Mechanism of Resveratrol Dimers Isolated from Grape Inhibiting 1O2 Induced DNA Damage by UHPLC-QTOF-MS2 and UHPLC-QQQ-MS2 Analyses
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Sample Preparation
2.3. Quantitative Analysis of Target Compounds by UHPLC-QQQ-MS2
2.4. Qualitative Detection and Collection of Target Compounds by UHPLC-QTOF-MS2
2.5. Data Analysis
3. Results
3.1. Qualitative Analysis of Reactants and Products
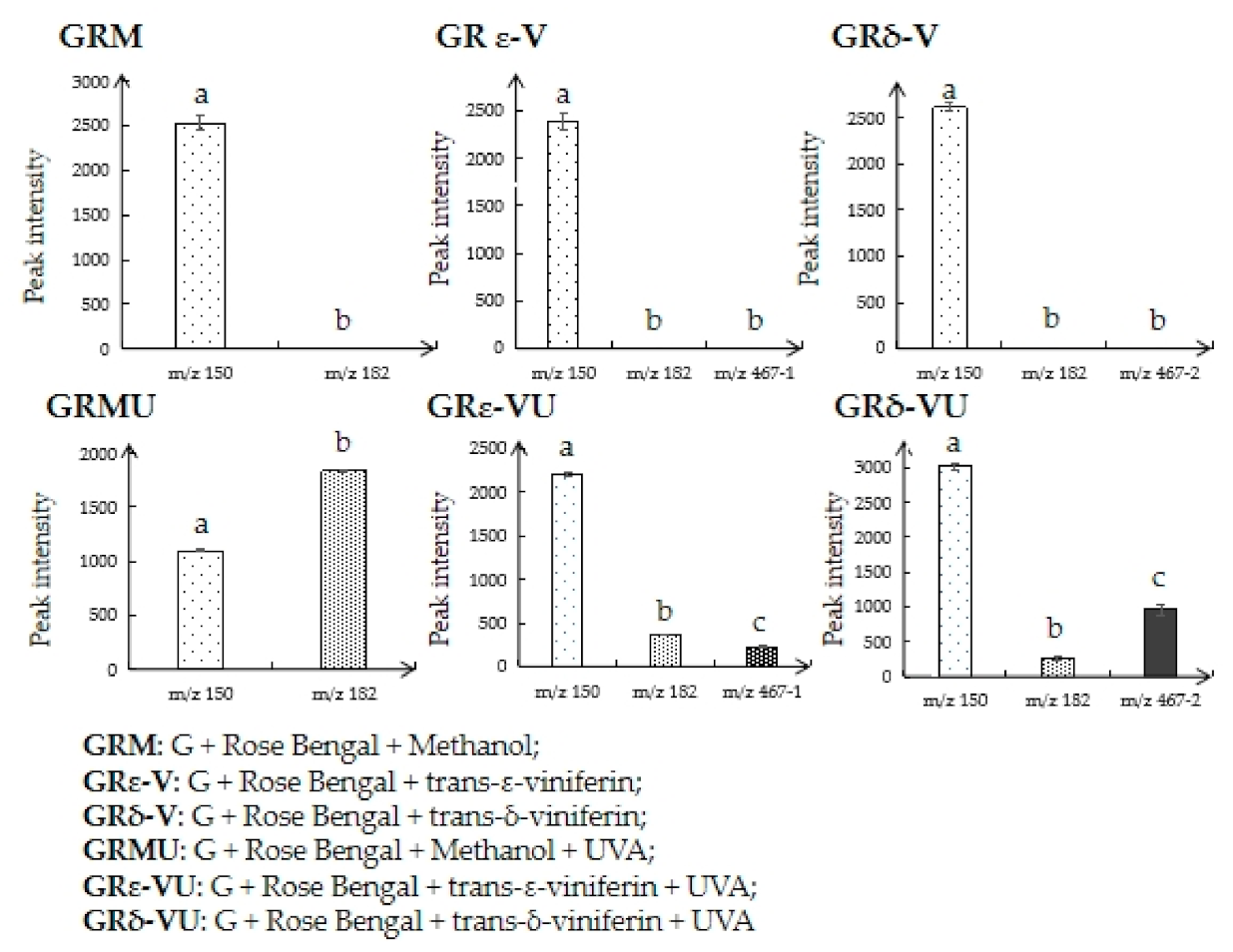
3.2. Quantitative Analysis of Reactants and Products (Characteristic Structure of Stilbenes Preventing 1O2 Induced DNA Damage)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative Stress and Antioxidant Defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef]
- DeFedericis, H.-C.; Patrzyc, H.B.; Rajecki, M.J.; Budzinski, E.E.; Iijima, H.; Dawidzik, J.B.; Evans, M.S.; Greene, K.F.; Box, H.C. Singlet Oxygen-Induced DNA Damage. Radiat. Res. 2006, 165, 445–451. [Google Scholar] [CrossRef]
- Zhang, Y.; Barnes, A.N.; Zhu, X.; Campbell, N.F.; Gao, R. Quantification of thiopurine/UVA-induced singlet oxygen production. J. Photochem. Photobiol. A Chem. 2011, 224, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Lemp, E.; Cañete, A.; Günther, G.; Pizarro, N.; Zanocco, A.L. Photosensitized generation of singlet molecular oxygen by aryloxazinones. J. Photochem. Photobiol. A Chem. 2008, 199, 345–352. [Google Scholar] [CrossRef]
- Stratton, S.P.; Liebler, D.C. Determination of Singlet Oxygen-Specific versus Radical-Mediated Lipid Peroxidation in Photosensitized Oxidation of Lipid Bilayers: Effect of β-Carotene and α-Tocopherol. Biochemistry 1997, 36, 12911–12920. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 2003, 305, 761–770. [Google Scholar] [CrossRef]
- Devasagayam, T.P.A.; Steenken, S.; Obendorf, M.S.W.; Schulz, W.A.; Sies, H. Formation of 8-hydroxy(deoxy)guanosine and generation of strand breaks at guanine residues in DNA by singlet oxygen. Biochemistry 1991, 30, 6283–6289. [Google Scholar] [CrossRef]
- Galano, A.; Tan, D.-X.; Reiter, R.J. Melatonin: A Versatile Protector against Oxidative DNA Damage. Molecules 2018, 23, 530. [Google Scholar] [CrossRef]
- Maynard, S.; Schurman, S.H.; Harboe, C.; De Souza-Pinto, N.C.; Bohr, V.A. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 2009, 30, 2–10. [Google Scholar] [CrossRef]
