Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient and Tissue Selection
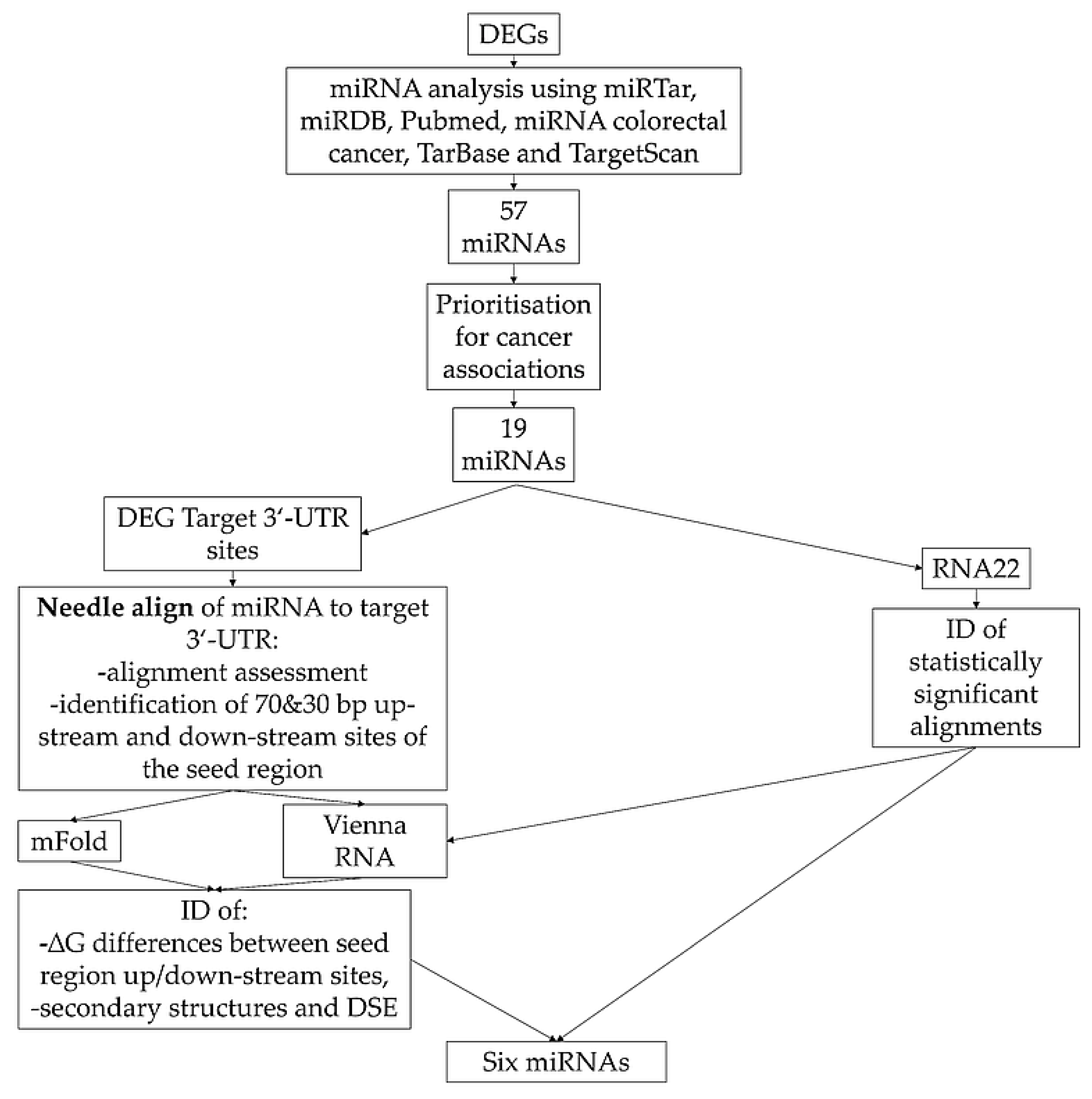
2.2. Target miRNAs Identification and Prioritization
2.3. RNA Isolation and Quality Assessment
2.4. Reverse Transcription (RT) and Pre-Amplification
2.5. Selection of Primers and Probes
2.6. Quantitative Real-Time PCR (qPCR)
2.7. Statistics
3. Results
3.1. Patient Characteristics
3.2. Differential Gene Expression
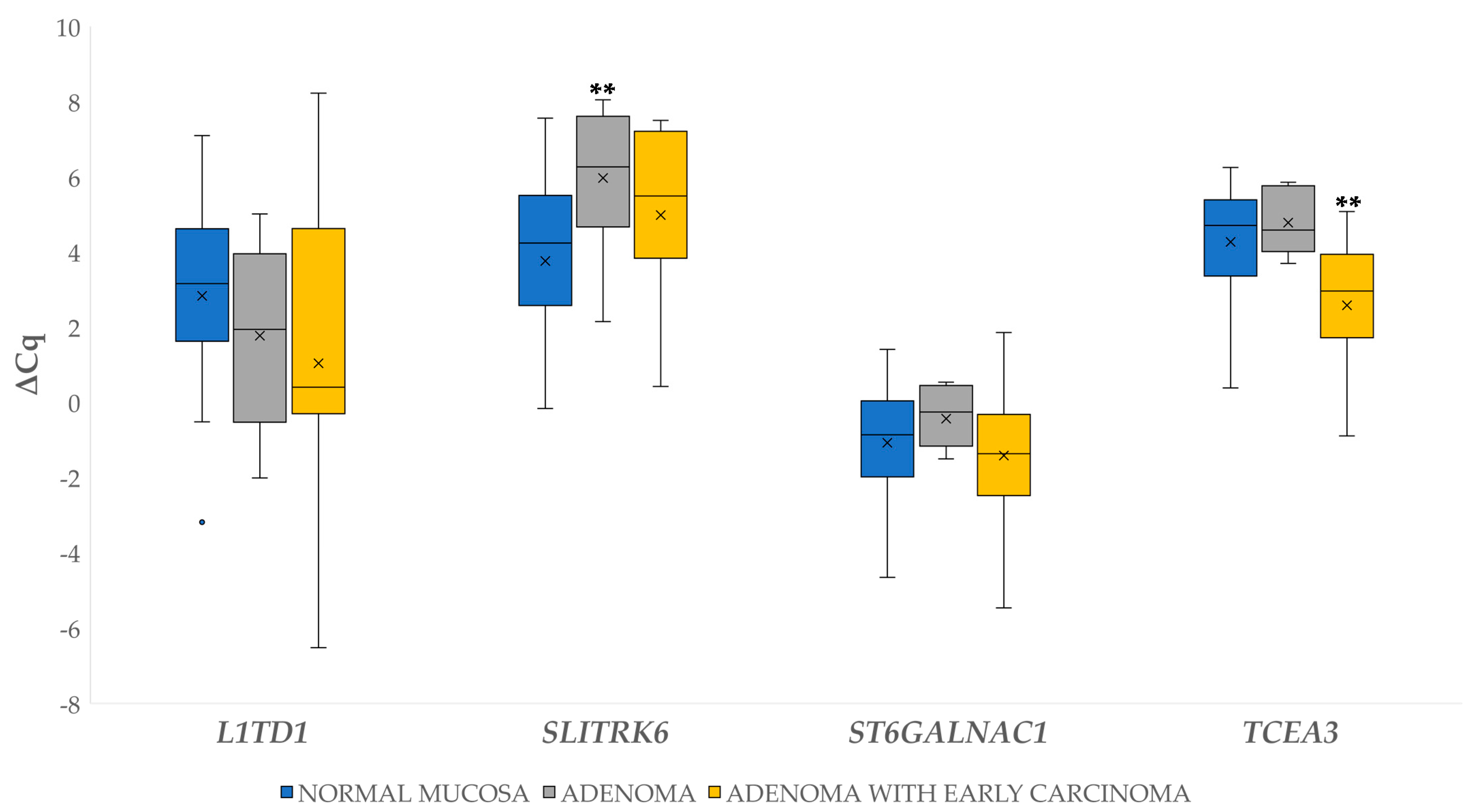
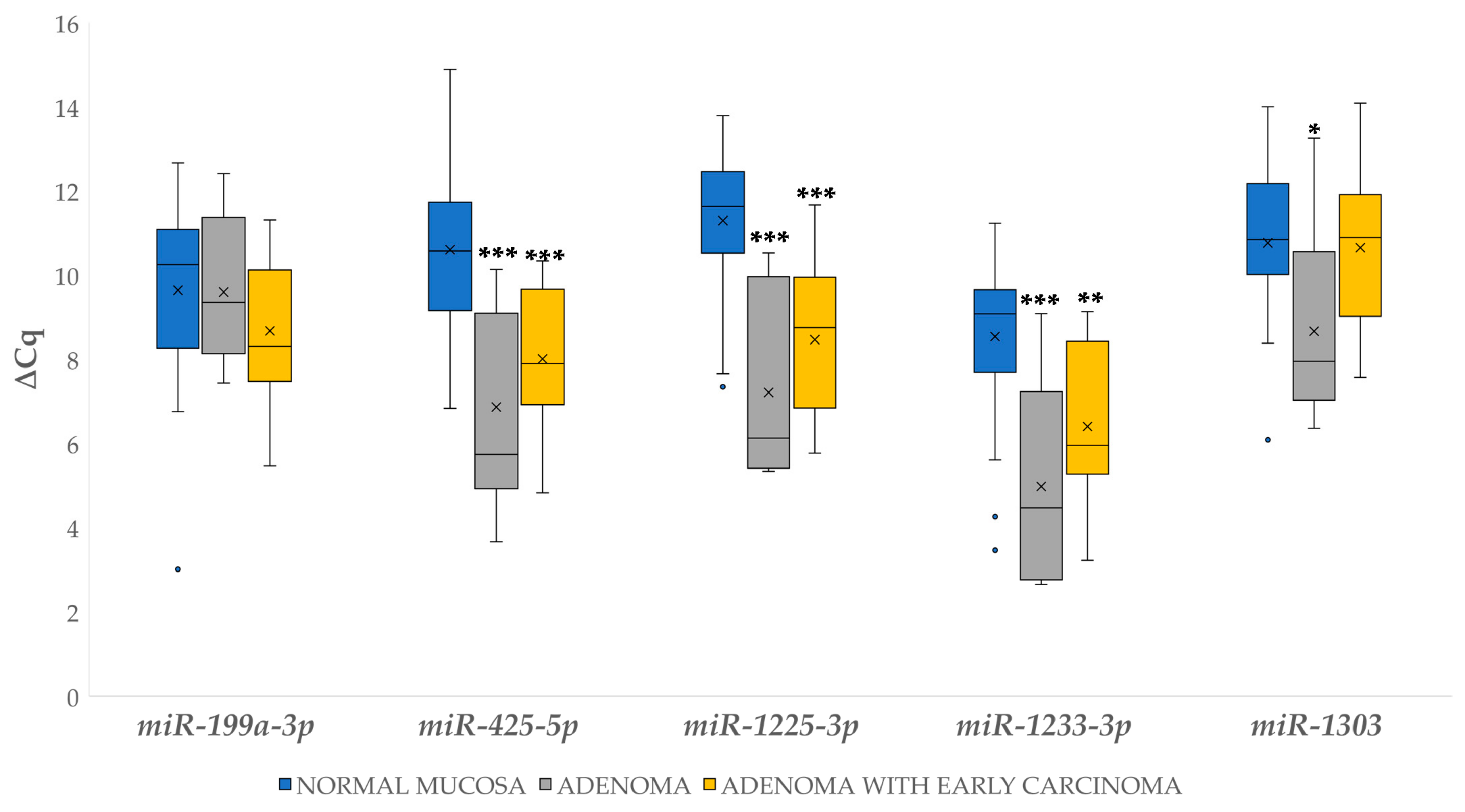
3.2.1. Differential Gene Expression in Adenoma and Adenoma with Early Carcinoma
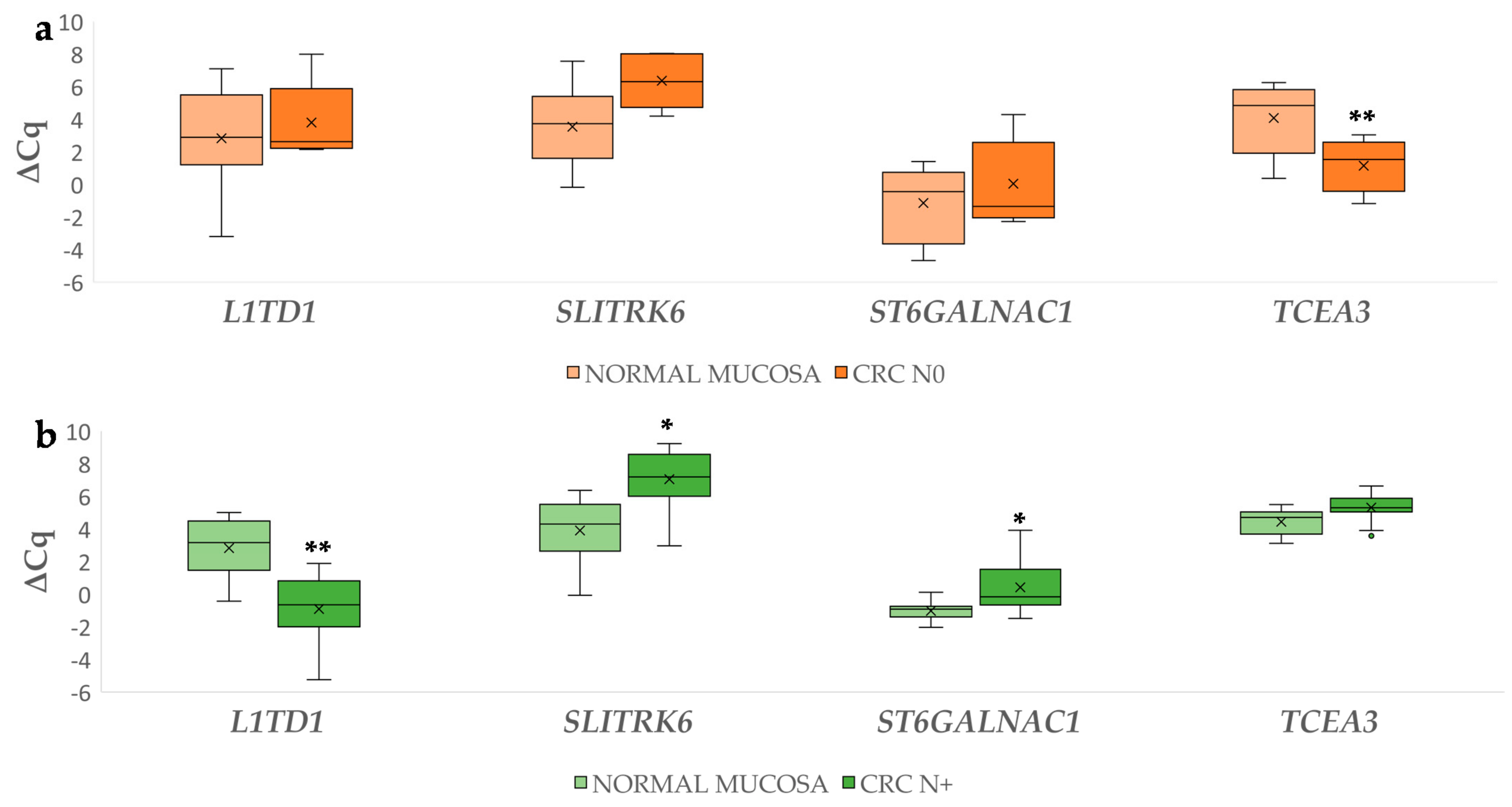
3.2.2. Differential Gene Expression in Carcinoma Compared to Normal Mucosa
3.2.3. Gene Expression in Carcinoma with Lymph Node Metastases Compared to Carcinoma without Lymph Node Metastases
3.3. Prioritization of Potential miRNA-Target Gene Associations
3.4. Differential miRNA Expression
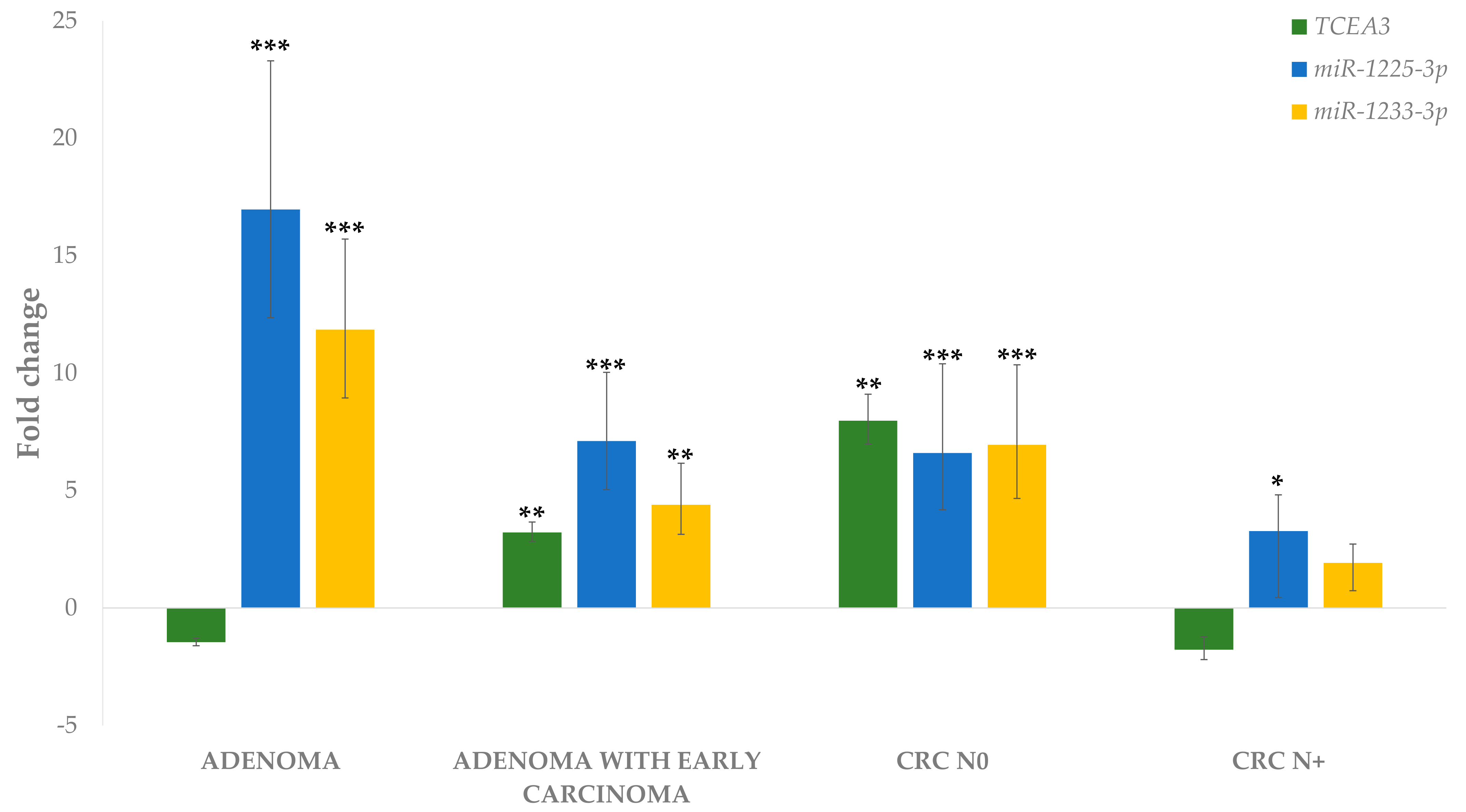
3.4.1. Differential Expression of miRNAs in Adenoma and Adenoma with Early Carcinoma
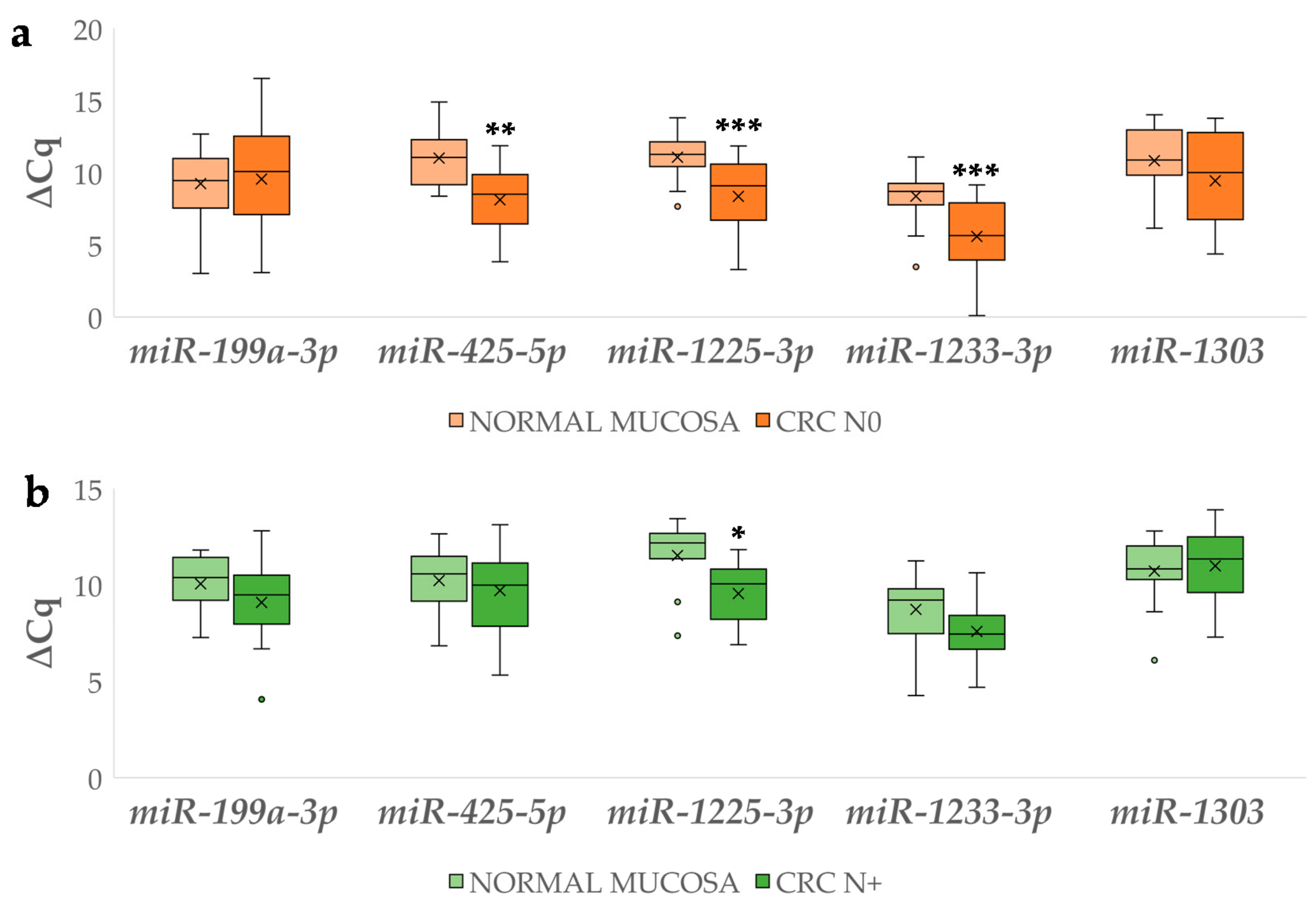
3.4.2. Differential miRNA Expression in Carcinoma with and without Lymph Node Metastases Compared to Corresponding Normal Mucosa
3.4.3. Differential Expression of miRNAs Between Carcinoma with and without Lymph Node Metastases
3.5. Correlation between Expression of Investigated Genes and Their Potentially Regulatory miRNAs
3.6. Gene and miRNA Correlation to the Level of Malignancy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kudryavtseva, A.V.; Lipatova, A.V.; Zaretsky, A.R.; Moskalev, A.A.; Fedorova, M.S.; Rasskazova, A.S.; Shibukhova, G.A.; Snezhkina, A.V.; Kaprin, A.D.; Alekseev, B.Y.; et al. Important molecular genetic markers of colorectal cancer. Oncotarget 2016, 7, 53959–53983. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, E.; Liu, H.; Wan, L.; Lai, M.-D. Epithelial–mesenchymal transition in colorectal cancer metastasis: A system review. Pathol. Res. Pr. 2015, 211, 557–569. [Google Scholar] [CrossRef]
- Balch, C.; Ramapuram, J.B.; Tiwari, A.K. The epigenomics of embryonic pathway signaling in colorectal cancer. Front. Pharmacol. 2017, 8, 267. [Google Scholar] [CrossRef]
- Sepulveda, A.R.; Portillo, A.J.D. Molecular basis of diseases of the gastrointestinal tract. In Molecular Pathology, 2nd ed.; Coleman, W.B., Tsongalis, G.J., Eds.; Elsevier BV: Amsterdam, The Netherlands, 2018; pp. 387–415. [Google Scholar] [CrossRef]
