Computational Study on Potential Novel Anti-Ebola Virus Protein VP35 Natural Compounds
Abstract
1. Introduction
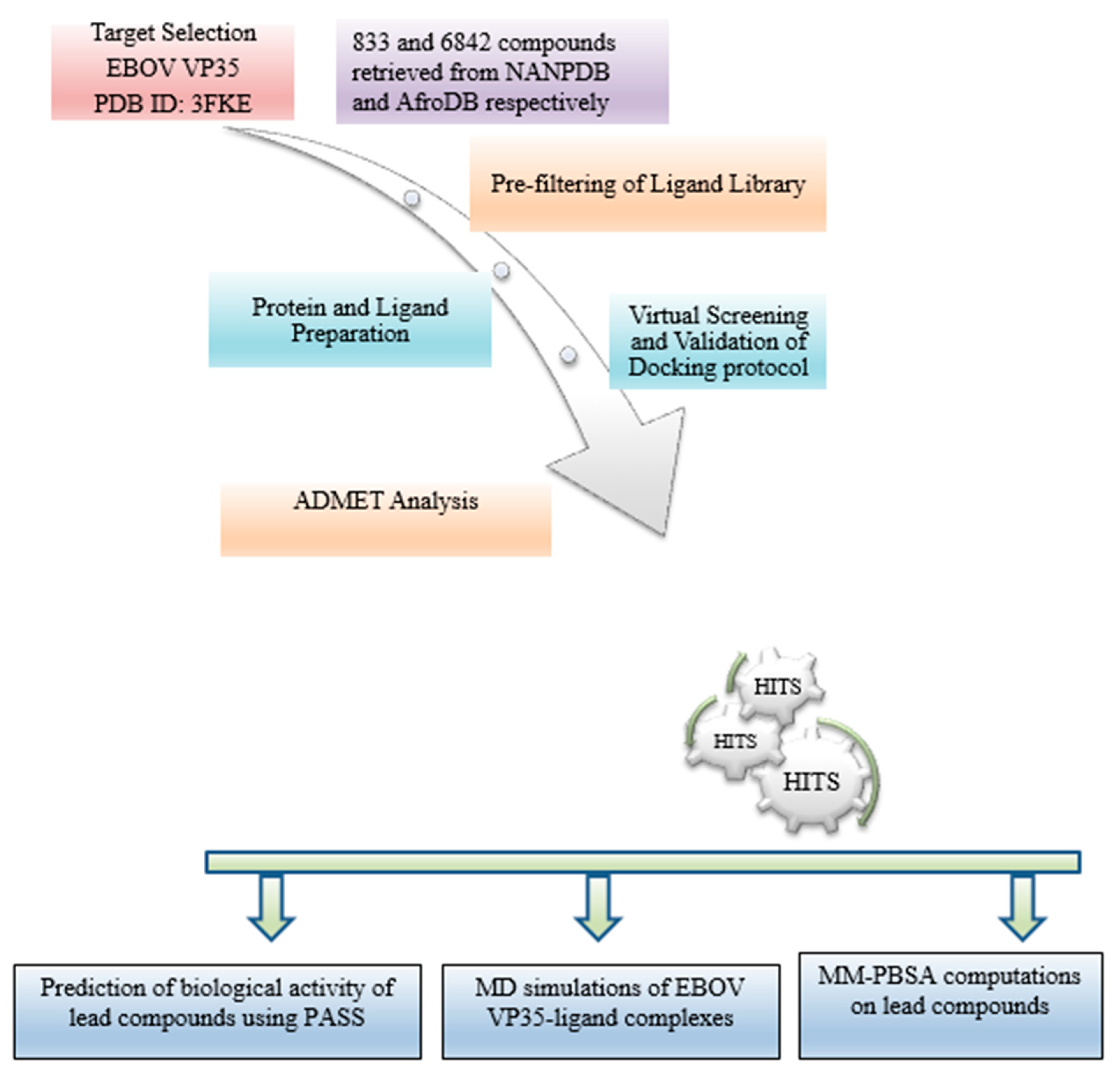
2. Materials and Methods
2.1. Protein Retrieval
2.2. Retrieval of Compounds from Natural Products Databases
2.3. Protein Active Site Evaluation
2.4. Pre-Filtering of Ligand Library
2.5. Protein and Ligand Preparation
2.6. Virtual Screening and Validation of Docking Protocol
2.7. Molecular Interaction Profiling
2.8. Pharmacokinetic Profiling
2.9. Prediction of Anti-Viral Activity of Lead Compounds
2.10. Quality and Efficiency of Evaluation of Potential Lead Compounds
2.11. Molecular Dynamic Simulations of Protein-Ligand Complexes
2.12. Binding Free Energy Calculations of Protein-Ligand Complexes by MM-PBSA
3. Results
3.1. Structural and Binding Site Analysis
3.2. Molecular Docking Studies
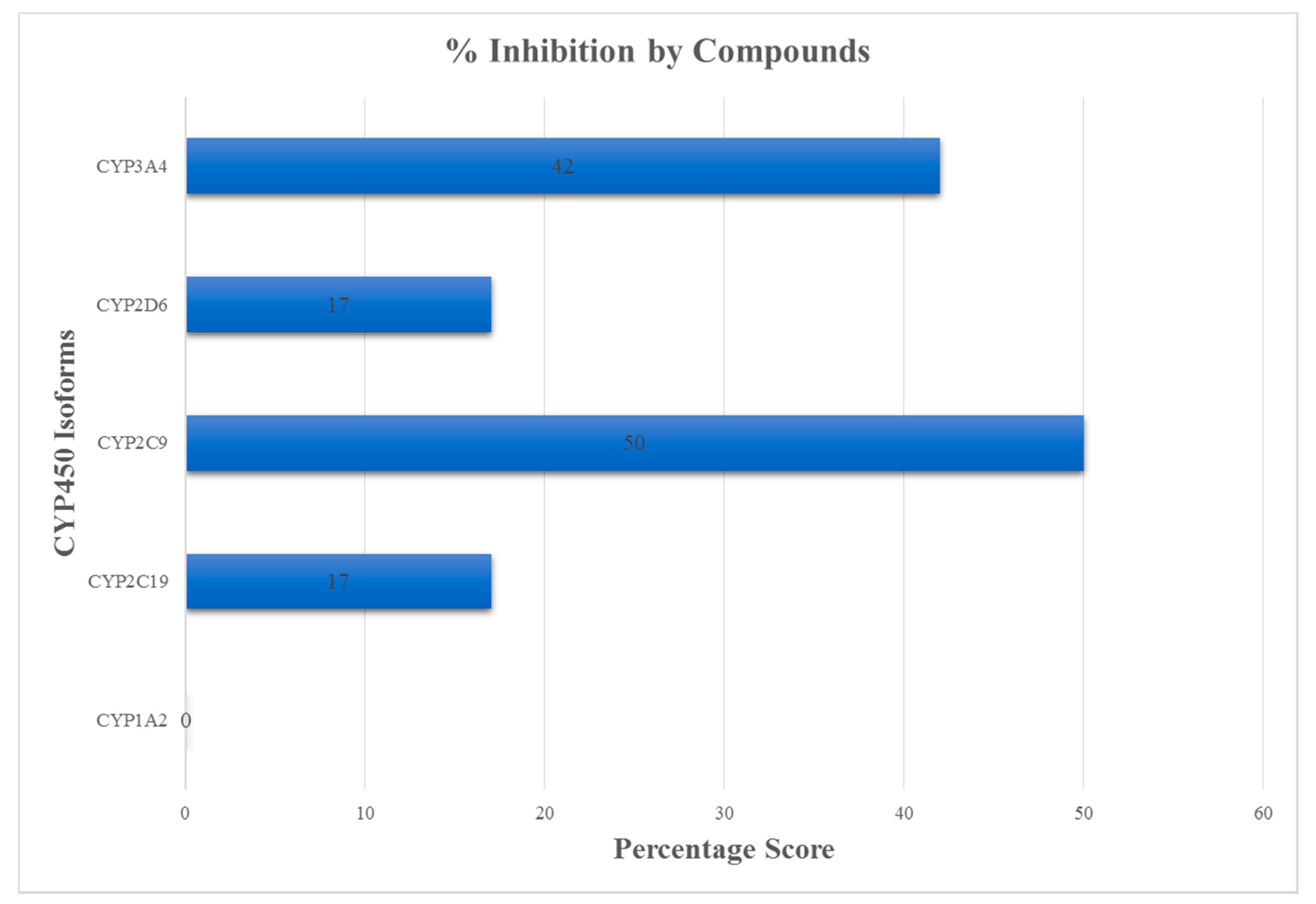
3.3. ADMET Profiling for Identification of Drug-Likeliness
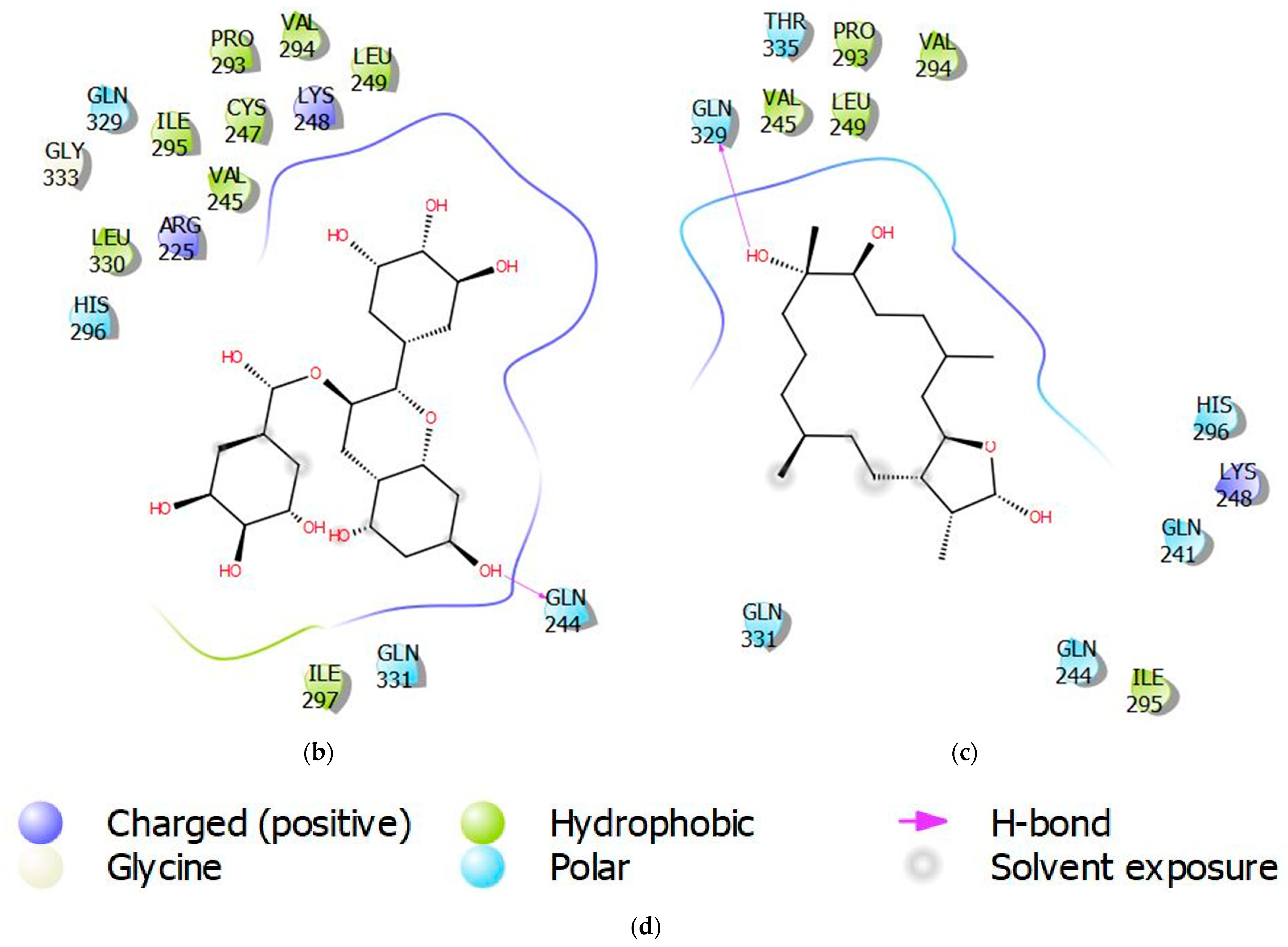
3.4. Molecular Interactions of Protein-Ligand Complexes
3.5. Biological Activity Predictions for Ligands
3.6. Assessment of Quality of Ligands
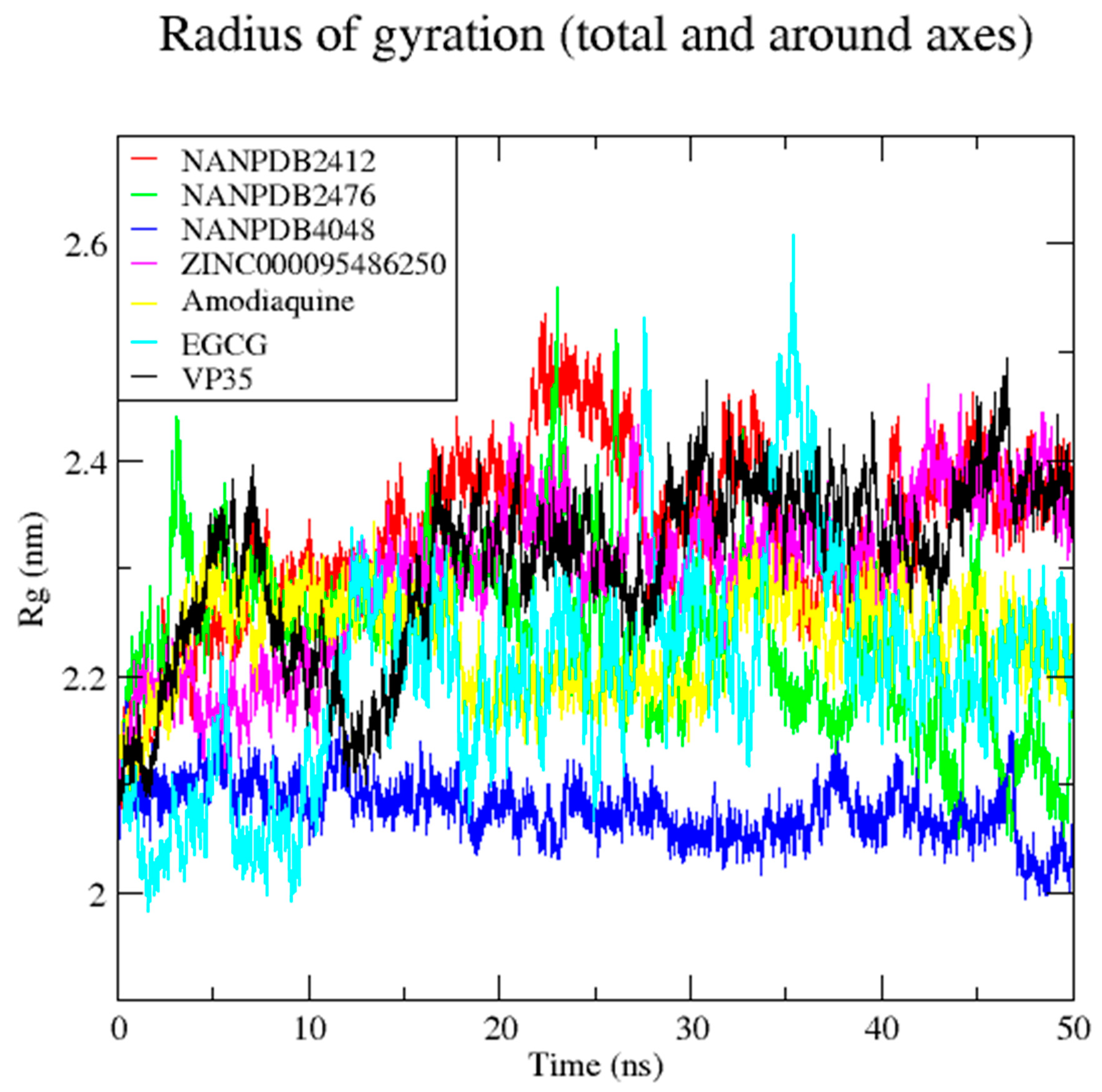
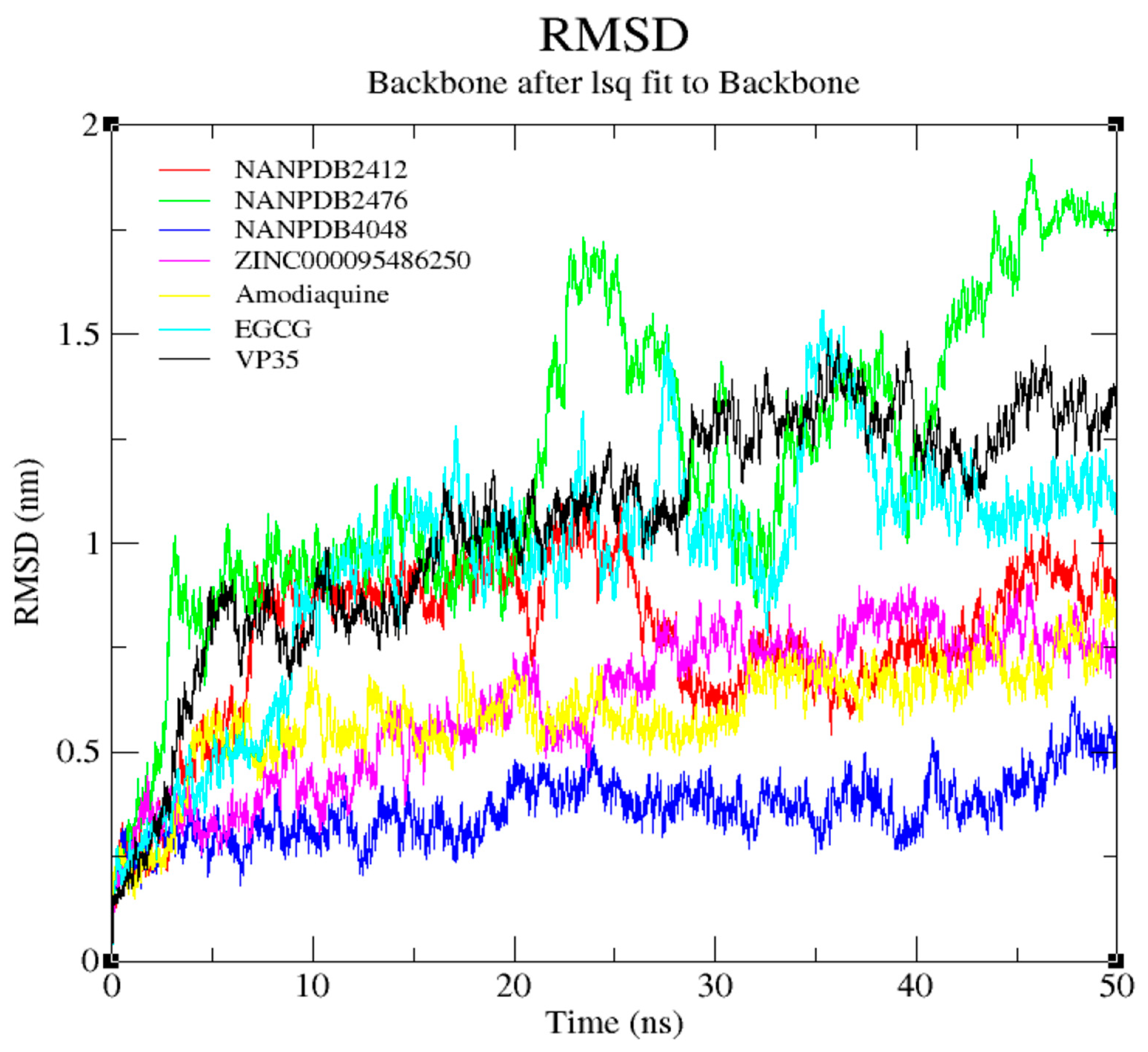
3.7. Molecular Dynamics Simulation of VP35-Ligand Complexes
3.8. MM-PBSA Computations on Potential Lead Compounds
3.9. Structural Similarity Search of Hits
| Compound ID | IUPAC Names | Two-Dimensional Structure |
|---|---|---|
| NANPDB2412 | (1R,2R,5S,7S,8S,13R,14R,17R)-2,7,14-trimethyl-16-oxapentacyclo[9.7.0.02,8.05,7.013,17]octadeca-3,10-diene-12,15-dione |  |
| NANPDB2476 | (1S,3R,10S,11R,14R,16R)-5,11,14-trimethyl-2,7-dioxapentacyclo[8.8.0.0¹,³.0⁴,⁸.011,16]octadeca-4,8-dien-6-one |  |
| NANPDB4048 | (1Z,2S,3aR,3bS,9aR,9bS,11aS)-1-ethylidene-2-hydroxy-9a,11a-dimethyl-1H,2H,3H,3aH,3bH,4H,5H,7H,8H,9H,9aH,9bH,11aH-cyclopenta[a]phenanthren-7-one | 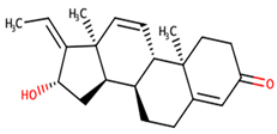 |
| ZINC000095486250 | (6aR,12aS)-6a,9,9,12a-tetramethyl-3H,4H,5H,6aH,7H,9H,10H,11H,12H,12aH-naphtho[2,1-b]oxocin-3-one | 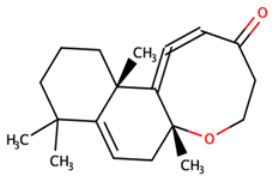 |
4. Implications and Future Prospects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajak, H.; Jain, D.K.; Singh, A.; Sharma, A.K.; Dixit, A. Ebola virus disease: Past, present and future. Asian Pac. J. Trop. Biomed. 2015, 5, 337–343. [Google Scholar] [CrossRef]
- Breman, J.G.; Heymann, D.L.; Lloyd, G.; McCormick, J.B.; Miatudila, M.; Murphy, F.A.; Muyembé-Tamfun, J.J.; Piot, P.; Ruppol, J.F.; Sureau, P.; et al. Discovery and Description of Ebola Zaire Virus in 1976 and Relevance to the West African Epidemic during 2013–2016. J. Infect. Dis. 2016, 214, S93–S101. [Google Scholar] [CrossRef]
- Schwartz, D.A. Maternal and Infant Death and the rVSV-ZEBOV Vaccine Through Three Recent Ebola Virus Epidemics-West Africa, DRC Équateur and DRC Kivu: 4 Years of Excluding Pregnant and Lactating Women and Their Infants from Immunization. Curr. Trop. Med. Rep. 2019, 6, 213–222. [Google Scholar] [CrossRef]
- Chowell, G.; Nishiura, H. Transmission dynamics and control of Ebola virus disease (EVD): A review. BMC Med. 2014, 12, 196. [Google Scholar] [CrossRef]
- Safari, S.; Baratloo, A.; Rouhipour, A.; Ghelichkhani, P.; Yousefifard, M. Ebola Hemorrhagic Fever as a Public Health Emergency of International Concern; a Review Article. Emergency 2015, 3, 3–7. [Google Scholar] [CrossRef]
- Vetter, P.; Fischer, W.A.; Schibler, M.; Jacobs, M.; Bausch, D.G.; Kaiser, L. Ebola Virus Shedding and Transmission: Review of Current Evidence. J. Infect. Dis. 2016, 214, S177–S184. [Google Scholar] [CrossRef]
- Dixit, D.; Masumbuko Claude, K.; Kjaldgaard, L.; Hawkes, M.T. Review of Ebola virus disease in children—How far have we come? Paediatr. Int. Child Health 2021, 41, 12–27. [Google Scholar] [CrossRef]
- Judson, S.; Prescott, J.; Munster, V. Understanding Ebola Virus Transmission. Viruses 2015, 7, 511–521. [Google Scholar] [CrossRef]
- To, K.K.W.; Chan, J.F.W.; Tsang, A.K.L.; Cheng, V.C.C.; Yuen, K.-Y. Ebola virus disease: A highly fatal infectious disease reemerging in West Africa. Microbes Infect. 2015, 17, 84–97. [Google Scholar] [CrossRef]
- Alexander, K.A.; Sanderson, C.E.; Marathe, M.; Lewis, B.L.; Rivers, C.M.; Shaman, J.; Drake, J.M.; Lofgren, E.; Dato, V.M.; Eisenberg, M.C.; et al. What Factors Might Have Led to the Emergence of Ebola in West Africa? PLoS Negl. Trop. Dis. 2015, 9, e0003652. [Google Scholar] [CrossRef]
- Goeijenbier, M.; van Kampen, J.J.A.; Reusken, C.B.E.M.; Koopmans, M.P.G.; van Gorp, E.C.M. Ebola virus disease: A review on epidemiology, symptoms, treatment and pathogenesis. Neth. J. Med. 2014, 72, 442–448. [Google Scholar]
- Maras, M.H.; Miranda, M.D. The weaponization of Ebola: A new risk in the wake of an outbreak? Comp. Strategy 2016, 35, 72–79. [Google Scholar] [CrossRef]
- St Claire, M.C.; Ragland, D.R.; Bollinger, L.; Jahrling, P.B. Animal Models of Ebolavirus Infection. Comp. Med. 2017, 67, 253–262. [Google Scholar]
