect of H
2O
2 on the stability of oxidized CISD2 cluster, the cuvette was prepared in a glove box (Jacomex, Dagneux, France) and closed with a septum. The 240–900 nm absorption spectra of the reaction mixture were recorded over time and corrected for baseline variations at 900 nm. For the cluster loss reaction, we paid attention to changes in absorbance at 460 nm, which is characteristic of oxidized CISD2 cluster. For cluster transfer reactions, we paid particular attention to changes in absorbance at 460 nm and 415 nm, which are characteristic of oxidized holo-CISD2 and holo-FDX, respectively. At time t, the extent of the loss of oxidized CISD2 cluster was determined using R(t) = A
460, whereas the extent of the cluster transfer was determined using the ratio R(t) = A
415/A
460. As previously described [
18], reaction progress at time t was estimated as (R(t)−R
initial)/(R
final−R
initial) with R
initial, the initial R value at time 0 and R
final the R value at the time necessary for reaction completion. If the transfer of Fe–S centers is not total, a theoretical value of R
final of 1.1 (ratio corresponding to oxidized holo-FDX) as theoretical maximum ratio was used.
4. Discussion
CISD2 and mitoNEET are two NEET proteins anchored to organelle membranes. While CISD2 is anchored at the ER membranes at the contact site with mitochondria, mitoNEET is localized at the outer mitochondrial membranes [
2]. However, without considering this difference in anchoring domain and resulting subcellular localization, mitoNEET and CISD2 appear to be very similar. Indeed, their cytosolic domains share high level of sequence identity and structural similarity [
2]. In the present study, we compared different characteristics of these two proteins to gain a better understanding of their specificity and mechanisms of action in the cell and highlighted for the first time major differences between these two closely related proteins.
We analyzed the protein level of CISD2 and mitoNEET in different mouse tissues (
Figure 2). Contradictory results came from two previous RT–PCR studies [
24,
25] and the CISD2 protein level had been measured only in a very limited number of organs [
26]. We found that, despite expression in many mouse tissues, neither CISD2 nor mitoNEET is ubiquitous. Moreover, their expression profile in tissues differs drastically. We speculate that such differences may reflect their distinct biological functions; notably in the pancreas, a high level of CISD2 expression is accompanied by a low level of mitoNEET. Since both proteins are involved in the regulation of mitochondrial calcium stores through distinct mechanisms [
27], the relative dependence of pancreatic cells on CISD2 for this essential regulation might help to understand why CISD2 deficiency in WFS2 patients leads to an early dysfunction of this tissue and diabetes. A similar expression profile in the spleen, where CISD2 also predominates over mitoNEET, suggests an as yet unanticipated possible involvement of CISD2 in immune cells. In contrast, mitoNEET is preferentially expressed in the liver, a key organ for the regulation of iron homeostasis. Interestingly, we previously proposed cytosolic aconitase as one physiological acceptor for the Fe–S cluster of mitoNEET [
18]. In fact, we previously established that the expression profile of mitoNEET in mouse tissues was similar to that of cytosolic aconitase. The iron-free form of cytosolic aconitase, IRP-1, controls the expression of different proteins involved in iron metabolism. Therefore, it makes sense to observe a strong expression of mitoNEET, deeply implicated in iron homeostasis, in the liver tissue. Remarkably, the expression profile of CISD2 drastically differs from that of cytosolic aconitase (compare
Figure 2 from this manuscript to
Figure 7 from reference [
18]), consistent with the idea that CISD2 is not a physiological Fe–S donor for cytosolic aconitase in vivo.
