Role of Kallikrein 7 in Body Weight and Fat Mass Regulation
Abstract
1. Introduction
2. Research Design and Methods
2.1. Animal Studies
2.2. Generation and Genotyping of Klk7−/− Mice
2.3. Phenotypic Characterization
2.4. RNA Isolation and Quantitative Real-Time PCR Analysis
2.5. Primary Adipocyte Cell Culture and Western Blot Analyses
2.6. Proteomics of AT Depots
2.7. Thyroid Gland Analysis
2.8. Statistical Analyses
3. Results
3.1. Generation of Klk7−/− Mice
3.2. Body Weight Gain and Body Fat Mass are Markedly Reduced in Klk7−/− Mice
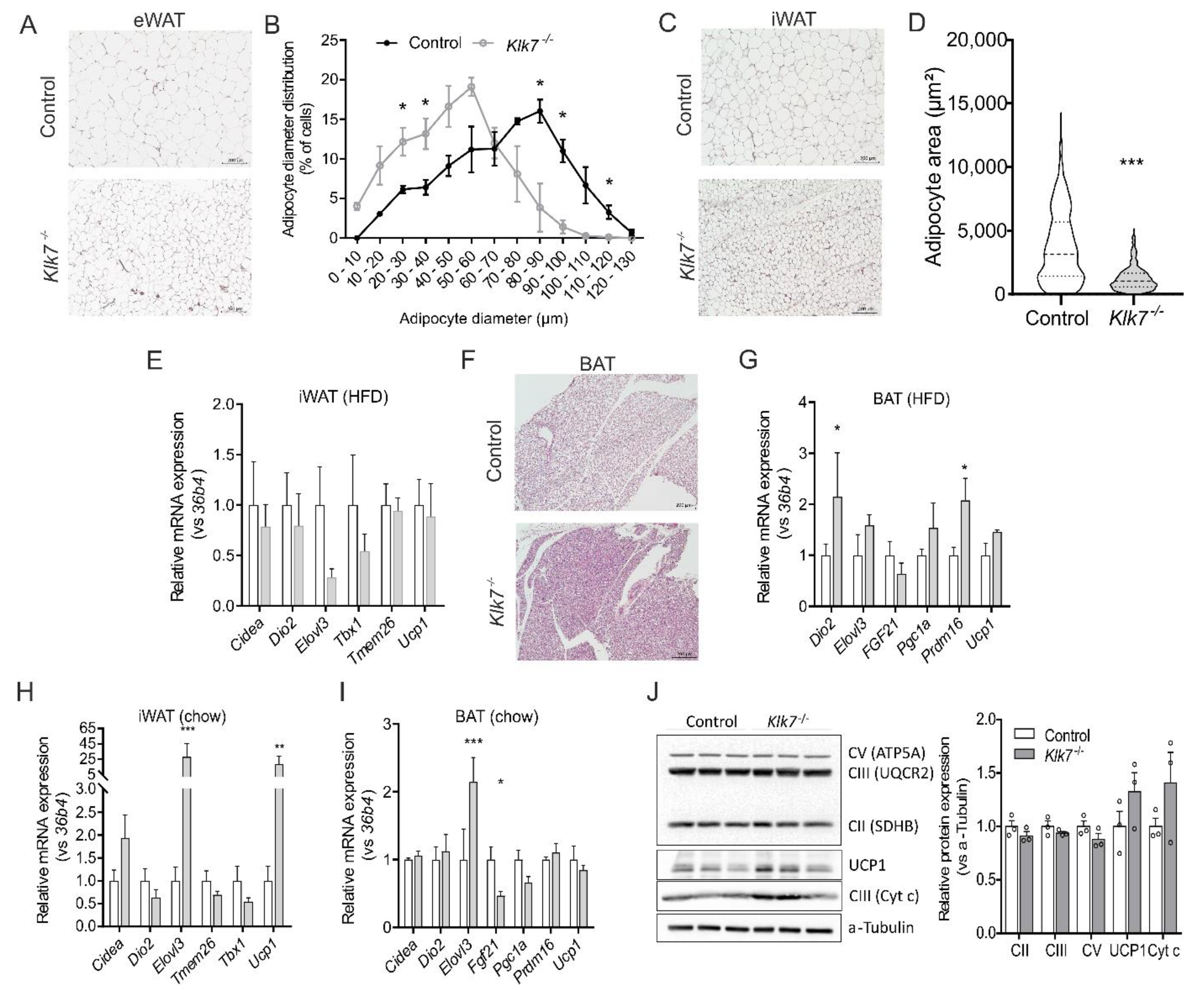
3.3. Consequences of Klk7−/− on Adipose Tissue
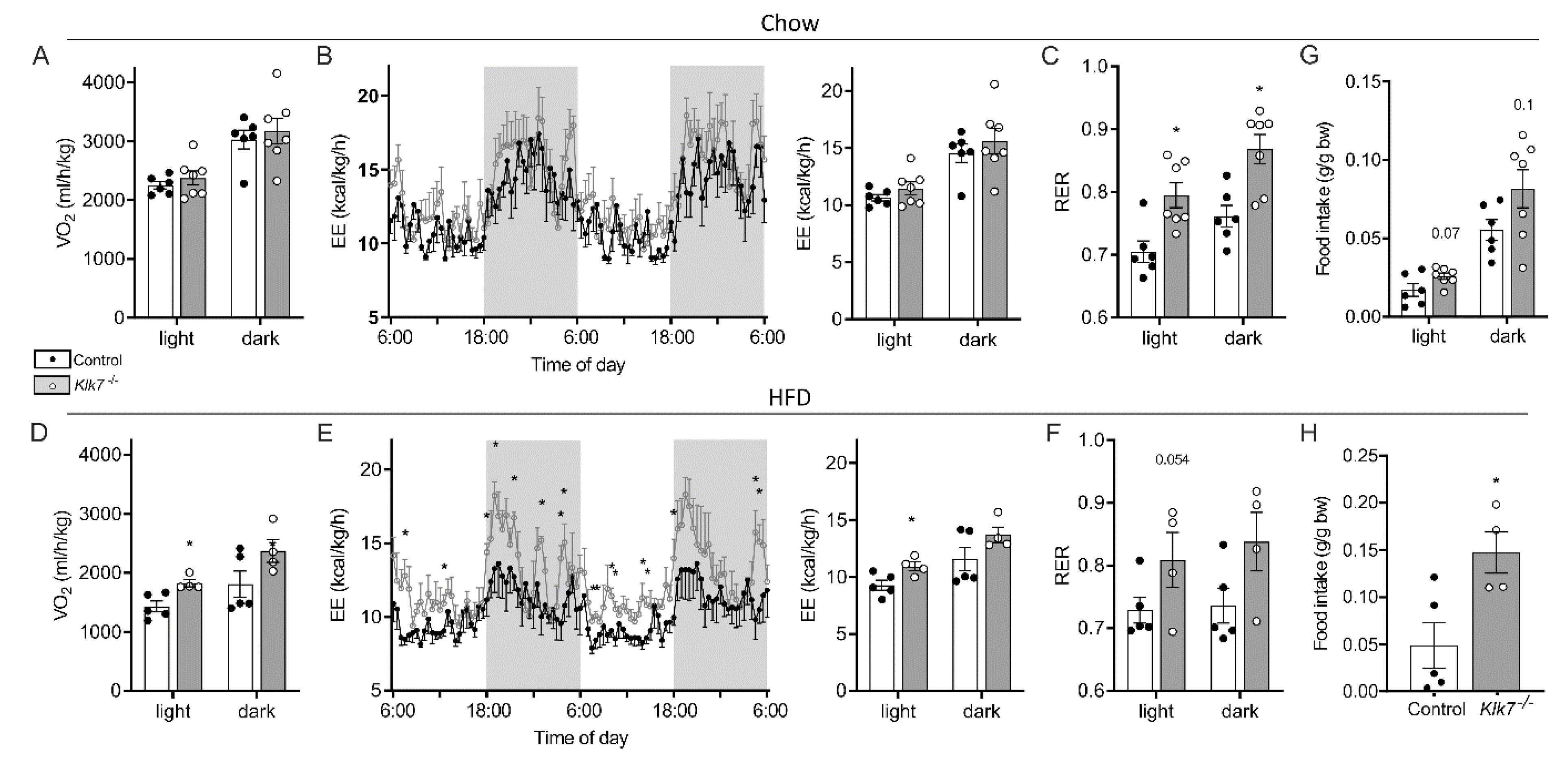
3.4. Klk7−/− Mice Exhibit Lower Body Temperature and Altered Circulating T3
3.5. Ablation of Klk7 Does Not Affect Circulating Vaspin and Chemerin
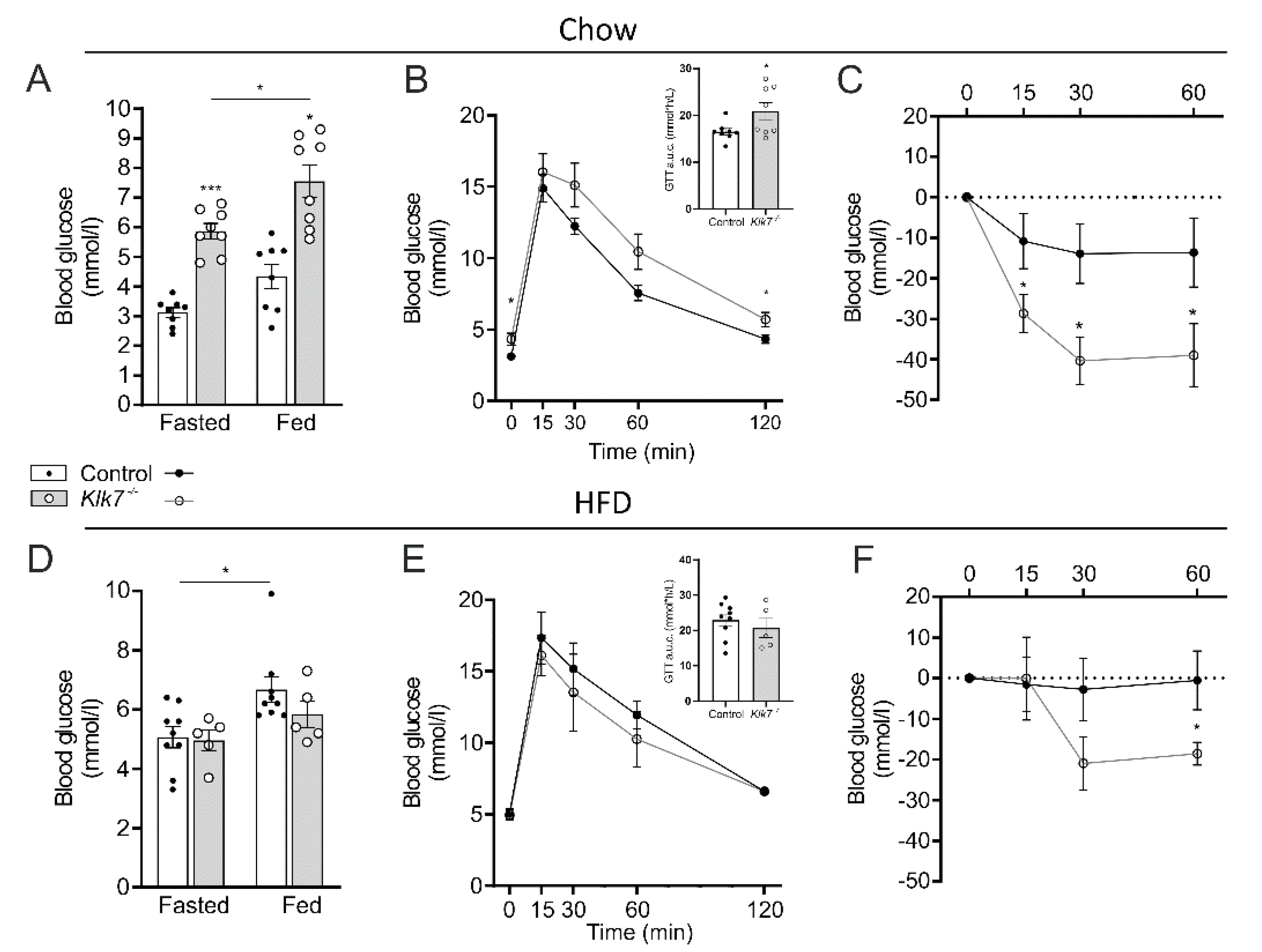
3.6. Klk7−/− Mice Exhibit Increased Sensitivity to Exogenous Insulin
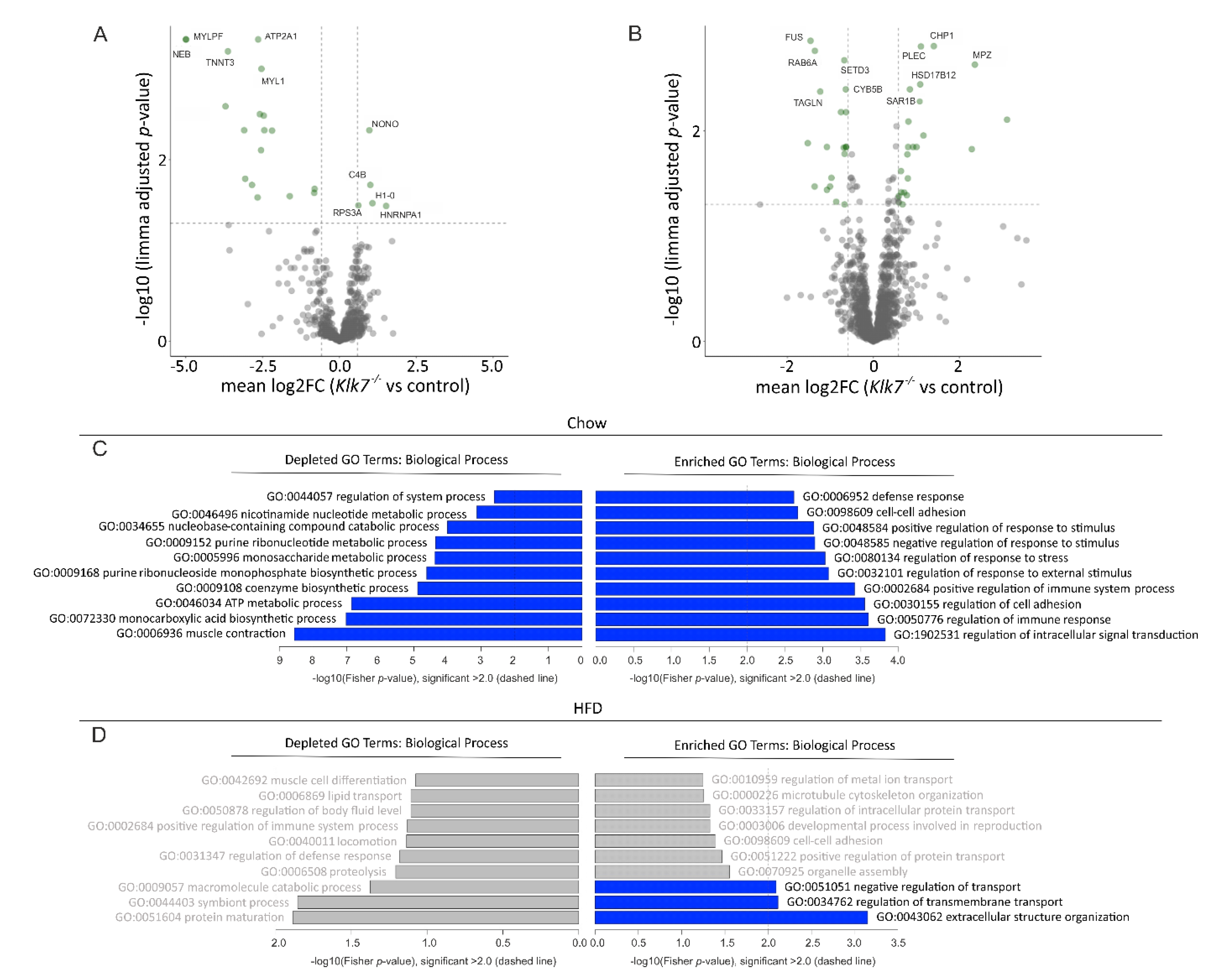
3.7. Klk7−/− Mice Have Depot-Specific Protein Expression Signatures in AT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKT | protein kinase B |
| AT | adipose tissue |
| a.u.c. | area under the curve |
| BAT | brown adipose tissue |
| bp | base pair |
| bw | body weight |
| Cidea | cell death inducing DFFA like effector a |
| CMV | cytomegalovirus |
| COL3A1 | collagen type 3A1 |
| Cre | causes/cyclic recombination |
| DEG | differentially expressed gene |
| Dio | deiodinase |
| DNAJC3 | DnaJ heat shock protein family (Hsp40) member C3 |
| EE | energy expenditure |
| Elovl3 | elongation of very long chain fatty acids like 3 |
| ER | endoplasmic reticulum |
| eWAT | epididymal white adipose tissue |
| Fc | fragment crystallizable |
| FC | fold change |
| FDR | false discovery rate |
| FFA | free fatty acid |
| FGF21 | fibroblast growth factor 21 |
| fT3 | free T3 |
| GIR | glucose infusion rate |
| GO | gene ontology |
| GTT | glucose tolerance tests |
| HC | heavy chain |
| HFD | high fat diet |
| HFE | homeostatic iron regulator |
| HGP | hepatic glucose production |
| HMW | high molecular weight |
| HRP | horseraddish peroxidase |
| i.p. | intraperitoneal |
| ITT | insulin tolerance tests |
| iWAT | inguinal white adipose tissue |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KLK7 | kallikrein-related peptidase 7–protein |
| KLK7 | kallikrein-related peptidase 7–human gene |
| Klk7 | kallikrein-related peptidase 7–mouse gene |
| Klk7−/− | mouse Klk7 gene knockout |
| Limma | linear models for microarray analysis |
| M1 | classically activated macrophage |
| M2 | alternatively activated macrophage |
| MCP-1 | monocyte chemotactic protein 1 |
| MRI | magnetic resonance imaging |
| MYH2 | myosin heavy chain 2 |
| MYLPF | myosin light chain |
| NCK1 | NCK adaptor protein 1 |
| NEB | nebulin |
| PDK4 | pyruvat dehydrogenase kinase 4 |