- Järvinen, H.J.; Mecklin, J.-P.; Sistonen, P. Screening reduces colorectal cancer rate in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 1995, 108, 1405–1411. [Google Scholar] [CrossRef]
- Brenner, H.; Jansen, L.; Ulrich, A.; Chang-Claude, J.; Hoffmeister, M. Survival of patients with symptom- and screening-detected colorectal cancer. Oncotarget 2016, 7, 44695–44704. [Google Scholar] [CrossRef]
- Fischer, J.; Walker, L.C.; Robinson, B.A.; Frizelle, F.A.; Church, J.M.; Eglinton, T.W. Clinical implications of the genetics of sporadic colorectal cancer. ANZ J. Surg. 2019, 89, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Blank, A.; Roberts, D.E.I.; Dawson, H.; Zlobec, I.; Lugli, A. Tumor heterogeneity in primary colorectal cancer and corresponding metastases. Does the apple fall far from the tree? Front. Med. 2018, 5, 234. [Google Scholar] [CrossRef]
- Dylla, S.J.; Beviglia, L.; Clarke, M.F.; Hoey, T.; Lewicki, J.; Gurney, A.L.; Park, I.-K.; Chartier, C.; Raval, J.; Ngan, L.; et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS ONE 2008, 3, e2428. [Google Scholar] [CrossRef]
- Saiki, Y.; Ishimaru, S.; Mimori, K.; Takatsuno, Y.; Nagahara, M.; Ishii, H.; Yamada, K.; Mori, M. Comprehensive analysis of the clinical significance of inducing pluripotent stemness-related gene expression in colorectal cancer cells. Ann. Surg. Oncol. 2009, 16, 2638–2644. [Google Scholar] [CrossRef]
- Munro, M.J.; Wickremesekera, S.K.; Peng, L.; Tan, S.T.; Itinteang, T. Cancer stem cells in colorectal cancer: A review. J. Clin. Pathol. 2018, 71, 110–116. [Google Scholar] [CrossRef]
- Hauptman, N.; Boštjančič, E.; Žlajpah, M.; Ranković, B.; Zidar, N. Bioinformatics analysis reveals most prominent gene candidates to distinguish colorectal adenoma from adenocarcinoma. BioMed Res. Int. 2018, 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.C.-B.; Ibrahim, A.; Fong, H.; Thompson, N.; Lock, L.F.; Donovan, P.J. L1TD1 is a marker for undifferentiated human embryonic stem cells. PLoS ONE 2011, 6, e19355. [Google Scholar] [CrossRef] [PubMed]
- Närvä, E.; Rahkonen, N.; Rao, A.; Lahesmaa, R.; Emani, M.R.; Lund, R.; Pursiheimo, J.-P.; Nästi, J.; Autio, R.; Rasool, O.; et al. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells 2011, 30, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Emani, M.R.; Närvä, E.; Stubb, A.; Chakroborty, D.; Viitala, M.; Rokka, A.; Rahkonen, N.; Moulder, R.; Denessiouk, K.; Trokovic, R.; et al. The L1TD1 protein interactome reveals the importance of post-transcriptional regulation in human pluripotency. Stem Cell Rep. 2015, 4, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Chakroborty, D.; Emani, M.; Klén, R.; Böckelman, C.; Hagström, J.; Haglund, C.; Ristimäki, A.; Lahesmaa, R.; Elo, L.L. L1TD1-a prognostic marker for colon cancer. BMC Cancer 2019, 19, 727. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, C.J.; Vik-Mo, E.O.; Behnan, J.; Helseth, E.; Langmoen, I.A. Transcriptional profiling of adult neural stem-like cells from the human brain. PLoS ONE 2014, 9, e114739. [Google Scholar] [CrossRef] [PubMed]
- Proenca, C.C.; Gao, P.; Shmelkov, S.V.; Rafii, S.; Lee, F.S. Slitrks as emerging candidate genes involved in neuropsychiatric disorders. Trends Neurosci. 2011, 34, 143–153. [Google Scholar] [CrossRef]
- Morrison, K.; Challita-Eid, P.M.; Verlinsky, A.; Aviña, H.; Doñate, F.; Joseph, I.B.; Pereira, D.S.; Morrison, K.; Stover, D.R.; Raitano, A.; et al. Development of ASG-15ME, a novel antibody-drug conjugate targeting SLITRK6, a new urothelial cancer biomarker. Mol. Cancer Ther. 2016, 15, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Farahani, E.; Patra, H.K.; Jangamreddy, J.R.; Rashedi, I.; Kawalec, M.; Pariti, R.K.R.; Batakis, P.; Wiechec, E. Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis 2014, 35, 747–759. [Google Scholar] [CrossRef]
- Ambriz, X.; De Lanerolle, P.; Ambrosio, J.R. The mechanobiology of the actin cytoskeleton in stem cells during differentiation and interaction with biomaterials. Stem Cells Int. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Condeelis, J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2007, 1773, 642–652. [Google Scholar] [CrossRef]
- Ogawa, T.; Hirohashi, Y.; Murai, A.; Nishidate, T.; Okita, K.; Wang, L.; Ikehara, Y.; Satoyoshi, T.; Usui, A.; Kubo, T.; et al. ST6GALNAC1 plays important roles in enhancing cancer stem phenotypes of colorectal cancer via the Akt pathway. Oncotarget 2017, 8, 112550–112564. [Google Scholar] [CrossRef]
- Park, K.-S.; Cha, Y.; Han, Y.-M.; Kim, J.; Song, J.; Kim, J.Y.; Tesar, P.J.; Lanza, R.; Lee, K.-A.; Kim, K.-S.; et al. Transcription elongation factor Tcea3 regulates the pluripotent differentiation potential of mouse embryonic stem cells, via the Lefty1-Nodal-Smad2 pathway. Stem Cells 2012, 31, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. (Eds.) TNM Classification of Malignant Tumours, 8th ed.; Wiley Blackwell: Oxford, UK, 2017. [Google Scholar]