It was observed that the extinction of CISD2 expression in mouse embryonic fibroblasts induced a very strong overexpression of mitoNEET, thus supporting the hypothesis that mitoNEET and CISD2 might share redundant functions [
28]. However, this assumption has been questioned because tissues from CISD2
−/− mice did not show higher mitoNEET protein levels [
25]. The same is true in human epithelial breast cancer cells, where the extinction of either one did not affect the level of the other protein [
6]. In line with these latter findings, we did not detect increased amounts of mitoNEET or CISD2 when knocking down CISD2 or mitoNEET respectively, in HeLa cells. Taken together, our analysis of CISD2 and mitoNEET expression in different mouse tissues, as well as previously published results from our group and others, substantiate the idea that CISD2 and mitoNEET are much more than just redundant backups in important cellular processes. However, it cannot be excluded that in some specific tissues their activities may overlap at least partially and compensatory mechanisms might exist between these two NEET proteins.
The proteins’ stability and their sensitivity to iron chelation are a second major point of divergence between CISD2 and mitoNEET. The half-life of a protein ranges from 30 min for the least stable to more than 200 h for the most stable [
29]. We established that the half-life of CISD2 is longer than 48 h, which is over 8-fold more stable than mitoNEET in unstressed cells. The low turnover of CISD2 probably reflects its greater importance compared to mitoNEET in fundamental cellular processes, such as calcium transfer from the endoplasmic reticulum to the mitochondria, regulation of autophagy and implication in cell proliferation [
3]. On the opposite, the relative instability of mitoNEET might help the protein to function in adaptive cellular responses, for instance under oxidative stress or iron deficiency conditions. In fact, our results show a strikingly different behavior of mitoNEET and CISD2 in response to iron deprivation. In the presence of iron chelators, mitoNEET protein levels fall very rapidly while those of CISD2 are only slightly affected. Therefore, the Fe–S cluster of mitoNEET might be its Achilles heel with regard its stability, meaning that proteases might rapidly eliminate apo-mitoNEET. In fact, we have previously shown that iron deprivation caused a degradation of mitoNEET that was partially blocked by the addition of the proteasomal inhibitor MG132, suggesting a role of the proteasome in the regulation of the apo-mitoNEET half-life [
11]. The much higher stability of CISD2 under conditions of iron deficiency might be understood as resulting from a stronger binding of the Fe–S cluster to the protein, from a greater stability of the apoprotein in cells, or from both of these possibilities. It is thus tempting to speculate that, in contrast to mitoNEET, the relative insensitivity of the half-life of CISD2 to intracellular iron levels might indicate that CISD2 has, at most, only a minor role in the adaptive response to iron chelation. On the other hand, as iron chelators are able to induce a strong mitophagy response [
30], the catabolism of mitochondrial components in a mitophagic process might also contribute to the loss of the mitochondrial mitoNEET following iron deprivation. Because CISD2 is located in the ER compartment and mitoNEET in the mitochondria, the difference in subcellular localization between CISD2 and mitoNEET might also explain the higher stability of CISD2 compared to mitoNEET in response to intracellular iron depletion.