| PE | phycoerythrin |
| PGC1A | PPAR gamma coactivator 1-alpha |
| PKG | protein kinase G |
| PPAR | peroxisome proliferator activated receptor |
| PRDM16 | PR/SET domain 16 |
| RARRES2 | retinoic acid receptor responder 2 |
| RER | respiratory exchange ratio |
| SC | single chain |
| SERCA | sarcoplasmic/endoplasmic reticulum calcium ATPase |
| SLC25A5 | ADP/ATP translocase 2 |
| SVF | stromal vascular fraction |
| Tbg | thyroxine-binding globulin |
| Tbx1 | T-box transcription factor 1 |
| TG | triglyceride |
| TH | thyroid hormone |
| TMEM26 | transmembrane protein 26 |
| TSH | thyroid stimulating hormone |
| TT4 | total T4 |
| TUB | tubulin |
| UCP1 | uncoupling protein 1 |
| Vaspin | visceral adipose tissue-derived serine protease inhibitor |
| VDAC1 | voltage-dependent anion-selective channel 1 |
References
- Bray, G.A. Medical consequences of obesity. J. Clin. Endocrinol. Metab. 2004, 89, 2583–2589. [Google Scholar] [CrossRef] [PubMed]
- Klöting, N.; Blüher, M. Adipocyte dysfunction, inflammation and metabolic syndrome. Rev. Endocr. Metab. Disord. 2014, 15, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Kusminski, C.M.; Bickel, P.E.; Scherer, P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat. Rev. Drug Discov. 2016, 15, 639–660. [Google Scholar] [CrossRef] [PubMed]
- Mesallamy, H.O.E.; Hamdy, N.M.; Mostafa, D.M.; Amin, A.I. The Serine Protease Granzyme B as an Inflammatory Marker, in Relation to the Insulin Receptor Cleavage in Human Obesity and Type 2 Diabetes Mellitus. J. Interf. Cytok. Res. 2014, 34, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-S.; Chen, J.-S.; Lin, L.-Y.; Schmid-Schönbein, G.W.; Chien, S.; Huang, P.-H.; Chen, J.-W.; Lin, S.-J. Inhibition of Serine Protease Activity Protects Against High Fat Diet-Induced Inflammation and Insulin Resistance. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Mansuy-Aubert, V.; Zhou, Q.L.; Xie, X.; Gong, Z.; Huang, J.-Y.; Khan, A.R.; Aubert, G.; Candelaria, K.; Thomas, S.; Shin, D.-J.; et al. Imbalance between Neutrophil Elastase and its Inhibitor α1-Antitrypsin in Obesity Alters Insulin Sensitivity, Inflammation, and Energy Expenditure. Cell Metab. 2013, 17, 534–548. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Y.; Pan, J.; Sun, J.; Liu, J.; Libby, P.; Sukhova, G.K.; Doria, A.; Katunuma, N.; Peroni, O.D.; et al. Cathepsin L activity controls adipogenesis and glucose tolerance. Nat. Cell Biol. 2007, 9, 970–977. [Google Scholar] [CrossRef]
- Funicello, M.; Novelli, M.; Ragni, M.; Vottari, T.; Cocuzza, C.; Soriano-Lopez, J.; Chiellini, C.; Boschi, F.; Marzola, P.; Masiello, P.; et al. Cathepsin K Null Mice Show Reduced Adiposity during the Rapid Accumulation of Fat Stores. PLoS ONE 2007, 2, e683. [Google Scholar] [CrossRef]