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Skok, D.J.; Hauptman, N.; Boštjančič, E.; Zidar, N. The integrative knowledge base for miRNA-mRNA expression in colorectal cancer. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA–target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Zhao, Y.; Samal, E.; Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nat. Cell Biol. 2005, 436, 214–220. [Google Scholar] [CrossRef]
- Boštjančič, E.; Zidar, N.; Glavač, D. MicroRNAs and cardiac sarcoplasmic reticulum calcium ATPase-2 in human myocardial infarction: Expression and bioinformatic analysis. BMC Genom. 2012, 13, 552. [Google Scholar] [CrossRef]
- Benias, P.C.; Wells, R.G.; Theise, N.D.; Sackey-Aboagye, B.; Klavan, H.; Reidy, J.; Buonocore, D.; Miranda, M.; Kornacki, S.; Wayne, M.; et al. Structure and distribution of an unrecognized interstitium in human tissues. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.F.; Zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G. Specificity of microRNA target selection in translational repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.-L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of microrna binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef]
- Latham, G.J. Normalization of microRNA quantitative RT-PCR data in reduced scale experimental designs. Methods Mol. Biol. 2010, 667, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.; Sillars-Hardebol, A.H.; Postma, C.; Mongera, S.; Droste, J.T.S.; Obulkasim, A.; Van De Wiel, M.; Van Criekinge, W.; Ylstra, B.; Fijneman, R.J.A.; et al. Colorectal adenoma to carcinoma progression is accompanied by changes in gene expression associated with ageing, chromosomal instability, and fatty acid metabolism. Cell. Oncol. 2012, 35, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Y.; Cao, Y.-X.; Zhou, X.; Wei, B.; Zhan, L.; Sun, S.-Y. Stimulative role of ST6GALNAC1 in proliferation, migration and invasion of ovarian cancer stem cells via the Akt signaling pathway. Cancer Cell Int. 2019, 19, 86. [Google Scholar] [CrossRef]
- Patani, N.; Jiang, W.; Mokbel, K. Prognostic utility of glycosyltransferase expression in breast cancer. Cancer Genom. Proteom. 2009, 5, 333–340. [Google Scholar]
- Julien, S.; Adriaenssens, E.; Ottenberg, K.; Furlan, A.J.; Courtand, G.; Vercoutter-Edouart, A.-S.; Hanisch, F.-G.; Delannoy, P.; Le Bourhis, X. ST6GalNAc I expression in MDA-MB-231 breast cancer cells greatly modifies their O-glycosylation pattern and enhances their tumourigenicity. Glycobiology 2005, 16, 54–64. [Google Scholar] [CrossRef]