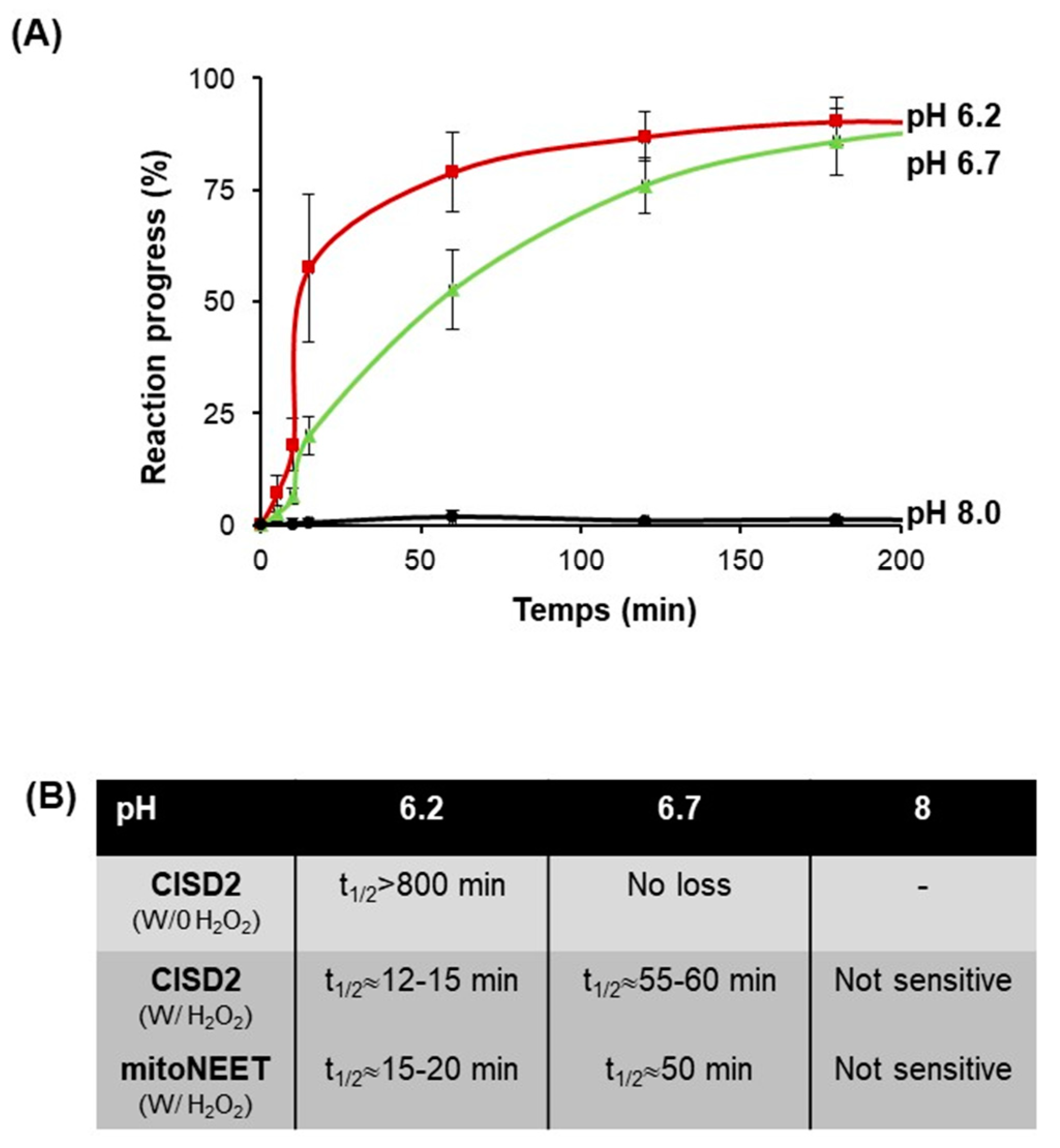
Our results with iron-deprived cells raised the question whether CISD2 stability in cells may be linked to the stability of its Fe–S cluster. We found that, like mitoNEET, CISD2 stability is significantly higher in anaerobiosis than in the presence of oxygen. Moreover, the oxidized Fe–S cluster has a high intrinsic stability (results under anaerobic conditions) even at acidic pHs, and its sensitivity to oxygen increases as the pH becomes more acidic. However, the stability of the oxidized mitoNEET cluster exhibited a much more pronounced pH dependency compared to CISD2. At pH 7 and above, both clusters were very stable but the mitoNEET cluster was the most stable. At pH 6.4, the Fe–S clusters of both proteins were equally stable in the presence of oxygen. Then, at pH below 6.4, the stability of the mitoNEET cluster dropped while that of CISD2 was only slightly affected. These two Fe–S clusters are similarly coordinated by 3Cys and 1His. However, cluster ligands are in turn stabilized by a complex network of hydrogen bonds with more distant residues that differs slightly between the two proteins [
2]. Investigations are underway to identify those residues in the secondary coordination sphere that might be involved in the stabilization of the CISD2 cluster in the presence of oxygen and its low pH dependency. Interestingly, induction of apoptosis in breast cancer cells leads to CISD2 interaction with the inhibitor of apoptosis-stimulating protein of p53 (iASPP) and a structural model of holo-CISD2 in interaction with iASPP has been proposed [
31]. Prohibition of this interaction by a iASPP-derived peptide also inhibits apoptosis in these cells. On the basis of our present results, we can consider that the pH–independent stability of CISD2 cluster would prevent CISD2 unfolding and help maintaining the interaction of CISD2 with iASPP, facilitating apoptosis despite cytosol acidification.