- Nakatsuka, A.; Wada, J.; Iseda, I.; Teshigawara, S.; Higashio, K.; Murakami, K.; Kanzaki, M.; Inoue, K.; Terami, T.; Katayama, A.; et al. Vaspin Is an Adipokine Ameliorating ER Stress in Obesity as a Ligand for Cell-Surface GRP78/MTJ-1 Complex. Diabetologia 2012, 61, 2823–2832. [Google Scholar] [CrossRef]
- Klöting, N.; Kovacs, P.; Kern, M.; Heiker, J.T.; Fasshauer, M.; Schön, M.R.; Stumvoll, M.; Beck-Sickinger, A.G.; Blüher, M. Central vaspin administration acutely reduces food intake and has sustained blood glucose-lowering effects. Diabetologia 2011, 54, 1819–1823. [Google Scholar] [CrossRef]
- Hida, K.; Wada, J.; Eguchi, J.; Zhang, H.; Baba, M.; Seida, A.; Hashimoto, I.; Okada, T.; Yasuhara, A.; Nakatsuka, A.; et al. Visceral adipose tissue-derived serine protease inhibitor: A unique insulin-sensitizing adipocytokine in obesity. Proc. Natl. Acad. Sci. USA 2005, 102, 10610–10615. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, L.; Di Nisio, C.; Recinella, L.; Chiavaroli, A.; Leone, S.; Ferrante, C.; Orlando, G.; Vacca, M. Effects of vaspin, chemerin and omentin-1 on feeding behavior and hypothalamic peptide gene expression in the rat. Peptides 2011, 32, 1866–1871. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, K.; Zhang, C.; Yang, G.; Yang, M.; Jia, Y.; Zhang, L.; Ma, Z.A.; Boden, G.; Li, L. Central administration of vaspin inhibits glucose production and augments hepatic insulin signaling in high-fat-diet-fed rat. Int. J. Obes. 2016, 40, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Zieger, K.; Weiner, J.; Krause, K.; Schwarz, M.; Kohn, M.; Stumvoll, M.; Bluher, M.; Heiker, J.T. Vaspin suppresses cytokine-induced inflammation in 3T3-L1 adipocytes via inhibition of NFkappaB pathway. Mol. Cell Endocrinol. 2018, 460, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Heiker, J.T.; Klöting, N.; Kovacs, P.; Kuettner, E.B.; Sträter, N.; Schultz, S.; Kern, M.; Stumvoll, M.; Blüher, M.; Beck-Sickinger, A.G. Vaspin inhibits kallikrein 7 by serpin mechanism. Cell. Mol. Life Sci. 2013, 70, 2569–2583. [Google Scholar] [CrossRef] [PubMed]
- Zieger, K.; Weiner, J.; Kunath, A.; Gericke, M.; Krause, K.; Kern, M.; Stumvoll, M.; Klöting, N.; Blüher, M.; Heiker, J.T. Ablation of kallikrein 7 (KLK7) in adipose tissue ameliorates metabolic consequences of high fat diet-induced obesity by counteracting adipose tissue inflammation in vivo. Cell. Mol. Life Sci. 2017, 75, 727–742. [Google Scholar] [CrossRef]
- Sang-Hyeop, L.N.-H.; Koh, B.-J.K.; Song, L.; Song-Cheol, K.; Sun, S.C. Putative Positive Role of Inflammatory Genes in Fat Deposition Supported by Altered Gene Expression in Purified Human Adipocytes and Preadipocytes from Lean and Obese Adipose Tissues. J. Transl. Med. 2020, 18, 433. [Google Scholar]
- Prassas, I.; Eissa, A.; Poda, G.; Diamandis, E.P. Erratum: Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat. Rev. Drug Discov. 2015, 14, 732. [Google Scholar] [CrossRef]