- Tamura, F.; Sato, Y.; Hirakawa, M.; Yoshida, M.; Ono, M.; Osuga, T.; Okagawa, Y.; Uemura, N.; Arihara, Y.; Murase, K.; et al. RNAi-mediated gene silencing of ST6GalNAc I suppresses the metastatic potential in gastric cancer cells. Gastric Cancer 2016, 19, 85–97. [Google Scholar] [CrossRef]
- Ozaki, H.; Matsuzaki, H.; Ando, H.; Kaji, H.; Nakanishi, H.; Ikehara, Y.; Narimatsu, H. Enhancement of metastatic ability by ectopic expression of ST6GalNAcI on a gastric cancer cell line in a mouse model. Clin. Exp. Metastasis 2012, 29, 229–238. [Google Scholar] [CrossRef]
- Cha, Y.; Kim, D.-K.; Hyun, J.; Kim, S.-J.; Park, K.-S. TCEA3 binds to TGF-beta receptor I and induces Smad-independent, JNK-dependent apoptosis in ovarian cancer cells. Cell. Signal. 2013, 25, 1245–1251. [Google Scholar] [CrossRef]
- Li, J.; Jin, Y.; Pan, S.; Chen, Y.; Wang, K.; Lin, C.; Jin, S.; Wu, J. TCEA3 attenuates gastric cancer growth by apoptosis induction. Med. Sci. Monit. 2015, 21, 3241–3246. [Google Scholar] [CrossRef]
- Guo, Y.; Bao, Y.; Ma, M.; Yang, W. Identification of key candidate genes and pathways in colorectal cancer by integrated bioinformatical analysis. Int. J. Mol. Sci. 2017, 18, 722. [Google Scholar] [CrossRef]
- Diaz, G.; Zamboni, F.; Tice, A.; Farci, P. Integrated ordination of miRNA and mRNA expression profiles. BMC Genom. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Liao, X.; Xue, H.; Wang, Y.-C.; Nazor, K.L.; Guo, S.; Trivedi, N.N.; Peterson, S.E.; Liu, Y.; Loring, J.F.; Laurent, L. Matched miRNA and mRNA signatures from an hESC-based in vitro model of pancreatic differentiation reveal novel regulatory interactions. J. Cell Sci. 2013, 126, 3848–3861. [Google Scholar] [CrossRef]
- Liu, R.; Liu, C.; Zhang, D.; Liu, B.; Chen, X.; Rycaj, K.; Jeter, C.; Calhoun-Davis, T.; Li, Y.; Yang, T.; et al. miR-199a-3p targets stemness-related and mitogenic signaling pathways to suppress the expansion and tumorigenic capabilities of prostate cancer stem cells. Oncotarget 2016, 7, 56628–56642. [Google Scholar] [CrossRef]