- Piette, J. New trends in photobiology: Biological consequences associated with DNA oxidation mediated by singlet oxygen. J. Photochem. Photobiol. B Biol. 1991, 11, 241–260. [Google Scholar] [CrossRef]
- Agnez-Lima, L.F.; Melo, J.T.; Silva, A.E.; Oliveira, A.H.S.; Timoteo, A.R.S.; Lima-Bessa, K.M.; Menck, C.F. DNA damage by singlet oxygen and cellular protective mechanisms. Mutat. Res. Rev. Mutat. Res. 2012, 751, 15–28. [Google Scholar] [CrossRef]
- Dumont, E.; Grüber, R.; Bignon, E.; Morell, C.; Moreau, Y.; Monari, A.; Ravanat, J.L. Probing the reactivity of singlet oxygen with purines. Nucleic Acids Res. 2016, 44, 56–62. [Google Scholar] [CrossRef]
- Lu, W.; Liu, J. Capturing transient endoperoxide in the singlet oxygen oxidation of guanine. Chem. Eur. J. 2016, 22, 3127–3138. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T.; Ravanat, J.L. Oxidatively generated damage to cellular DNA by UVB and UVA radiation. Photochem. photobiol. 2015, 91, 140–155. [Google Scholar] [CrossRef]
- Kino, K.; Morikawa, M.; Kobayashi, T.; Kobayashi, T.; Komori, R.; Sei, Y.; Miyazawa, H. The oxidation of 8-oxo-7, 8-dihydroguanine by iodine. Bioorgan. Med. Chem. Lett. 2010, 20, 3818–3820. [Google Scholar] [CrossRef]