- Egelrud, T. Purification and Preliminary Characterization of Stratum Corneum Chymotryptic Enzyme: A Proteinase That May Be Involved in Desquamation. J. Investig. Dermatol. 1993, 101, 200–204. [Google Scholar] [CrossRef]
- Sondell, B.; Thornell, L.E.; Egelrud, T. Evidence that stratum corneum chymotryptic enzyme is transported to the stratum corneum extracellular space via lamellar bodies. J. Investig. Dermatol. 1995, 104, 819–823. [Google Scholar] [CrossRef]
- Ekholm, E.; Egelrud, T. Stratum corneum chymotryptic enzyme in psoriasis. Arch. Dermatol. Res. 1999, 291, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Di Nardo, A.; Bardan, A.; Murakami, M.; Ohtake, T.; Coda, A.; Dorschner, R.A.; Bonnart, C.; Descargues, P.; Hovnanian, A.; et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat. Med. 2007, 13, 975–980. [Google Scholar] [CrossRef]
- Kasparek, P.; Ileninova, Z.; Zbodakova, O.; Kanchev, I.; Benada, O.; Chalupský, K.; Brattsand, M.; Beck, I.M.; Sedlacek, R. KLK5 and KLK7 Ablation Fully Rescues Lethality of Netherton Syndrome-Like Phenotype. PLoS Genet. 2017, 13, e1006566. [Google Scholar] [CrossRef] [PubMed]
- Samaras, P.; Schmidt, T.; Frejno, M.; Gessulat, S.; Reinecke, M.; Jarzab, A.; Zecha, J.; Mergner, J.; Giansanti, P.; Ehrlich, H.-C.; et al. ProteomicsDB: A multi-omics and multi-organism resource for life science research. Nucleic Acids Res. 2019, 48, D1153–D1163. [Google Scholar] [CrossRef] [PubMed]
- Borgoño, C.A.; Diamandis, E.P. The emerging roles of human tissue kallikreins in cancer. Nat. Rev. Cancer 2004, 4, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, F.; Baron, U.; Rajewsky, K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995, 23, 5080–5081. [Google Scholar] [CrossRef] [PubMed]
- Föll, M.; Fahrner, M.; Oria, V.O.; Kühs, M.; Biniossek, M.L.; Werner, M.; Bronsert, P.; Schilling, O. Reproducible proteomics sample preparation for single FFPE tissue slices using acid-labile surfactant and direct trypsinization. Clin. Proteom. 2018, 15, 1–15. [Google Scholar] [CrossRef]
- Oria, V.O.; Bronsert, P.; Thomsen, A.; Föll, M.; Zamboglou, C.; Hannibal, L.; Behringer, S.; Biniossek, M.; Schreiber, C.; Grosu, A.; et al. Proteome Profiling of Primary Pancreatic Ductal Adenocarcinomas Undergoing Additive Chemoradiation Link ALDH1A1 to Early Local Recurrence and Chemoradiation Resistance. Transl. Oncol. 2018, 11, 1307–1322. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Kammers, K.; Cole, R.N.; Tiengwe, C.; Ruczinski, I. Detecting significant changes in protein abundance. EuPA Open Proteom. 2015, 7, 11–19. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. TopGo: Enrichment Analysis for Gene Ontology; R Package Version 2.42.0. 2020. Available online: https://bioconductor.org/packages/release/bioc/html/topGO.html (accessed on 27 January 2021). [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; McInnes, J.; Kizilirmak, C.; Rehders, M.; Qatato, M.; Wirth, E.K.; Schweizer, U.; Verrey, F.; Heuer, H.; Brix, K. Interdependence of thyroglobulin processing and thyroid hormone export in the mouse thyroid gland. Eur. J. Cell Biol. 2017, 96, 440–456. [Google Scholar] [CrossRef] [PubMed]