- Falasca, L.; Agrati, C.; Petrosillo, N.; Di Caro, A.; Capobianchi, M.R.; Ippolito, G.; Piacentini, M. Molecular mechanisms of Ebola virus pathogenesis: Focus on cell death. Cell Death Differ. 2015, 22, 1250–1259. [Google Scholar] [CrossRef]
- Weyer, J.; Grobbelaar, A.; Blumberg, L. Ebola Virus Disease: History, Epidemiology and Outbreaks. Curr. Infect. Dis. Rep. 2015, 17, 21. [Google Scholar] [CrossRef]
- Hewlett, A.; Vasa, A.M.; Cieslak, T.J.; Lowe, J.J.; Schwedhelm, S. Viral Hemorrhagic Fever Preparedness. In Infection Prevention; Springer International Publishing: Cham, Switzerland, 2018; pp. 197–211. ISBN 9783319609805. [Google Scholar]
- Takamatsu, Y.; Kolesnikova, L.; Becker, S. Ebola virus proteins NP, VP35, and VP24 are essential and sufficient to mediate nucleocapsid transport. Proc. Natl. Acad. Sci. USA 2018, 115, 1075–1080. [Google Scholar] [CrossRef]
- Hume, A.J.; Mühlberger, E. Distinct Genome Replication and Transcription Strategies within the Growing Filovirus Family. J. Mol. Biol. 2019, 431, 4290–4320. [Google Scholar] [CrossRef]
- Balmith, M.; Soliman, M.E.S. Potential Ebola drug targets-filling the gap: A critical step forward towards the design and discovery of potential drugs. Biologia 2017, 72, 1–13. [Google Scholar] [CrossRef]
- Takada, A.; Kawaoka, Y. The pathogenesis of Ebola hemorrhagic fever. Trends Microbiol. 2001, 9, 506–511. [Google Scholar] [CrossRef]
- Olukitibi, T.A.; Ao, Z.; Mahmoudi, M.; Kobinger, G.A.; Yao, X. Dendritic Cells/Macrophages-Targeting Feature of Ebola Glycoprotein and its Potential as Immunological Facilitator for Antiviral Vaccine Approach. Microorganisms 2019, 7, 402. [Google Scholar] [CrossRef]
- Jasenosky, L.D.; Cadena, C.; Mire, C.E.; Borisevich, V.; Haridas, V.; Ranjbar, S.; Nambu, A.; Bavari, S.; Soloveva, V.; Sadukhan, S.; et al. The FDA-Approved Oral Drug Nitazoxanide Amplifies Host Antiviral Responses and Inhibits Ebola Virus. iScience 2019, 19, 1279–1290. [Google Scholar] [CrossRef]
- Kimberlin, C.R.; Bornholdt, Z.A.; Li, S.; Woods, V.L.; MacRae, I.J.; Saphire, E.O. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. USA 2010, 107, 314–319. [Google Scholar] [CrossRef]
- Cárdenas, W.B.; Loo, Y.-M.; Gale, M.; Hartman, A.L.; Kimberlin, C.R.; Martínez-Sobrido, L.; Saphire, E.O.; Basler, C.F. Ebola Virus VP35 Protein Binds Double-Stranded RNA and Inhibits Alpha/Beta Interferon Production Induced by RIG-I Signaling. J. Virol. 2006, 80, 5168–5178. [Google Scholar] [CrossRef]
- Leung, D.W.; Basler, C.F.; Amarasinghe, G.K. Molecular mechanisms of viral inhibitors of RIG-I-like receptors. Trends Microbiol. 2012, 20, 139–146. [Google Scholar] [CrossRef]
- Seesuay, W.; Jittavisutthikul, S.; Sae-lim, N.; Sookrung, N.; Sakolvaree, Y.; Chaicumpa, W. Human transbodies that interfere with the functions of Ebola virus VP35 protein in genome replication and transcription and innate immune antagonism. Emerg. Microbes Infect. 2018, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ekins, S.; Freundlich, J.S.; Coffee, M. A common feature pharmacophore for FDA-approved drugs inhibiting the Ebola virus. F1000Research 2014, 3, 277. [Google Scholar] [CrossRef] [PubMed]
- Pleško, S. In Silico Study of Plant Polyphenols’ Interactions with VP24–Ebola Virus Matrix Protein. Acta Chim. Slov. 2015, 62, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Raj, U.; Varadwaj, P.K. Flavonoids as Multi-target Inhibitors for Proteins Associated with Ebola Virus: In Silico Discovery Using Virtual Screening and Molecular Docking Studies. Interdiscip. Sci. Comput. Life Sci. 2016, 8, 132–141. [Google Scholar] [CrossRef]
- Saxena, D.; Kaul, G.; Dasgupta, A.; Chopra, S. Atoltivimab/maftivimab/odesivimab (Inmazeb) combination to treat infection caused by Zaire ebolavirus. Drugs Today 2021, 57, 483. [Google Scholar] [CrossRef]
- Lane, T.; Anantpadma, M.; Freundlich, J.S.; Davey, R.A.; Madrid, P.B.; Ekins, S. The Natural Product Eugenol Is an Inhibitor of the Ebola Virus In Vitro. Pharm. Res. 2019, 36, 104. [Google Scholar] [CrossRef]
- Catarino, L.; Romeiras, M.M. Biodiversity of Vegetation and Flora in Tropical Africa. Diversity 2020, 12, 369. [Google Scholar] [CrossRef]
- Glanzer, J.G.; Byrne, B.M.; McCoy, A.M.; James, B.J.; Frank, J.D.; Oakley, G.G. In silico and in vitro methods to identify ebola virus VP35-dsRNA inhibitors. Bioorg. Med. Chem. 2016, 24, 5388–5392. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Koulouridi, E.; Valli, M.; Ntie-Kang, F.; Bolzani, V.D.S. A primer on natural product-based virtual screening. Phys. Sci. Rev. 2019, 4, 251–290. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Telukunta, K.K.; Döring, K.; Simoben, C.V.; Moumbock, A.F.A.; Malange, Y.I.; Njume, L.E.; Yong, J.N.; Sippl, W.; Günther, S. NANPDB: A Resource for Natural Products from Northern African Sources. J. Nat. Prod. 2017, 80, 2067–2076. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Ekins, S.; Freundlich, J.S.; Reynolds, R.C. Fusing Dual-Event Data Sets for Mycobacterium tuberculosis Machine Learning Models and Their Evaluation. J. Chem. Inf. Model. 2013, 53, 3054–3063. [Google Scholar] [CrossRef]