- Shen, Z.-L.; Wang, B.; Jiang, K.-W.; Ye, C.-X.; Cheng, C.; Yan, Y.-C.; Zhang, J.-Z.; Yang, Y.; Gao, Z.-D.; Ye, Y.-J.; et al. Downregulation of miR-199b is associated with distant metastasis in colorectal cancer via activation of SIRT1 and inhibition of CREB/KISS1 signaling. Oncotarget 2016, 7, 35092–35105. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, D.; Kuai, J.; Cheng, M.; Fang, X.; Li, G. Upregulation of miR-199a/b contributes to cisplatin resistance via Wnt/β-catenin-ABCG2 signaling pathway in ALDHA1+ colorectal cancer stem cells. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Miao, X.; Zhu, K.; Cui, S.; Meng, Q.; Sun, J.; Wang, T. Micro RNA-425-5p regulates chemoresistance in colorectal cancer cells via regulation of Programmed Cell Death 10. J. Cell. Mol. Med. 2016, 20, 360–369. [Google Scholar] [CrossRef]
- Nishida, N.; Nagahara, M.; Mori, M.; Sato, T.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Sugihara, K.; et al. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic mirna clusters. Clin. Cancer Res. 2012, 18, 3054–3070. [Google Scholar] [CrossRef]
- Yuan, Z.; Xiu, C.; Liu, D.; Zhou, G.; Yang, H.; Pei, R.; Ding, C.; Cui, X.; Sun, J.; Song, K. Long noncoding RNA LINC-PINT regulates laryngeal carcinoma cell stemness and chemoresistance through miR-425-5p/PTCH1/SHH axis. J. Cell. Physiol. 2019, 234, 23111–23122. [Google Scholar] [CrossRef]
- Liu, J.; Ma, L.; Wang, Z.; Wang, L.; Liu, C.; Chen, R.; Zhang, J. MicroRNA expression profile of gastric cancer stem cells in the MKN-45 cancer cell line. Acta Biochim. Biophys. Sin. 2014, 46, 92–99. [Google Scholar] [CrossRef][Green Version]
- Marques, F.Z.; Campain, A.E.; Tomaszewski, M.; Zukowska-Szczechowska, E.; Yang, Y.H.J.; Charchar, F.J.; Morris, B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 2011, 58, 1093–1098. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Ding, Y.; Fan, Z.; Zhang, J.; Zhang, H.; Jiang, B.; Zhu, Y. Serum MicroRNA profile in patients with colon adenomas or cancer. BMC Med. Genom. 2017, 10, 23. [Google Scholar] [CrossRef]
- El-Murr, N.; Abidi, Z.; Wanherdrick, K.; Svrcek, M.; Gaub, M.-P.; Flejou, J.-F.; Hamelin, R.; Duval, A.; Lesuffleur, T. MiRNA genes constitute new targets for microsatellite instability in colorectal cancer. PLoS ONE 2012, 7, e31862. [Google Scholar] [CrossRef]
- Giampieri, R.; Scartozzi, M.; Cecchini, L.; Guerrieri, M.; Bearzi, I.; Cascinu, S.; Loretelli, C.; Piva, F.; Mandolesi, A.; Lezoche, G.; et al. Cancer stem cell gene profile as predictor of relapse in high risk stage II and stage III, radically resected colon cancer patients. PLoS ONE 2013, 8, e72843. [Google Scholar] [CrossRef]