- Qatato, M.; Szumska, J.; Skripnik, V.; Rijntjes, E.; Köhrle, J.; Brix, K. Canonical TSH Regulation of Cathepsin-Mediated Thyroglobulin Processing in the Thyroid Gland of Male Mice Requires Taar1 Expression. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Dauth, S.; Rakov, H.; Sîrbulescu, R.F.; Ilies, I.; Weber, J.; Dugershaw, B.B.; Braun, D.; Rehders, M.; Wirth, E.K.; Führer, D.; et al. Function of Cathepsin K in the Central Nervous System of Male Mice is Independent of Its Role in the Thyroid Gland. Cell. Mol. Neurobiol. 2020, 40, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.L.; Diamandis, E.P. Distribution of 15 human kallikreins in tissues and biological fluids. Clin Chem 2007, 53, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Prassas, I.; Dimitromanolakis, A.; Diamandis, E.P. Novel Biological Substrates of Human Kallikrein 7 Identified through Degradomics. J. Biol. Chem. 2015, 290, 17762–17775. [Google Scholar] [CrossRef] [PubMed]
- Mullur, R.; Liu, Y.-Y.; Brent, G.A. Thyroid Hormone Regulation of Metabolism. Physiol. Rev. 2014, 94, 355–382. [Google Scholar] [CrossRef]
- Weiner, J.; Hankir, M.K.; Heiker, J.T.; Fenske, W.; Krause, K. Thyroid hormones and browning of adipose tissue. Mol. Cell. Endocrinol. 2017, 458, 156–159. [Google Scholar] [CrossRef]
- Clements, J.A.; Matheson, B.; Funder, J. Tissue-specific regulation of the expression of rat kallikrein gene family members by thyroid hormone. Biochem. J. 1990, 267, 745–750. [Google Scholar] [CrossRef]
- Friedrichs, B.; Tepel, C.; Reinheckel, T.; Deussing, J.; Von Figura, K.; Herzog, V.; Peters, C.; Saftig, P.; Brix, K. Thyroid functions of mouse cathepsins B, K, and L. J. Clin. Investig. 2003, 111, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Brix, K.; Szumska, J.; Weber, J.; Qatato, M.; Venugopalan, V.; Al-Hashimi, A.; Rehders, M. Auto-Regulation of the Thyroid Gland Beyond Classical Pathways. Exp. Clin. Endocrinol. Diabetes 2020, 128, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Kralova, L.I.; Kralova, A.; Cejkova, S.; Fronek, J.; Petras, M.; Sekerkova, A.; Thieme, F.; Janousek, L.; Poledne, R. Characterisation and comparison of adipose tissue macrophages from human subcutaneous, visceral and perivascular adipose tissue. J. Transl. Med. 2016, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Haider, N.; Dusseault, J.; Larose, L. Nck1 Deficiency Impairs Adipogenesis by Activation of PDGFRalpha in Preadipocytes. Science 2018, 6, 22–37. [Google Scholar]