- Thapa, B.; Munk, B.H.; Burrows, C.J.; Schlegel, H.B. Computational Study of Oxidation of Guanine by Singlet Oxygen (1Δg) and Formation of Guanine: Lysine Cross-Links. Chem. Eur. J. 2017, 23, 5804–5813. [Google Scholar] [CrossRef]
- Moriwaki, S.; Takahashi, Y.; Shimizu, H.; Inoue, M.; Sugiyama, Y.; Inoue, S. Decreased repair of singlet oxygen-induced DNA damage in xeroderma pigmentosum group A cells determined by plasmid host cell reactivation. J. Dermatol. Sci. 2012, 66, 242–244. [Google Scholar] [CrossRef] [PubMed]
- Rünger, T.M.; Möller, K.; Epe, B. Repair of ultraviolet B and singlet oxygen-induced DNA damage in xeroderma pigmentosum cells. J. Investig. Dermatol. 1995, 104, 68–73. [Google Scholar] [CrossRef]
- Hong, E.H.; Heo, E.Y.; Song, J.H.; Kwon, B.E.; Lee, J.Y.; Park, Y.; Ko, H.J. Trans-scirpusin A showed antitumor effects via autophagy activation and apoptosis induction of colorectal cancer cells. Oncotarget 2017, 8, 41401. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhou, Q.; Li, P.; Wang, Z.; Liu, S.; He, C.; Xiao, P. Update on phytochemistry and pharmacology of naturally occurring resveratrol oligomers. Molecules 2017, 22, 2050. [Google Scholar] [CrossRef] [PubMed]