- Merlos-Suárez, A.; Barriga, F.M.; Clevers, H.; Sancho, E.; Mangues, R.; Batlle, E.; Jung, P.; Iglesias, M.; Céspedes, M.V.; Rossell, D.; et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011, 8, 511–524. [Google Scholar] [CrossRef]
- Colak, S.; Zimberlin, C.D.; Fessler, E.; Hogdal, L.J.; Prasetyanti, P.R.; Grandela, C.; Letai, A.; Medema, J.P. Decreased mitochondrial priming determines chemoresistance of colon cancer stem cells. Cell Death Differ. 2014, 21, 1170–1177. [Google Scholar] [CrossRef]
- Lombardo, Y.; Scopelliti, A.; Cammareri, P.; Todaro, M.; Iovino, F.; Ricci–Vitiani, L.; Gulotta, G.; Dieli, F.; De Maria, R.; Stassi, G. Bone morphogenetic protein 4 induces differentiation of colorectal cancer stem cells and increases their response to chemotherapy in mice. Gastroenterology 2011, 140, 297–309.e6. [Google Scholar] [CrossRef]
- Lotti, F.; Jarrar, A.M.; Pai, R.K.; Hitomi, M.; Lathia, J.; Mace, A.; Gantt, G.A.; Sukhdeo, K.; DeVecchio, J.; Vasanji, A.; et al. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. J. Exp. Med. 2013, 210, 2851–2872. [Google Scholar] [CrossRef]
- Zeuner, A.; Todaro, M.; Stassi, G.; De Maria, R. Colorectal cancer stem cells: From the crypt to the clinic. Cell Stem Cell 2014, 15, 692–705. [Google Scholar] [CrossRef]
- Wang, V.; Wu, W. MicroRNA-based therapeutics for cancer. BioDrugs 2009, 23, 15–23. [Google Scholar] [CrossRef]
- Sanz-Pamplona, R.; Berenguer, A.; Cordero, D.; Molleví, D.G.; Crous-Bou, M.; Sole, X.; Paré-Brunet, L.; Guino, E.; Salazar, R.; Santos, C.; et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol. Cancer 2014, 13, 46. [Google Scholar] [CrossRef]
- Polley, A.C.; Mulholland, F.; Pin, C.; Williams, E.A.; Bradburn, D.M.; Mills, S.J.; Mathers, J.C.; Johnson, I.T. Proteomic analysis reveals field-wide changes in protein expression in the morphologically normal mucosa of patients with colorectal neoplasia. Cancer Res. 2006, 66, 6553–6562. [Google Scholar] [CrossRef]




| Gene/miRNA | Assay ID | Sequence (Probe Sequence or Mature miRNA Sequence) |
|---|---|---|
| B2M | Hs99999907_m1 | GTTAAGTGGGATCGAGACATGTAAG |
| IPO8 | Hs00183533_m1 | GGGGAATTGATCAGTGCATTCCACT |
| L1TD1 | Hs00219458_m1 | TTTTTCGCCAGGCACCAAGGCACAG |
| SLITRK6 | Hs00536106_s1 | TTTCCATGGACTGGAAAACCTGGAA |
| ST6GALNAC1 | Hs01027885_m1 | AGGAGGCCTTCAGACGACTTGCCCT |
| TCEA3 | Hs00957468_m1 | GAAATCGAAGATCATATCTACCAAG |
| hsa-miR-199a-3p | 002304 | ACAGUAGUCUGCACAUUGGUUA |
| hsa-miR-335-5p | 000546 | UCAAGAGCAAUAACGAAAAAUGU |