- Hubler, M.J.; Peterson, K.R.; Hasty, A.H. Iron homeostasis: A new job for macrophages in adipose tissue? Trends Endocrinol. Metab. 2015, 26, 101–109. [Google Scholar] [CrossRef]
- Sharma, N.K.; Das, S.K.; Mondal, A.K.; Hackney, O.G.; Chu, W.S.; Kern, P.A.; Rasouli, N.; Spencer, H.J.; Yao-Borengasser, A.; Elbein, S.C. Endoplasmic Reticulum Stress Markers Are Associated with Obesity in Nondiabetic Subjects. J. Clin. Endocrinol. Metab. 2008, 93, 4532–4541. [Google Scholar] [CrossRef]
- Giacchetti, G.; Faloia, E.; Sardu, C.; Camilloni, M.A.; Mariniello, B.; Gatti, C.; Garrapa, G.; Guerrieri, M.; Mantero, F. Gene expression of angiotensinogen in adipose tissue of obese patients. Int. J. Obes. 2000, 24, S142–S143. [Google Scholar] [CrossRef]
- Kim, S.; Urs, S.; Massiera, F.; Wortmann, P.; Joshi, R.; Heo, Y.-R.; Andersen, B.; Kobayashi, H.; Teboul, M.; Ailhaud, G.P.; et al. Effects of High-Fat Diet, Angiotensinogen (agt) Gene Inactivation, and Targeted Expression to Adipose Tissue on Lipid Metabolism and Renal Gene Expression. Horm. Metab. Res. 2002, 34, 721–725. [Google Scholar] [CrossRef]
- Kurokawa, J.; Nagano, H.; Ohara, O.; Kubota, N.; Kadowaki, T.; Arai, S.; Miyazaki, T. Apoptosis inhibitor of macrophage (AIM) is required for obesity-associated recruitment of inflammatory macrophages into adipose tissue. Proc. Natl. Acad. Sci. USA 2011, 108, 12072–12077. [Google Scholar] [CrossRef]
- Schultz, S.; Saalbach, A.; Heiker, J.T.; Meier, R.; Zellmann, T.; Simon, J.C.; Beck-Sickinger, A.G. Proteolytic activation of prochemerin by kallikrein 7 breaks an ionic linkage and results in C-terminal rearrangement. Biochem. J. 2013, 452, 271–280. [Google Scholar] [CrossRef]
- Kralisch, S.; Weise, S.; Sommer, G.; Lipfert, J.; Lossner, U.; Bluher, M.; Stumvoll, M.; Fasshauer, M. Interleukin-1beta induces the novel adipokine chemerin in adipocytes in vitro. Regul. Pept. 2009, 154, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Takahashi, Y.; Takahashi, K.; Zolotaryov, F.N.; Hong, K.S.; Kitazawa, R.; Iida, K.; Okimura, Y.; Kaji, H.; Kitazawa, S.; et al. Chemerin enhances insulin signaling and potentiates insulin-stimulated glucose uptake in 3T3-L1 adipocytes. FEBS Lett. 2008, 582, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Helfer, G.; Wu, Q.-F. Chemerin: A multifaceted adipokine involved in metabolic disorders. J. Endocrinol. 2018, 238, R79–R94. [Google Scholar] [CrossRef] [PubMed]
- Skytt, A.; Stromqvist, M.; Egelrud, T. Primary Substrate Specificity of Recombinant Human Stratum Corneum Chymotryptic Enzyme. Biochem. Biophys. Res. Commun. 1995, 211, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.P.C.; Freitas, R.F.; De Melo, L.S.; Barros, T.G.; Santos, J.A.N.; Juliano, M.A.; Pinheiro, S.; Blaber, M.; Juliano, L.; Muri, E.M.F.; et al. Isomannide-Based Peptidomimetics as Inhibitors for Human Tissue Kallikreins 5 and 7. ACS Med. Chem. Lett. 2013, 5, 128–132. [Google Scholar] [CrossRef]