- Serafim, M.S.M.; Kronenberger, T.; Oliveira, P.R.; Poso, A.; Honório, K.M.; Mota, B.E.F.; Maltarollo, V.G. The application of machine learning techniques to innovative antibacterial discovery and development. Expert Opin. Drug Discov. 2020, 15, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.; Atanasova, M.; Valkova, I.; Stavrakov, G.; Philipova, I.; Zhivkova, Z.; Zheleva-Dimitrova, D.; Konstantinov, S.; Dimitrov, I. Novel hits for acetylcholinesterase inhibition derived by docking-based screening on ZINC database. J. Enzym. Inhib. Med. Chem. 2018, 33, 768–776. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Alam, S.; Khan, F. 3D-QSAR studies on Maslinic acid analogs for Anticancer activity against Breast Cancer cell line MCF-7. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Hussain, W.; Qaddir, I.; Mahmood, S.; Rasool, N. In silico targeting of non-structural 4B protein from dengue virus 4 with spiropyrazolopyridone: Study of molecular dynamics simulation, ADMET and virtual screening. VirusDisease 2018, 29, 147–156. [Google Scholar] [CrossRef]
- Haghighi, O.; Davaeifar, S.; Zahiri, H.S.; Maleki, H.; Noghabi, K.A. Homology Modeling and Molecular Docking Studies of Glutamate Dehydrogenase (GDH) from Cyanobacterium Synechocystis sp. PCC 6803. Int. J. Pept. Res. Ther. 2020, 26, 783–793. [Google Scholar] [CrossRef]
- Konidala, K.K.; Bommu, U.D.; Yeguvapalli, S.; Pabbaraju, N. In silico insights into prediction and analysis of potential novel pyrrolopyridine analogs against human MAPKAPK-2: A new SAR-based hierarchical clustering approach. 3 Biotech 2018, 8, 385. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; De Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Réau, M.; Langenfeld, F.; Zagury, J.F.; Lagarde, N.; Montes, M. Decoys selection in benchmarking datasets: Overview and perspectives. Front. Pharmacol. 2018, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cruz, A.; Ramsey, S.; Dickson, C.J.; Duca, J.S.; Hornak, V.; Koes, D.R.; Kurtzman, T. Hidden bias in the DUD-E dataset leads to misleading performance of deep learning in structure-based virtual screening. PLoS ONE 2019, 14, e0220113. [Google Scholar] [CrossRef]
- Goksuluk, D.; Korkmaz, S.; Zararsiz, G.; Karaagaoglu, A.E. easyROC: An Interactive Web-tool for ROC Curve Analysis Using R Language Environment. R J. 2016, 8, 213. [Google Scholar] [CrossRef]
- Shamsara, J. Correlation between Virtual Screening Performance and Binding Site Descriptors of Protein Targets. Int. J. Med. Chem. 2018, 2018, 3829307. [Google Scholar] [CrossRef] [PubMed]
- Biovia, D. Discovery Studio Modeling Environment, Release 2017, San Diego: DassaultSystèmes, 2016. Adres. 2016. Available online: http//accelrys.com/products/collaborative-science/biovia-discoverystudio/visualizationdownload.php (accessed on 6 May 2020).
- Kumavath, R.; Azad, M.; Devarapalli, P.; Tiwari, S.; Kar, S.; Barh, D.; Azevedo, V.; Kumar, A.P. Novel aromatase inhibitors selection using induced fit docking and extra precision methods: Potential clinical use in ER-alpha-positive breast cancer. Bioinformation 2016, 12, 324–331. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Al Wasidi, A.S.; Hassan, A.S.; Naglah, A.M. In vitro cytotoxicity and druglikeness of pyrazolines and pyridines bearing benzofuran moiety. J. Appl. Pharm. Sci. 2020, 10, 142–148. [Google Scholar] [CrossRef]
- Zafar, F.; Gupta, A.; Thangavel, K.; Khatana, K.; Sani, A.A.; Ghosal, A.; Tandon, P.; Nishat, N. Physicochemical and Pharmacokinetic Analysis of Anacardic Acid Derivatives. ACS Omega 2020, 5, 6021–6030. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Filimonov, D.A.; Druzhilovskiy, D.S.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Dmitriev, A.V.; Pogodin, P.V.; Poroikov, V.V. Computer-aided prediction of biological activity spectra for chemical compounds: Opportunities and limitation. Biomed. Chem. Res. Methods 2018, 1, e00004. [Google Scholar] [CrossRef]
- Tarasova, O.; Biziukova, N.; Kireev, D.; Lagunin, A.; Ivanov, S.; Filimonov, D.; Poroikov, V. A computational approach for the prediction of treatment history and the effectiveness or failure of antiretroviral therapy. Int. J. Mol. Sci. 2020, 21, 748. [Google Scholar] [CrossRef]
- Rajput, A.; Kumar, M. Anti-Ebola: An initiative to predict Ebola virus inhibitors through machine learning. Mol. Divers. 2021, 1, 1–10. [Google Scholar] [CrossRef]
- Kenny, P.W.; Leitão, A.; Montanari, C.A. Ligand efficiency metrics considered harmful. J. Comput.-Aided Mol. Des. 2014, 28, 699–710. [Google Scholar] [CrossRef]