| hsa-miR-425-5p | 001516 | AAUGACACGAUCACUCCCGUUGA |
| hsa-miR-1225-3p | 002766 | UGAGCCCCUGUGCCGCCCCCAG |
| hsa-miR-1233-3p | 002768 | UGAGCCCUGUCCUCCCGCAG |
| hsa-miR-1274b | 002884 | UCCCUGUUCGGGCGCCA |
| hsa-miR-1303 | 002792 | UUUAGAGACGGGGUCUUGCUCU |
| RNU6B | 001093 | CGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT |
| Patients | Adenoma | Adenoma with Early Carcinoma | CRC without Lymph Node Metastases | CRC with Lymph Node Metastases |
|---|---|---|---|---|
| M:F | 10:1 | 9:4 | 4:6 | 9:4 |
| Age | 62.3 ± 10.7 | 64.9 ± 5.7 | 72.7 ± 11.6 | 73.2 ± 11.8 |
| Gene | miRNA | Association with at Least Two Databases | Folding Free Energy Constraints | RNA22 | Direct Validation |
|---|---|---|---|---|---|
| L1TD1 | hsa-miR-1303 | + | + | − | − |
| SLITRK6 | hsa-miR-199a-3p | + | − | + | − |
| hsa-miR-425-5p | − | + | − | + | |
| ST6GALNAC1 | hsa-miR-335-5p | + | + | − | − |
| TCEA3 | hsa-miR-335-5p | + | + | − | − |
| hsa-miR-1225-3p | + | + | − | − | |
| hsa-miR-1233-3p | + | + | + * | − |
| Gene and miRNA | Correlation Coefficient | Significance (2-Tailed) | |
|---|---|---|---|
| L1TD1 | miR-1303 | −0.024 | 0.862 |
| SLITRK6 | miR-425-5p | −0.187 | 0.176 |
| miR-199a-3p | −0.323 | 0.017 * | |
| TCEA3 | miR-1233-3p | 0.116 | 0.360 |
| miR-1225-3p | 0.056 | 0.660 | |
| Gene and miRNA | Correlation Coefficient | Significance (2-Tailed) |
|---|---|---|
| L1TD1 | 0.336 | 0.011 |
| SLITRK6 | −0.433 | <0.001 |
| ST6GALNAC1 | −0.186 | 0.141 |
| TCEA3 | 0.102 | 0.419 |
| miR-199a-3p | 0.128 | 0.291 |
| miR-425-5p | 0.209 | 0.083 |
| miR-1225-3p | 0.345 | 0.003 |
| miR-1233-3p | 0.276 | 0.021 |
| miR-1303 | 0.014 | 0.912 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urh, K.; Žlajpah, M.; Zidar, N.; Boštjančič, E. Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis. Biomedicines 2021, 9, 179. https://doi.org/10.3390/biomedicines9020179
Urh K, Žlajpah M, Zidar N, Boštjančič E. Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis. Biomedicines. 2021; 9(2):179. https://doi.org/10.3390/biomedicines9020179
Chicago/Turabian StyleUrh, Kristian, Margareta Žlajpah, Nina Zidar, and Emanuela Boštjančič. 2021. "Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis" Biomedicines 9, no. 2: 179. https://doi.org/10.3390/biomedicines9020179
APA StyleUrh, K., Žlajpah, M., Zidar, N., & Boštjančič, E. (2021). Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis. Biomedicines, 9(2), 179. https://doi.org/10.3390/biomedicines9020179