- Arama, D.P.; Soualmia, F.; Lisowski, V.; Longevial, J.-F.; Bosc, E.; Maillard, L.T.; Martinez, J.; Masurier, N.; El Amri, C. Pyrido-imidazodiazepinones as a new class of reversible inhibitors of human kallikrein 7. Eur. J. Med. Chem. 2015, 93, 202–213. [Google Scholar] [CrossRef]
- Tan, X.; Soualmia, F.; Furio, L.; Renard, J.-F.; Kempen, I.; Qin, L.; Pagano, M.; Pirotte, B.; El Amri, C.; Hovnanian, A.; et al. Toward the First Class of Suicide Inhibitors of Kallikreins Involved in Skin Diseases. J. Med. Chem. 2014, 58, 598–612. [Google Scholar] [CrossRef]
- Jendrny, C.; Beck-Sickinger, A.G. Inhibition of Kallikrein-Related Peptidases 7 and 5 by Grafting Serpin Reactive-Center Loop Sequences onto Sunflower Trypsin Inhibitor-1 (SFTI-1). ChemBioChem 2016, 17, 719–726. [Google Scholar] [CrossRef]
- De Veer, S.J.; Furio, L.; Swedberg, J.E.; Munro, C.A.; Brattsand, M.; Clements, J.A.; Hovnanian, A.; Harris, J.M. Selective Substrates and Inhibitors for Kallikrein-Related Peptidase 7 (KLK7) Shed Light on KLK Proteolytic Activity in the Stratum Corneum. J. Investig. Dermatol. 2017, 137, 430–439. [Google Scholar] [CrossRef]
- Murafuji, H.; Sakai, H.; Goto, M.; Imajo, S.; Sugawara, H.; Muto, T. Discovery and structure–activity relationship study of 1,3,6-trisubstituted 1,4-diazepane-7-ones as novel human kallikrein 7 inhibitors. Bioorganic Med. Chem. Lett. 2017, 27, 5272–5276. [Google Scholar] [CrossRef]
- De Souza, A.S.; Pacheco, B.D.C.; Pinheiro, S.; Muri, E.M.F.; Dias, L.R.S.; Lima, C.H.S.; Garrett, R.; de Moraes, M.B.M.; de Souza, B.E.G.; Puzer, L. 3-Acyltetramic acids as a novel class of inhibitors for human kallikreins 5 and 7. Bioorg. Med. Chem. Lett. 2019, 29, 1094–1098. [Google Scholar] [CrossRef]
- Murafuji, H.; Muto, T.; Goto, M.; Imajo, S.; Sugawara, H.; Oyama, Y.; Minamitsuji, Y.; Miyazaki, S.; Murai, K.; Fujioka, H. Discovery and structure-activity relationship of imidazolinylindole derivatives as kallikrein 7 inhibitors. Bioorganic Med. Chem. Lett. 2019, 29, 334–338. [Google Scholar] [CrossRef]
- Chen, Q.-Y.; Luo, D.; Seabra, G.M.; Luesch, H. Ahp-Cyclodepsipeptides as tunable inhibitors of human neutrophil elastase and kallikrein 7: Total synthesis of tutuilamide A, serine protease selectivity profile and comparison with lyngbyastatin7. Bioorganic Med. Chem. 2020, 28, 115756. [Google Scholar] [CrossRef] [PubMed]
- Hanke, S.; Tindall, C.A.; Pippel, J.; Ulbricht, D.; Pirotte, B.; Ravaux, M.R.-; Heiker, J.T.; Sträter, N. Structural Studies on the Inhibitory Binding Mode of Aromatic Coumarinic Esters to Human Kallikrein-Related Peptidase 7. J. Med. Chem. 2020, 63, 5723–5733. [Google Scholar] [CrossRef] [PubMed]