- Vion, E.; Page, G.; Bourdeaud, E.; Paccalin, M.; Guillard, J.; Bilan, A.R. Trans ε-viniferin is an amyloid-β disaggregating and anti-inflammatory drug in a mouse primary cellular model of Alzheimer’s disease. Mol. Cell. Neurosci. 2018, 88, 1–6. [Google Scholar] [CrossRef]
- Kong, Q.; Ren, X.; Jiang, L.; Pan, Y.; Sun, C. Scirpusin A, a hydroxystilbene dimer from Xinjiang wine grape, acts as an effective singlet oxygen quencher and DNA damage protector. J. Sci. Food Agric. 2010, 90, 823–828. [Google Scholar] [CrossRef]
- Kong, Q.; Ren, X.; Qi, J.; Yu, J.; Lu, J.; Wang, S. Carbon-Carbon Double Bond and Resorcinol in Resveratrol and Its Analogues: What Is the Characteristic Structure in Quenching Singlet Oxygen? Biomolecules 2019, 9, 268. [Google Scholar] [CrossRef]
- Kong, Q.; Yin, X.; Yu, J.; Ren, X. Mechanistic processes of resveratrol in inhibiting the oxidative damage of guanine, as evidenced by UHPLC-MS2. J. Chromatogr. B 2018, 1093, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Xueyan, R.; Jia, Y.; Xuefeng, Y.; Lidan, T.; Qingjun, K. Isolation and purification of five phenolic compounds from the Xinjiang wine grape (Vitis Vinifera) and determination of their antioxidant mechanism at cellular level. Eur. Food Res. Technol. 2018, 244, 1569–1579. [Google Scholar] [CrossRef]