- Reynolds, C.H.; Reynolds, R.C. Group Additivity in Ligand Binding Affinity: An Alternative Approach to Ligand Efficiency. J. Chem. Inf. Model. 2017, 57, 3086–3093. [Google Scholar] [CrossRef]
- Cavalluzzi, M.M.; Mangiatordi, G.F.; Nicolotti, O.; Lentini, G. Ligand efficiency metrics in drug discovery: The pros and cons from a practical perspective. Expert Opin. Drug Discov. 2017, 12, 1087–1104. [Google Scholar] [CrossRef]
- Islam, M.A.; Pillay, T.S. Identification of promising anti-DNA gyrase antibacterial compounds using de novo design, molecular docking and molecular dynamics studies. J. Biomol. Struct. Dyn. 2019, 38, 1798–1809. [Google Scholar] [CrossRef]
- Selvaraj, C.; Sakkiah, S.; Tong, W.; Hong, H. Molecular dynamics simulations and applications in computational toxicology and nanotoxicology. Food Chem. Toxicol. 2018, 112, 495–506. [Google Scholar] [CrossRef]
- Zhu, S. Validation of the Generalized Force Fields GAFF, CGenFF, OPLS-AA, and PRODRGFF by Testing against Experimental Osmotic Coefficient Data for Small Drug-Like Molecules. J. Chem. Inf. Model. 2019, 59, 4239–4247. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Viet, M.H.; Li, M.S. Effects of water models on binding affinity: Evidence from all-atom simulation of binding of tamiflu to A/H5N1 neuraminidase. Sci. World J. 2014, 2014, 536084. [Google Scholar] [CrossRef]
- Kumar, D.; Kumari, K.; Jayaraj, A.; Kumar, V.; Kumar, R.V.; Dass, S.K.; Chandra, R.; Singh, P. Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. J. Biomol. Struct. Dyn. 2020, 39, 2659–2672. [Google Scholar] [CrossRef]
- Childers, M.C.; Daggett, V. Validating Molecular Dynamics Simulations against Experimental Observables in Light of Underlying Conformational Ensembles. J. Phys. Chem. B 2018, 122, 6673–6689. [Google Scholar] [CrossRef]
- Martínez, L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE 2015, 10, e0119264. [Google Scholar] [CrossRef]
- Ul Hasnain, M.J.; Shoaib, M.; Qadri, S.; Afzal, B.; Anwar, T.; Abbas, S.H.; Sarwar, A.; Malik, H.M.T.; Pervez, M.T. Computational analysis of functional single nucleotide polymorphisms associated with SLC26A4 gene. PLoS ONE 2020, 15, e0225368. [Google Scholar] [CrossRef]
- Paissoni, C.; Spiliotopoulos, D.; Musco, G.; Spitaleri, A. GMXPBSA 2.1: A GROMACS tool to perform MM/PBSA and computational alanine scanning. Comput. Phys. Commun. 2015, 186, 105–107. [Google Scholar] [CrossRef]
- Alkarkhi, A.F.M.; Alqaraghuli, W.A.A. R Statistical Software. In Applied Statistics for Environmental Science with R; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Leung, D.W.; Ginder, N.D.; Fulton, D.B.; Nix, J.; Basler, C.F.; Honzatko, R.B.; Amarasinghe, G.K. Structure of the Ebola VP35 interferon inhibitory domain. Proc. Natl. Acad. Sci. USA 2009, 106, 411–416. [Google Scholar] [CrossRef]
- Dilley, K.A.; Voorhies, A.A.; Luthra, P.; Puri, V.; Stockwell, T.B.; Lorenzi, H.; Basler, C.F.; Shabman, R.S. The Ebola virus VP35 protein binds viral immunostimulatory and host RNAs identified through deep sequencing. PLoS ONE 2017, 12, e0178717. [Google Scholar] [CrossRef]
- Prins, K.C.; Binning, J.M.; Shabman, R.S.; Leung, D.W.; Amarasinghe, G.K.; Basler, C.F. Basic Residues within the Ebolavirus VP35 Protein Are Required for Its Viral Polymerase Cofactor Function. J. Virol. 2010, 84, 10581–10591. [Google Scholar] [CrossRef]
- Banerjee, A.; Mitra, P. Ebola Virus VP35 Protein: Modeling of the Tetrameric Structure and an Analysis of Its Interaction with Human PKR. J. Proteome Res. 2020, 19, 4533–4542. [Google Scholar] [CrossRef]
- Brown, C.S.; Lee, M.S.; Leung, D.W.; Wang, T.; Xu, W.; Luthra, P.; Anantpadma, M.; Shabman, R.S.; Melito, L.M.; MacMillan, K.S.; et al. In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity. J. Mol. Biol. 2014, 426, 2045–2058. [Google Scholar] [CrossRef]
- Leung, D.W.; Prins, K.C.; Borek, D.M.; Farahbakhsh, M.; Tufariello, J.M.; Ramanan, P.; Nix, J.C.; Helgeson, L.A.; Otwinowski, Z.; Honzatko, R.B.; et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol. 2010, 17, 165–172. [Google Scholar] [CrossRef]
- Mirza, M.U.; Ikram, N. Integrated computational approach for virtual hit identification against ebola viral proteins VP35 and VP40. Int. J. Mol. Sci. 2016, 17, 1748. [Google Scholar] [CrossRef]
- Kashyap, S. Comparative Insillico Studies on Phytochemicals of Ocimum as Natural Inhibitors of Ebola Vp-35 Protein. Indo Am. J. Pharm. Sci. 2019, 10, 489–511. [Google Scholar] [CrossRef]
- Empereur-mot, C.; Guillemain, H.; Latouche, A.; Zagury, J.-F.; Viallon, V.; Montes, M. Predictiveness curves in virtual screening. J. Cheminform. 2015, 7, 52. [Google Scholar] [CrossRef]
- Li, F.; He, H. Assessing the Accuracy of Diagnostic Tests. Shanghai Arch. Psychiatry 2018, 30, 207–212. [Google Scholar] [CrossRef]
- Mohan, R.R.; Wilson, M.; Gorham, R.D.; Harrison, R.E.S.; Morikis, V.A.; Kieslich, C.A.; Orr, A.A.; Coley, A.V.; Tamamis, P.; Morikis, D. Virtual Screening of Chemical Compounds for Discovery of Complement C3 Ligands. ACS Omega 2018, 3, 6427–6438. [Google Scholar] [CrossRef] [PubMed]