- Laureano, A.F.S.; Zani, M.B.; Sant’Ana, A.M.; Tognato, R.C.; Lombello, C.B.; Nascimento, M.H.M.D.; Helmsing, S.; Fühner, V.; Hust, M.; Puzer, L. Generation of recombinant antibodies against human tissue kallikrein 7 to treat skin diseases. Bioorganic Med. Chem. Lett. 2020, 30, 127626. [Google Scholar] [CrossRef] [PubMed]



| Chow | HFD | |||
|---|---|---|---|---|
| Control (n) | Klk7−/− (n) | Control (n) | Klk7−/− (n) | |
| Body weight (g) | 29.25 ± 0.62 (8) | 26.83 ± 0.73 (8) * | 39.97 ± 1.38 (9) | 34.24 ± 0.89 (5) * |
| Body length (cm) | 9.95 ± 0.73 (8) | 9.76 ± 0.11 (8) | 9.86 ± 0.09 (9) | 10.00 ± 0.10 (5) |
| Body temp. (°C) | 35.81 ± 0.37 (10) | 34.09 ± 0.71 (8) * | ||
| Serum lipids | ||||
| Triglycerides (mmol/L) | 1.10 ± 0.05 (8) | 1.27 ± 0.14 (3) | 1.48 ± 0.05 (9) | 1.34 ± 0.11 (5) |
| Cholesterol (mmol/L) | 2.32 ± 0.04 (8) | 2.33 ± 0.08 (3) | 4.60 ± 0.28 (9) | 4.14 ± 0.24 (5) |
| FFA (mmol/L) | 1.71 ± 0.10 (8) | 2.06 ± 0.23 (3) | 2.39 ± 0.05 (9) | 2.84 ± 0.20 (5) * |
| Glycerol (nmol/mL) | 670.21 ± 30.49 (8) | 722.59 ± 51.94 (7) | 889.81 ± 44.83 (7) | 848.92 ± 29.95 (2) |
| Adipokines | ||||
| Adiponectin (µg/mL) | 47.68 ± 1.22 (8) | 54.77 ± 1.96 (7) ** | 48.49 ± 1.02 (8) | 49.52 ± 1.39 (5) |
| Leptin (ng/mL) | 13.90 ± 2.69 (8) | 6.16 ± 2.41 (7) ** | 178.59 ± 28.59 (8) | 49.45 ± 19.48 (5) * |
| Vaspin (pg/mL) | 65.18 ± 11.15 (7) | 58.65 ± 6.18 (8) | 122.17 ± 22.57 (4) | 136.91 ± 16.43 (4) |
| Chemerin (ng/mL) | 128.52 ± 6.58 (7) | 111.56 ± 4.89 (6) | 168.69 ± 15.21 (7) | 156.29 ± 14.45 (4) |
| MCP-1 (pg/mL) | 215.90 ± 43.91 (7) | 141.52 ± 19.81 (8) * | 147.35 ± 18.90 (6) | 108.55 ± 18.53 (4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kunath, A.; Weiner, J.; Krause, K.; Rehders, M.; Pejkovska, A.; Gericke, M.; Biniossek, M.L.; Dommel, S.; Kern, M.; Ribas-Latre, A.; et al. Role of Kallikrein 7 in Body Weight and Fat Mass Regulation. Biomedicines 2021, 9, 131. https://doi.org/10.3390/biomedicines9020131
Kunath A, Weiner J, Krause K, Rehders M, Pejkovska A, Gericke M, Biniossek ML, Dommel S, Kern M, Ribas-Latre A, et al. Role of Kallikrein 7 in Body Weight and Fat Mass Regulation. Biomedicines. 2021; 9(2):131. https://doi.org/10.3390/biomedicines9020131
Chicago/Turabian StyleKunath, Anne, Juliane Weiner, Kerstin Krause, Maren Rehders, Anastasija Pejkovska, Martin Gericke, Martin L. Biniossek, Sebastian Dommel, Matthias Kern, Aleix Ribas-Latre, and et al. 2021. "Role of Kallikrein 7 in Body Weight and Fat Mass Regulation" Biomedicines 9, no. 2: 131. https://doi.org/10.3390/biomedicines9020131
APA StyleKunath, A., Weiner, J., Krause, K., Rehders, M., Pejkovska, A., Gericke, M., Biniossek, M. L., Dommel, S., Kern, M., Ribas-Latre, A., Schilling, O., Brix, K., Stumvoll, M., Klöting, N., Heiker, J. T., & Blüher, M. (2021). Role of Kallikrein 7 in Body Weight and Fat Mass Regulation. Biomedicines, 9(2), 131. https://doi.org/10.3390/biomedicines9020131