- Kong, Q.; Ren, X.; Hu, R.; Yin, X.; Jiang, G.; Pan, Y. Isolation and purification of two antioxidant isomers of resveratrol dimer from the wine grape by counter-current chromatography. J. Sep. Sci. 2016, 39, 2374–2379. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Yu, J.; Kong, Q.; Ren, X. Mechanism of isomers and analogues of resveratrol dimers selectively quenching singlet oxygen by UHPLC-ESI-MS2. Food Chem. 2017, 237, 1101–1111. [Google Scholar] [CrossRef]
- Slade, P.G.; Priestley, N.D.; Sugden, K.D. Spiroiminodihydantoin as an Oxo-Atom Transfer Product of 8-Oxo-2’-deoxyguanosine Oxidation by Chromium (V). Org. Lett. 2007, 9, 4411–4414. [Google Scholar] [CrossRef] [PubMed]
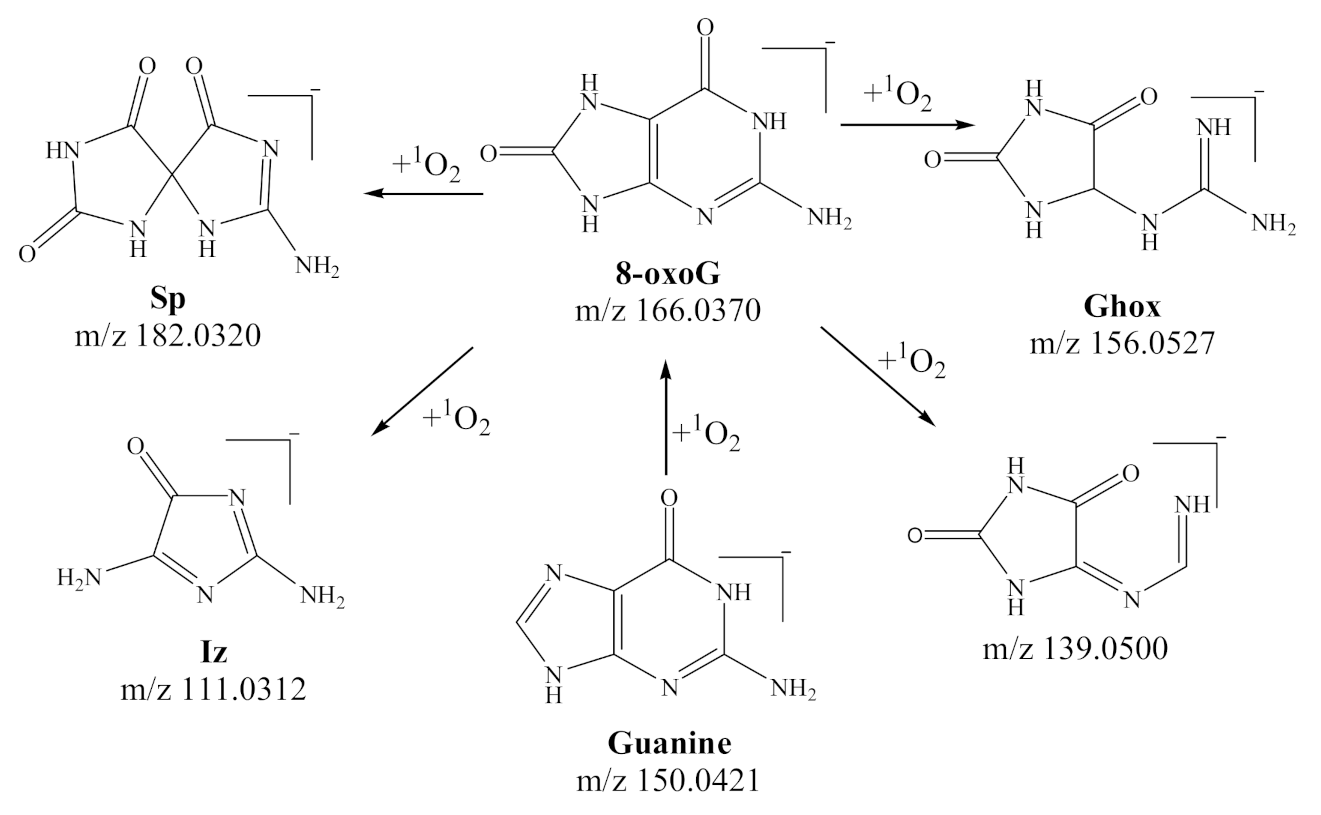
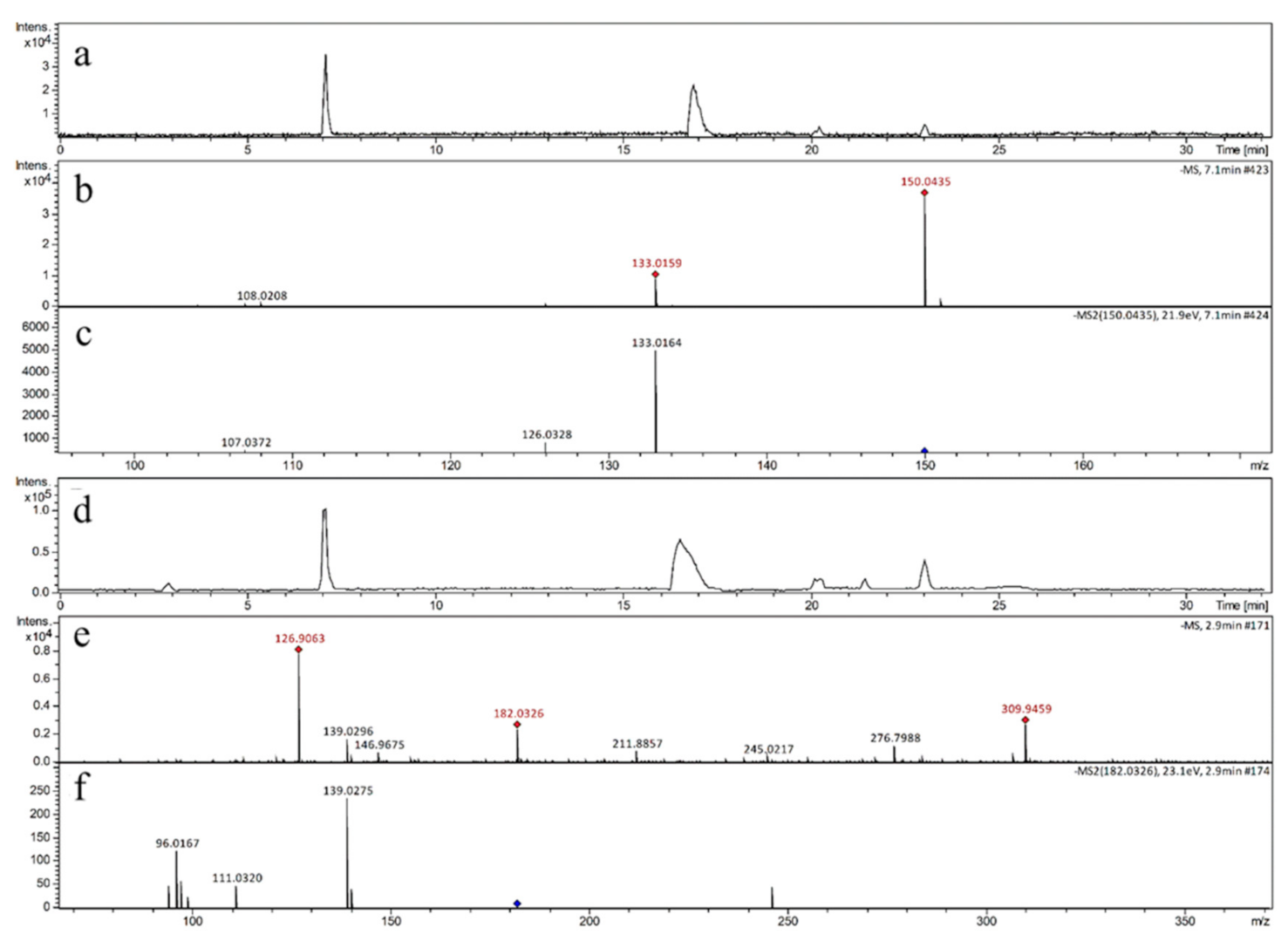
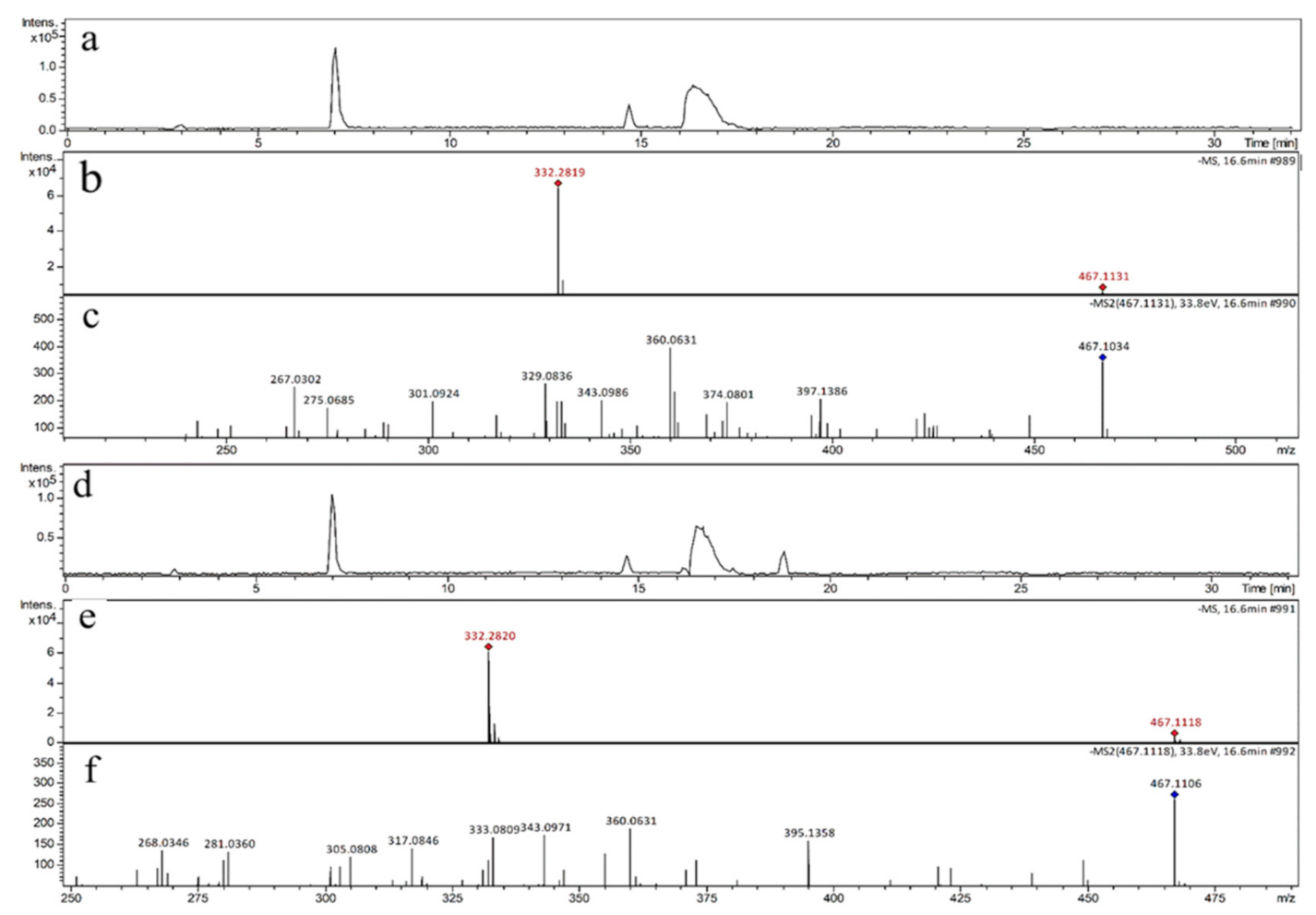

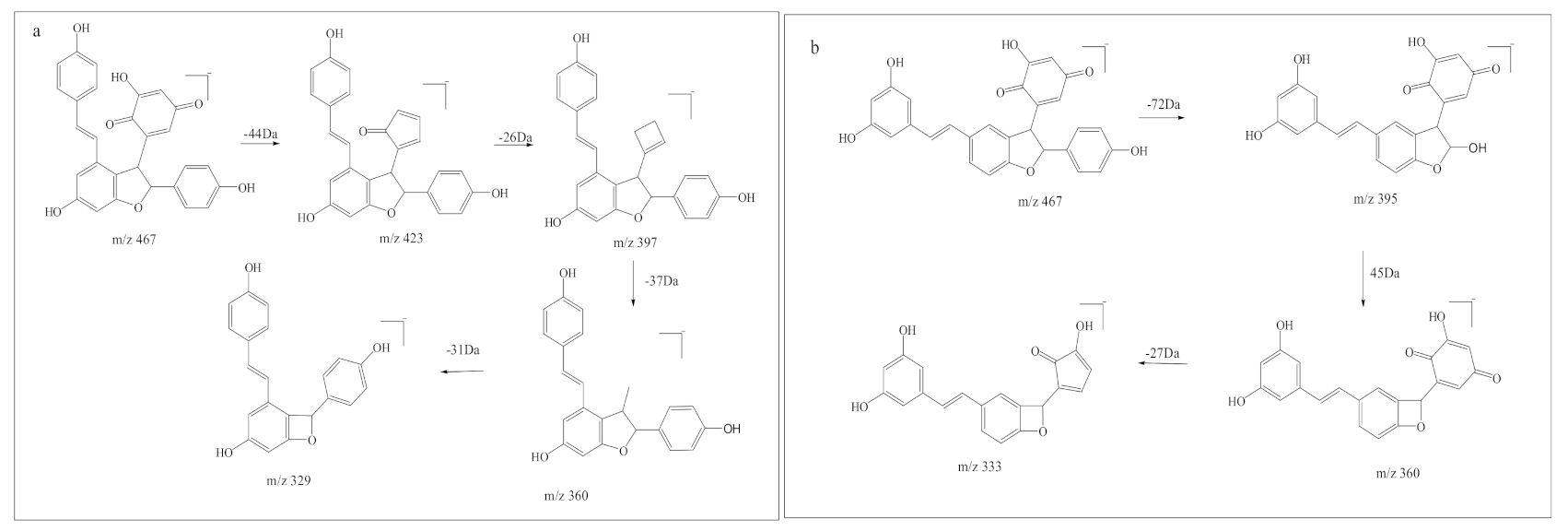
| Precursor Ion | Product Ion | Fragmentor (V) | Collision Energy (eV) |
|---|---|---|---|
| m/z 150 | m/z 133 | 60 | 10 |
| m/z 182 | m/z 139 | 60 | 6 |
| m/z 467-1 | m/z 360 | 165 | 20 |