- Palacio-Rodríguez, K.; Lans, I.; Cavasotto, C.N.; Cossio, P. Exponential consensus ranking improves the outcome in docking and receptor ensemble docking. Sci. Rep. 2019, 9, 5142. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, K.O.; Kolapo, T.U.; Onawole, A.T.; Islam, M.A.; Adegoke, R.O.; Badmus, S.O. Molecular dynamics and combined docking studies for the identification of Zaire ebola virus inhibitors. J. Biomol. Struct. Dyn. 2019, 9, 5142. [Google Scholar] [CrossRef]
- Zerroug, A.; Belaidi, S.; BenBrahim, I.; Sinha, L.; Chtita, S. Virtual screening in drug-likeness and structure/activity relationship of pyridazine derivatives as Anti-Alzheimer drugs. J. King Saud Univ.-Sci. 2019, 31, 595–601. [Google Scholar] [CrossRef]
- El-Kattan, A.; Varm, M. Oral Absorption, Intestinal Metabolism and Human Oral Bioavailability. In Topics on Drug Metabolism; BoD—Books on Demand: Norderstedt, Germany, 2012. [Google Scholar]
- Bowen, L.; Smith, B.; Steinbach, S.; Billioux, B.; Summers, A.; Azodi, S.; Ohayon, J.; Schindler, M.; Nath, A. Survivors of Ebola Virus Disease Have Persistent Neurological Deficits (Abstract S53.003). In Proceedings of the American Academy of Neurology Annual Meeting, Vancouver, BC, Canada, 15–21 April 2016. [Google Scholar]
- Billioux, B.J.; Smith, B.; Nath, A. Neurological Complications of Ebola Virus Infection. Neurotherapeutics 2016, 13, 461–470. [Google Scholar] [CrossRef]
- Sagui, E.; Janvier, F.; Baize, S.; Foissaud, V.; Koulibaly, F.; Savini, H.; Maugey, N.; Aletti, M.; Granier, H.; Carmoi, T. Severe Ebola Virus Infection with Encephalopathy: Evidence for Direct Virus Involvement. Clin. Infect. Dis. 2015, 61, 1627–1628. [Google Scholar] [CrossRef]
- de Greslan, T.; Billhot, M.; Rousseau, C.; Mac Nab, C.; Karkowski, L.; Cournac, J.-M.; Bordes, J.; Gagnon, N.; Dubrous, P.; Duron, S.; et al. Ebola Virus–Related Encephalitis: Table 1. Clin. Infect. Dis. 2016, 63, 1076–1078. [Google Scholar] [CrossRef][Green Version]
- Wong, G.; Qiu, X.; Bi, Y.; Formenty, P.; Sprecher, A.; Jacobs, M.; Gao, G.F.; Kobinger, G. More Challenges from Ebola: Infection of the Central Nervous System. J. Infect. Dis. 2016, 214, S294–S296. [Google Scholar] [CrossRef] [PubMed]
- Harder, B.G.; Blomquist, M.R.; Wang, J.; Kim, A.J.; Woodworth, G.F.; Winkles, J.A.; Loftus, J.C.; Tran, N.L. Developments in Blood-Brain Barrier Penetrance and Drug Repurposing for Improved Treatment of Glioblastoma. Front. Oncol. 2018, 8, 462. [Google Scholar] [CrossRef]
- Karthika, C.; Sureshkumar, R. P-Glycoprotein Efflux Transporters and Its Resistance Its Inhibitors and Therapeutic Aspects. In Creatinine—A Comprehensive Update [Working Title]; IntechOpen: London, UK, 2020. [Google Scholar]
- Ma, J.D.; Tsunoda, S.M.; Bertino, J.S.; Trivedi, M.; Beale, K.K.; Nafziger, A.N. Evaluation of in vivo P-glycoprotein phenotyping probes: A need for validation. Clin. Pharmacokinet. 2010, 49, 223–237. [Google Scholar] [CrossRef]
- Dutkiewicz, Z.; Mikstacka, R. Structure-Based Drug Design for Cytochrome P450 Family 1 Inhibitors. Bioinorg. Chem. Appl. 2018, 2018, 3924608. [Google Scholar] [CrossRef]
- Egieyeh, S.A.; Syce, J.; Malan, S.F.; Christoffels, A. Prioritization of anti-malarial hits from nature: Chemo-informatic profiling of natural products with in vitro antiplasmodial activities and currently registered anti-malarial drugs. Malar. J. 2016, 15, 50. [Google Scholar] [CrossRef]
- Ren, J.X.; Zhang, R.T.; Zhang, H.; Cao, X.S.; Liu, L.K.; Xie, Y. Identification of novel VP35 inhibitors: Virtual screening driven new scaffolds. Biomed. Pharmacother. 2016, 84, 199–207. [Google Scholar] [CrossRef]
- Baikerikar, S. Curcumin and natural derivatives inhibit Ebola viral proteins: An in silico approach. Pharmacognosy Res. 2017, 9, 15. [Google Scholar] [CrossRef]
- Parasuraman, S. Prediction of activity spectra for substances. J. Pharmacol. Pharmacother. 2011, 2, 52–53. [Google Scholar] [CrossRef]
- Tarasova, O.; Filimonov, D.; Poroikov, V. PASS-based approach to predict HIV-1 reverse transcriptase resistance. J. Bioinform. Comput. Biol. 2017, 15, 1650040. [Google Scholar] [CrossRef] [PubMed]
- Bixler, S.L.; Duplantier, A.J.; Bavari, S. Discovering Drugs for the Treatment of Ebola Virus. Curr. Treat. Options Infect. Dis. 2017, 9, 299–317. [Google Scholar] [CrossRef]
- Mirza, M.U.; Vanmeert, M.; Ali, A.; Iman, K.; Froeyen, M.; Idrees, M. Perspectives towards antiviral drug discovery against Ebola virus. J. Med. Virol. 2019, 91, 2029–2048. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Elvati, P.; Violi, A. Antiviral Drug-Membrane Permeability: The Viral Envelope and Cellular Organelles. arXiv 2020, arXiv:2007.14965. [Google Scholar]
- Mazzon, M.; Marsh, M. Targeting viral entry as a strategy for broad-spectrum antivirals [version 1; peer review: 3 approved]. F1000Research 2019, 8. [Google Scholar] [CrossRef]