We showed that under anaerobic conditions at pH 8, CISD2 and mitoNEET are not affected by the addition of H
2O
2 (
Figure 7) while under the same conditions the clusters of the bacterial IscU, involved in Fe–S cluster maturation, and of SufB, also involved in Fe–S maturation but under stress conditions, are quickly degraded (t
1/2 lower than 5 and 10 min, respectively) [
22]. The resistance of the CISD2 cluster to H
2O
2 compared to other Fe–S proteins suggests that CISD2 functions involving its prosthetic group are likely to be preserved during oxidative stress. However, we noticed that at lower pHs, the CISD2 cluster became more sensitive to H
2O
2. Therefore, we cannot exclude that, under specific pathophysiological conditions leading to a strong decrease of the cytosolic pH, CISD2 cluster could be more sensitive to high concentrations of intracellular H
2O
2, resulting in either an increased degradation of the cluster or its transfer to a receiving protein.
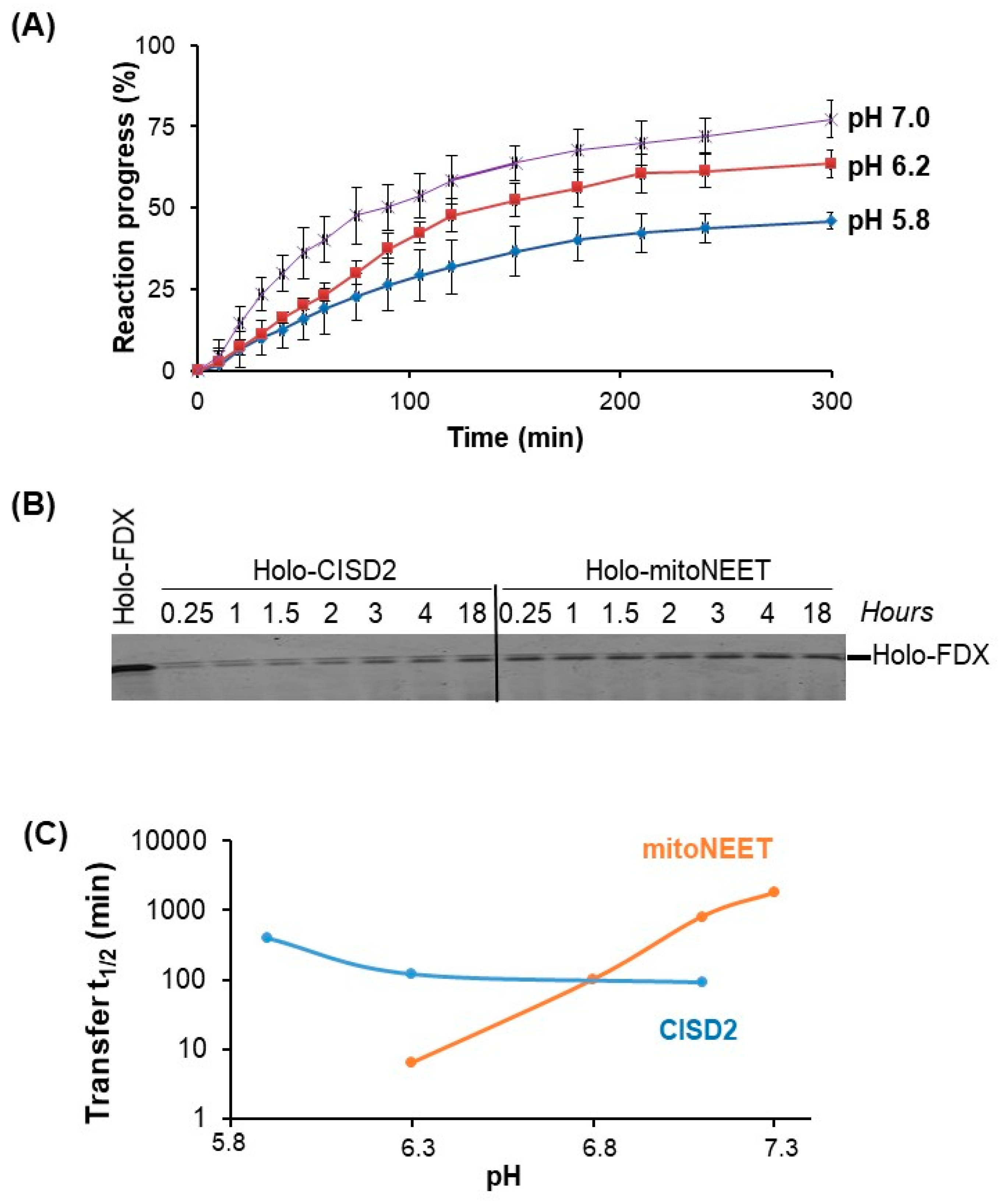
Finally, we investigated the ability of CISD2 to transfer its Fe–S cluster to a model apo-acceptor protein in vitro. Unlike mitoNEET, we found that the transfer rate was only moderately affected by the pH. Under our experimental conditions, we never obtained a complete transfer even after 300 min of incubation. At pH 7, the transfer rate was about 360 M
−1·min
−1 at room temperature, which is of the same order of magnitude as those published for the transfer of CISD2 cluster to an hyperthermophilic ferredoxin [
23] or to anamorsin [
17] at 37 °C. Comparing these values to those of common Fe–S cluster transfer proteins involved in Fe–S cluster biogenesis such as Grx5, IscU, NfuA, IscA, reveals that the kinetics of the transfer using CISD2 is very slow, noting that the in vitro transfer rate is typically between 1000 and 50,000 M