| m/z 467-2 | m/z 360 | 165 | 25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, Q.; Zeng, Q.; Yu, J.; Xiao, H.; Lu, J.; Ren, X. Mechanism of Resveratrol Dimers Isolated from Grape Inhibiting 1O2 Induced DNA Damage by UHPLC-QTOF-MS2 and UHPLC-QQQ-MS2 Analyses. Biomedicines 2021, 9, 271. https://doi.org/10.3390/biomedicines9030271
Kong Q, Zeng Q, Yu J, Xiao H, Lu J, Ren X. Mechanism of Resveratrol Dimers Isolated from Grape Inhibiting 1O2 Induced DNA Damage by UHPLC-QTOF-MS2 and UHPLC-QQQ-MS2 Analyses. Biomedicines. 2021; 9(3):271. https://doi.org/10.3390/biomedicines9030271
Chicago/Turabian StyleKong, Qingjun, Qingzhi Zeng, Jia Yu, Hongxi Xiao, Jun Lu, and Xueyan Ren. 2021. "Mechanism of Resveratrol Dimers Isolated from Grape Inhibiting 1O2 Induced DNA Damage by UHPLC-QTOF-MS2 and UHPLC-QQQ-MS2 Analyses" Biomedicines 9, no. 3: 271. https://doi.org/10.3390/biomedicines9030271
APA StyleKong, Q., Zeng, Q., Yu, J., Xiao, H., Lu, J., & Ren, X. (2021). Mechanism of Resveratrol Dimers Isolated from Grape Inhibiting 1O2 Induced DNA Damage by UHPLC-QTOF-MS2 and UHPLC-QQQ-MS2 Analyses. Biomedicines, 9(3), 271. https://doi.org/10.3390/biomedicines9030271