- Jamkhande, P.G.; Pathan, S.K.; Wadher, S.J. In silico PASS analysis and determination of antimycobacterial, antifungal, and antioxidant efficacies of maslinic acid in an extract rich in pentacyclic triterpenoids. Int. J. Mycobacteriol. 2016, 5, 417–425. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Broni, E.; Teye, J.; Quansah, E.; Issah, I.; Wilson, M.D.; Miller, W.A.; Tiburu, E.K.; Bonney, J.H.K. Pharmacoinformatics-based identification of potential bioactive compounds against Ebola virus protein VP24. Comput. Biol. Med. 2019, 113, 103414. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L.; Keserü, G.M.; Leeson, P.D.; Rees, D.C.; Reynolds, C.H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discov. 2014, 13, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.H. Ligand efficiency metrics: Why all the fuss? Future Med. Chem. 2015, 7, 1363–1365. [Google Scholar] [CrossRef]
- Laraia, L.; McKenzie, G.; Spring, D.R.; Venkitaraman, A.R.; Huggins, D.J. Overcoming Chemical, Biological, and Computational Challenges in the Development of Inhibitors Targeting Protein-Protein Interactions. Chem. Biol. 2015, 22, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Arnott, J.A.; Planey, S.L. The influence of lipophilicity in drug discovery and design. Expert Opin. Drug Discov. 2012, 7, 863–875. [Google Scholar] [CrossRef]
- Leeson, P.D.; Springthorpe, B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 2007, 6, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.Y.; Coumar, M.S.; Shiao, H.Y.; Wang, W.C.; Chen, C.W.; Song, J.S.; Chen, C.H.; Lin, W.H.; Wu, S.H.; Hsu, J.T.A.; et al. Ligand efficiency based approach for efficient virtual screening of compound libraries. Eur. J. Med. Chem. 2014, 83, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Arnott, J.A.; Kumar, R.; Planey, S.L. Lipophilicity Indices for Drug Development. J. Appl. Biopharm. Pharmacokinet. 2013, 1, 31–36. [Google Scholar]
- Xue, X.; Bao, G.; Zhang, H.Q.; Zhao, N.Y.; Sun, Y.; Zhang, Y.; Wang, X.L. An application of fit quality to screen MDM2/p53 protein-protein interaction inhibitors. Molecules 2018, 23, 3174. [Google Scholar] [CrossRef]
- Bembenek, S.D.; Tounge, B.A.; Reynolds, C.H. Ligand efficiency and fragment-based drug discovery. Drug Discov. Today 2009, 14, 278–283. [Google Scholar] [CrossRef]
- Islam, R.; Parves, M.R.; Paul, A.S.; Uddin, N.; Rahman, M.S.; Al Mamun, A.; Hossain, M.N.; Ali, M.A.; Halim, M.A. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 39, 3213–3224. [Google Scholar] [CrossRef] [PubMed]
- Calero-Rubio, C.; Paik, B.; Jia, X.; Kiick, K.L.; Roberts, C.J. Predicting unfolding thermodynamics and stable intermediates for alanine-rich helical peptides with the aid of coarse-grained molecular simulation. Biophys. Chem. 2016, 217, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.H.; Chen, K.B.; Lee, W.Y.; Sun, M.F.; Lee, C.C.; Chen, C.Y.C. Ligand-based and structure-based investigation for Alzheimer’s disease from traditional Chinese medicine. Evid.-Based Complement. Altern. Med. 2014, 2014, 364819. [Google Scholar] [CrossRef]
- Karthick, V.; Nagasundaram, N.; Doss, C.G.P.; Chakraborty, C.; Siva, R.; Lu, A.; Zhang, G.; Zhu, H. Virtual screening of the inhibitors targeting at the viral protein 40 of Ebola virus. Infect. Dis. Poverty 2016, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Kokubo, H. Exploring the Stability of Ligand Binding Modes to Proteins by Molecular Dynamics Simulations: A Cross-docking Study. J. Chem. Inf. Model. 2017, 57, 2514–2522. [Google Scholar] [CrossRef]
- Goyal, S.; Binnington, B.; McCarthy, S.D.S.; Desmaële, D.; Férrié, L.; Figadère, B.; Loiseau, P.M.; Branch, D.R. Inhibition of in vitro Ebola infection by anti-parasitic quinoline derivatives. F1000Research 2020, 9, 268. [Google Scholar] [CrossRef]
- Jawad, B.; Poudel, L.; Podgornik, R.; Steinmetz, N.F.; Ching, W.Y. Molecular mechanism and binding free energy of doxorubicin intercalation in DNA. Phys. Chem. Chem. Phys. 2019, 21, 3877–3893. [Google Scholar] [CrossRef]
- Zhou, H.X.; Pang, X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef]
- Poli, G.; Granchi, C.; Rizzolio, F.; Tuccinardi, T. Application of MM-PBSA methods in virtual screening. Molecules 2020, 25, 1971. [Google Scholar] [CrossRef]
- Shen, C.; Liu, H.; Wang, X.; Lei, T.; Wang, E.; Xu, L.; Yu, H.; Li, D.; Yao, X. Importance of incorporating protein flexibility in molecule modeling: A theoretical study on type I1/2 NIK inhibitors. Front. Pharmacol. 2019, 10, 345. [Google Scholar] [CrossRef]