−1·min
−1 with the vast majority greater than 5000 M
−1·min
−1 at room temperature [
32]. We wondered if the transfer of the Fe–S cluster of CISD2, which was not completed after 5 h of reaction in our conditions, has any physiological significance. We have previously demonstrated that mitoNEET can repair a damaged cluster of cytosolic aconitase in cellulo after oxidative stress [
11]. Thus, the transfer of the mitoNEET cluster can occur in cellulo although the transfer of this cluster in vitro was also slow at a pH close to that of the cytosol (pH 7.4), while fast at a pH slightly below 6 [
15]. Thus, either the chaperone proteins might facilitate this process at cytosolic near neutral pH or the local pH near mitoNEET in cells might be slightly acidic, at least under certain physiological conditions favorable for the transfer of the cluster. During the induction of apoptosis, for example, the cytosolic pH can drop to around 6.3 [
33]. In the case of CISD2, the in vitro cluster transfer is not efficient at any pH suggesting that CISD2 could be only a weak cluster donor in cellulo. However, we cannot exclude the possibility that the slow reaction in vitro is due to an inappropriate and non-physiological acceptor protein (no acceptor protein of the Fe–S cluster of CISD2 has been identified in cellulo to date) or alternatively that this transfer requires the involvement of facilitating chaperone proteins. In conclusion, while they are very similar in sequence and structure, this study revealed unexpected major differences between CISD2 and mitoNEET regarding their expression in tissues and their cellular and biochemical features. Further investigations are required to decipher in detail their respective biological functions.