- Asiedu, S.O.; Kwofie, S.K.; Broni, E.; Wilson, M.D. Computational Identification of Potential Anti-Inflammatory Natural Compounds Targeting the p38 Mitogen-Activated Protein Kinase (MAPK): Implications for COVID-19-Induced Cytokine Storm. Biomolecules 2021, 11, 653. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Chang, F.R.; Chen, S.R.; Wu, Y.H.Y.C.; Hu, H.C.; Wu, Y.H.Y.C.; Backlund, A.; Cheng, Y. Bin Anti-dengue virus constituents from Formosan zoanthid Palythoa mutuki. Mar. Drugs 2016, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, C.; Jomori, T.; Tanaka, J.; Senba, M.; Mori, N. Peridinin, a carotenoid, inhibits proliferation and survival of HTLV-1-infected T-cell lines. Int. J. Oncol. 2016, 49, 1713–1721. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; He, L.; Xie, H.; Wu, S.; Li, Y.; Zhang, P.; Yang, Z.; Huang, X. Antiviral activity of Carbenoxolone disodium against dengue virus infection. J. Med. Virol. 2017, 89, 571–581. [Google Scholar] [CrossRef]
- Haga, I.R.; Simpson, J.L.; Hawes, P.C.; Beard, P.M. Carbenoxolone-mediated cytotoxicity inhibits Vaccinia virus replication in a human keratinocyte cell line. Sci. Rep. 2018, 8, 16956. [Google Scholar] [CrossRef]
- Dargan, D.J.; Subak-Sharpe, J.H. The antiviral activity against herpes simplex virus of the triterpenoid compounds carbenoxolone sodium and cicloxolone sodium. J. Antimicrob. Chemother. 1986, 18, 185–200. [Google Scholar] [CrossRef]
- Kim, J.; Park, K.E.; Jeong, Y.S.; Kim, Y.M.; Park, H.; Nam, J.H.; Jung, K.; Son, W.S.; Jung, H.S.; Lee, J.H.; et al. Therapeutic efficacy of ABN401, a highly potent and selective MET inhibitor, based on diagnostic biomarker test in MET-addicted cancer. Cancers 2020, 12, 1575. [Google Scholar] [CrossRef]
- Awadh Ali, N.; Al Sokari, S.; Gushash, A.; Anwar, S.; Al-Karani, K.; Al-Khulaidi, A. Ethnopharmacological survey of medicinal plants in Albaha Region, Saudi Arabia. Pharmacognosy Res. 2017, 9, 401–407. [Google Scholar] [CrossRef]
- Islam, M.A.; Pillay, T.S. Pharmacoinformatics-based identification of chemically active molecules against Ebola virus. J. Biomol. Struct. Dyn. 2018, 37, 4104–4119. [Google Scholar] [CrossRef]
- Setlur, A.S.; Naik, S.Y.; Skariyachan, S. Herbal Lead as Ideal Bioactive Compounds Against Probable Drug Targets of Ebola Virus in Comparison with Known Chemical Analogue: A Computational Drug Discovery Perspective. Interdiscip. Sci. Comput. Life Sci. 2017, 9, 254–277. [Google Scholar] [CrossRef] [PubMed]
- Tambunan, U.S.F.; Alkaff, A.H.; Nasution, M.A.F. Bioinformatics Approach to Screening and Developing Drug against Ebola. In Advances in Ebola Control; BoD—Books on Demand: Norderstedt, Germany, 2018. [Google Scholar]
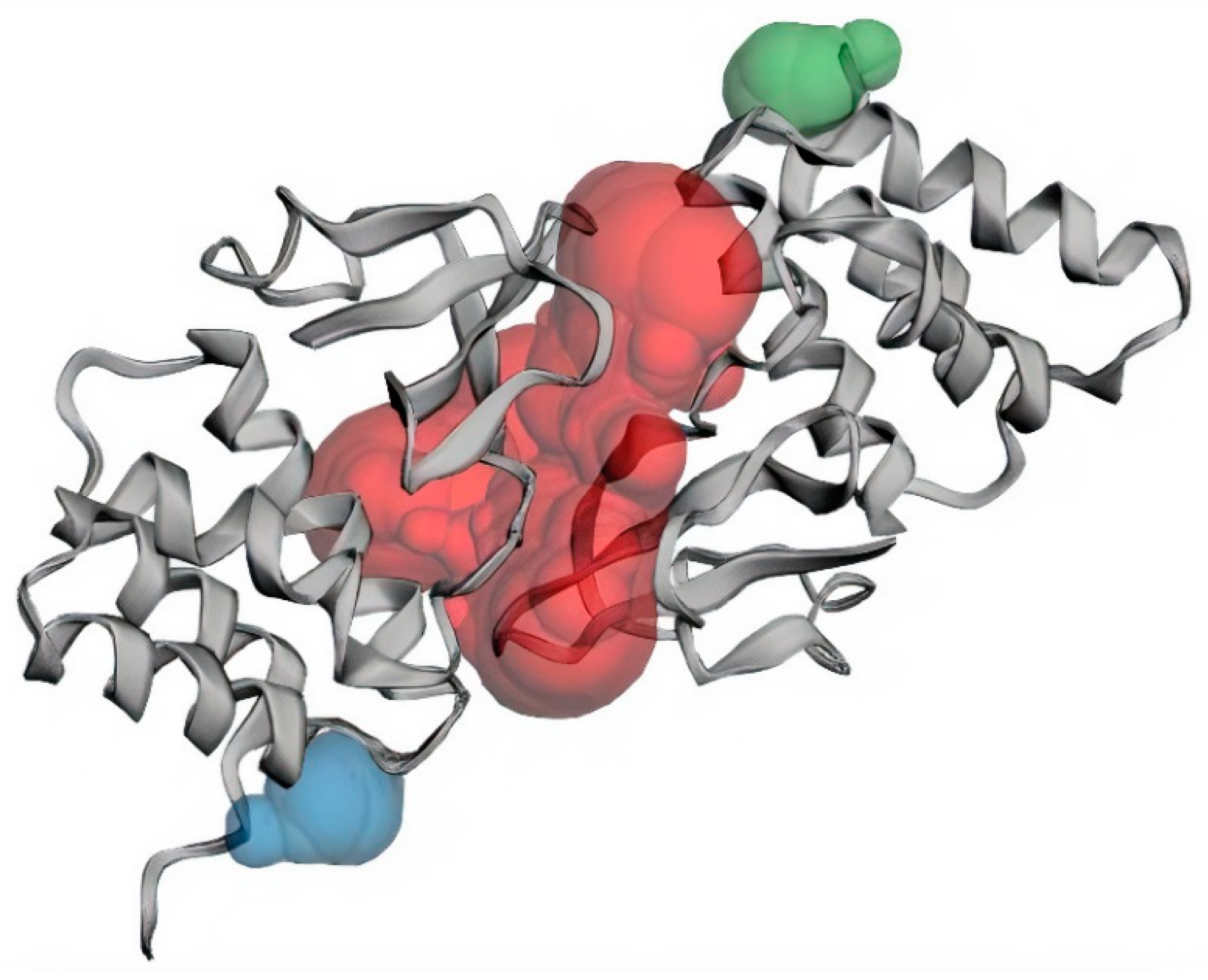
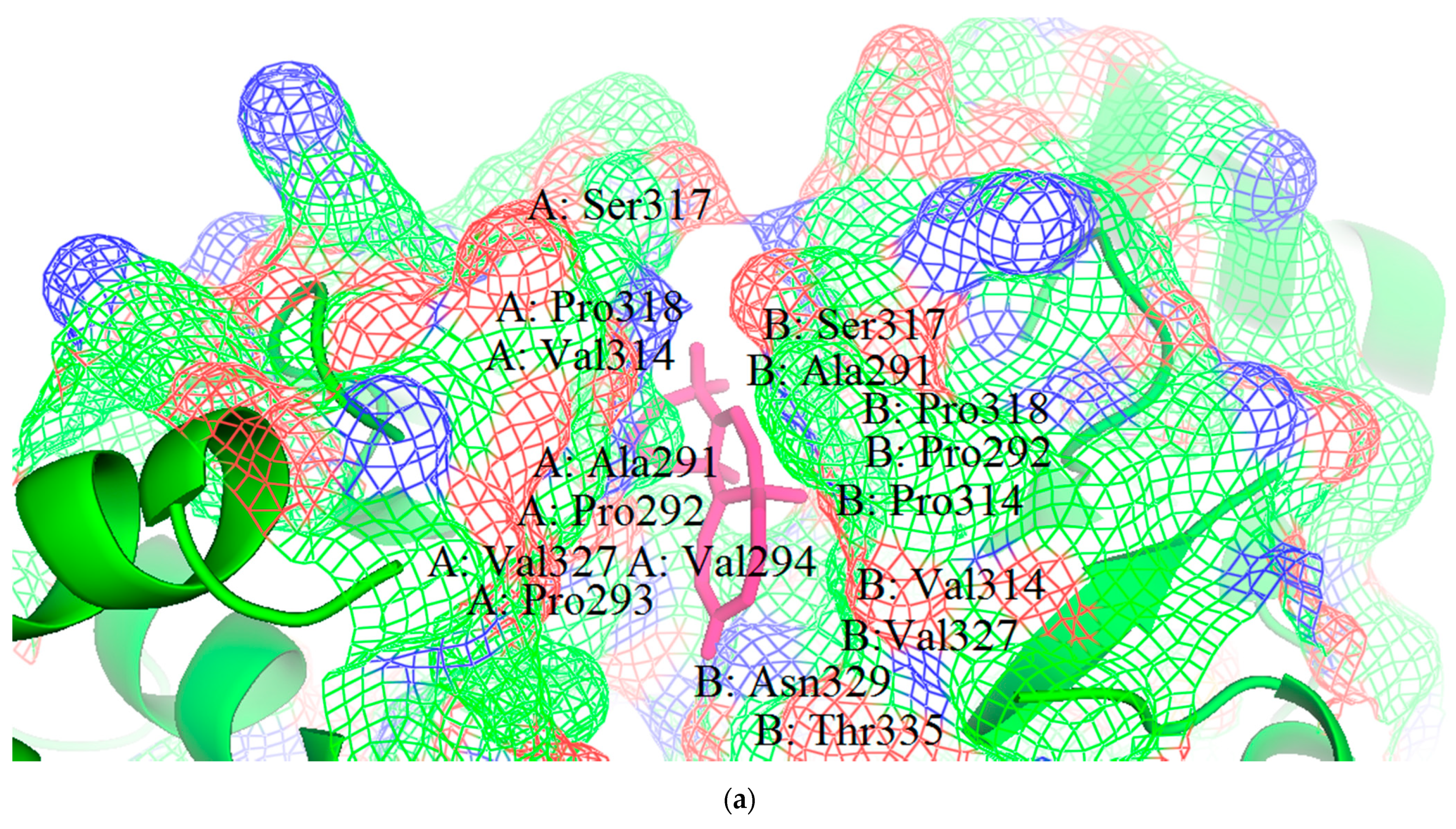
| Binding Sites | Chain | Amino Acid Residues | Surface Area (SA)/Å2 | Volume/Å3 |
|---|---|---|---|---|
| Pocket 1 | A | Val245, Lys248, Leu249, Asp252, Ser253, Ile286, Phe287, Gln288, Asp289, Ala290, Ala291, Pro292, Pro293, Val294, Ile295, His296, Ile297, Arg298, Val314, Pro315, Pro316, Ser317, Pro318, Lys319, Val327, Gln329, Leu330, Gln331, Gly333, Thr335. | 1155.05 | 1078.689 |
| B | Gln241, Gln244, Val245, Lys248, Leu249, Asp252, Ser253, Ile286, Gln288, Asp289, Ala290, Ala291, Pro292, Pro293, Val294, Ile295, His296, Val314, Pro315, Pro316, Ser317, Pro318, Lys319, Val327, Gln329, Leu330, Gln331, Gly333, Lys334, Thr335. | |||
| Pocket 2 | B | Asp218, Ile219, Asn254, Leu256, Asp257 | 48.092 | 35.5916 |
| Pocket 3 | A | Asp218, Ile219, Asn254, Leu256, Asp257 | 52.040 | 34.782 |
| Compound ID | Binding Energy (kcal/mol) | Number of Hydrogen Bonds | Hydrogen Bond Residues | Hydrogen Bond Length (Å) | Hydrophobic Contacts |
|---|---|---|---|---|---|
| NANPDB86 | −8.5 | 1 | Gln329 | 2.0 | Val245, Leu249, Pro293, Val294, Ile295 |
| NANPDB95 | −8.1 | 0 | - | - | Pro316, Ala291, Pro292, Leu249, Pro293, Val294, Val327, Ile286, Ala290, Pro315, Pro318, Val314 |
| NANPDB142 | −8.0 | 0 | - | - | Pro318, Ala291, Pro315, Pro316, Ala290, Val294, Val327, Val314, Leu249 |
| NANPDB205 | −8.3 | 0 | - | - | Leu249, Pro293, Val245, Ile295 |
| NANPDB397 | −8.1 | 0 | - | - | Pro318, Val314, Ala291, Pro292, Pro293, Val327, Val294 |
| NANPDB2412 | −8.2 | 0 | - | - | Pro318, Pro316, Ala290, Pro315, Ala291, Val314, Pro292, Val294, Pro293, Val327 |
| NANPDB2476 | −8.0 | 0 | - | - | Pro316, Ala291, Pro315, Pro318, Pro292, Val314, Val327, Val294 |
| NANPDB3355 | −8.2 | 0 | - | - | Pro316, Ala290, Ala291, Pro292, Val314, Pro318, Val294, Val327 |
| NANPDB4048 | −8.2 | 0 | - | - | Pro318, Ala291, Val314, Pro292, Pro293, Leu249, Val294, Val327 |
| ZINC000014612849 | −8.1 | 0 | - | - | Val314, Pro292, Ala291, Pro318, Pro315, Val327, Val294 |
| ZINC000033831303 | −8.0 | 0 | - | - | Pro293, Leu249, Ile295, Val245, Val294 |
| ZINC000095486250 | −8.1 | 0 | - | - | Ala291, Pro318, Pro292, Val314, Pro293, Val327, Val294 |
| Amodiaquine | −7.0 | 0 | - | - | Ala291, Pro318, Ala291, Pro315, Val327, Val294, Pro292, Val314 |
| Chloroquine | −5.9 | 0 | - | - | Pro318, Val314, Val327, Pro292, Ala291, Val294, Pro293 |
| EGCG | −8.1 | 1 | Gln244 | 2.01 | Val294, Pro293, Leu249, Val245, Cys247, Ile297, Leu330 |
| Gossypetin | −7.5 | 1 | Leu330 | 1.97 | Ile295, Val294, Pro293, Leu249, Val245 |
| Taxifolin | −7.4 | 0 | - | - | Val314, Ala290, Ala291, Pro318, Val294, Val327, Pro292, Leu249 |
| Compound ID | Estimated Solubility Log S | Estimated Solubility Class | GI Absorption | BBB Permeant | P-glycoprotein Substrate |
|---|---|---|---|---|---|
| NANPDB86 | −3.79 | Soluble | High | Yes | No |
| NANPDB95 | −3.57 | Soluble | High | Yes | No |
| NANPDB142 | −3.77 | Soluble | High | Yes | No |
| NANPDB205 | −2.61 | Soluble | High | Yes | No |
| NANPDB397 | −3.09 | Soluble | High | Yes | No |
| NANPDB2412 | −3.99 | Soluble | High | Yes | No |
| NANPDB2476 | −3.89 | Soluble | High | Yes | No |
| NANPDB3355 | −3.25 | Soluble | High | Yes | No |
| NANPDB4048 | −3.73 | Soluble | High | Yes | No |
| ZINC000014612849 | −3.00 | Soluble | High | Yes | No |
| ZINC000033831303 | −3.89 | Soluble | High | Yes | No |
| ZINC000095486250 | −3.41 | Soluble | High | Yes | No |
| Amodiaquine | −5.9 | Moderately soluble | High | Yes | No |
| Chloroquine | −4.55 | Moderately soluble | High | Yes | No |
| EGCG | −3.56 | Soluble | Low | No | No |
| Gossypetin | −3.40 | Soluble | Low | No | No |
| Taxifolin | −2.66 | Soluble | High | No | No |
| Compound ID | Mutagenic | Tumorigenic | Reproductive Effect | Irritant |
|---|---|---|---|---|
| NANPDB86 | None | None | None | None |
| NANPDB95 | None | None | None | None |
| NANPDB142 | None | None | None | None |
| NANPDB205 | None | None | High | None |
| NANPDB397 | None | None | None | None |
| NANPDB2412 | None | None | None | None |
| NANPDB2476 | None | None | None | High |
| NANPDB3355 | None | High | None | High |
| NANPDB4048 | None | None | High | None |
| ZINC000014612849 | Low | None | None | None |
| ZINC000033831303 | High | High | None | High |
| ZINC000095486250 | None | None | None | None |
| Amodiaquine | High | None | High | High |
| Chloroquine | High | None | None | High |
| EGCG | None | None | None | None |
| Gossypetin | High | None | None | None |
| Taxifolin | None | None | None | None |
| Compound ID | Biological Activity | Pa | Pi |
|---|---|---|---|
| NANPDB86 | Rhinovirus | 0.444 | 0.052 |
| Herpes | 0.334 | 0.069 | |
| Protein synthesis inhibitor | 0.467 | 0.008 | |
| Transcription factor inhibitor | 0.39 | 0.026 | |
| RNA synthesis inhibitor | 0.287 | 0..63 | |
| NANPDB95 | Herpes | 0.394 | 0.038 |
| Picornavirus | 0.337 | 0.173 | |
| Transcription factor inhibitor | 0.557 | 0.008 | |
| Protein synthesis inhibitor | 0.493 | 0.007 | |
| RNA synthesis inhibitor | 0.331 | 0.038 | |
| NANPDB142 | Rhinovirus | 0.413 | 0.078 |
| Herpes | 0.332 | 0.071 | |
| Picornavirus | 0.352 | 0.156 | |
| DNA polymerase 1 inhibitor | 0.625 | 0.003 | |
| RNA synthesis inhibitor | 0.285 | 0.065 | |
| NANPDB205 | Adenovirus | 0.222 | 0.176 |
| Protein synthesis inhibitor | 0.238 | 0.041 | |
| RNA synthesis inhibitor | 0.251 | 0.100 | |
| DNA synthesis inhibitor | 0.207 | 0.141 | |
| NANPDB397 | - | - | - |
| NANPDB2412 | Herpes | 0.410 | 0.031 |
| Rhinovirus | 0.345 | 0.167 | |
| Transcription factor inhibitor | 0.283 | 0.013 | |
| DNA synthesis inhibitor | 0.232 | 0.101 | |
| RNA synthesis inhibitor | 0.231 | 0.125 | |
| NANPDB2476 | Influenza | 0.476 | 0.027 |
| Rhinovirus | 0.381 | 0.114 | |
| Protein synthesis inhibitor | 0.376 | 0.019 | |
| RNA synthesis inhibitor | 0.277 | 0.072 | |
| NANPDB3355 | Rhinovirus | 0.552 | 0.012 |
| Protein synthesis inhibitor | 0.353 | 0.022 | |
| Transcription factor inhibitor | 0.240 | 0.093 | |
| RNA synthesis inhibitor | 0.241 | 0.111 | |
| NANPDB4048 | Influenza | 0.621 | 0.011 |
| Rhinovirus | 0.362 | 0.140 | |
| Membrane permeability inhibitor | 0.753 | 0.020 | |
| RNA synthesis inhibitor | 0.484 | 0.009 | |
| ZINC000014612849 | - | - | - |
| ZINC000033831303 | RNA synthesis inhibitor | 0.281 | 0.069 |
| ZINC000095486250 | Influenza | 0.399 | 0.047 |
| Herpes | 0.273 | 0.111 | |
| RNA synthesis inhibitor | 0.298 | 0.056 | |
| DNA polymerase I inhibitor | 0.275 | 0.098 | |
| Amodiaquine | - | - | - |
| Chloroquine | - | - | - |
| EGCG | Influenza | 0.771 | 0.003 |
| Rhinovirus | 0.514 | 0.020 | |
| Herpes | 0.480 | 0.012 | |
| HIV | 0.300 | 0.008 | |
| Hepatitis B | 0.316 | 0.029 | |
| Transcription factor inhibitor | 0.404 | 0.007 | |
| RNA synthesis inhibitor | 0.318 | 0.044 | |
| DNA polymerase I inhibitor | 0.294 | 0.070 | |
| Gossypetin | Hepatitis B | 0.498 | 0.005 |
| Influenza | 0.415 | 0.042 | |
| Membrane permeability inhibitor | 0.953 | 0.002 | |
| RNA synthesis inhibitor | 0.358 | 0.029 | |
| DNA polymerase I inhibitor | 0.331 | 0.040 | |
| Taxifolin | Influenza | 0.620 | 0.011 |
| Herpes | 0.492 | 0.010 | |
| Rhinovirus | 0.503 | 0.023 | |
| Hepatitis B | 0.399 | 0.015 | |
| Membrane permeability inhibitor | 0.850 | 0.005 | |
| Transcription factor inhibitor | 0.413 | 0.022 | |
| DNA polymerase I inhibitor | 0.329 | 0.041 | |
| RNA synthesis inhibitor | 0.394 | 0.021 |
| Compound ID | Number of Heavy Atoms | Log P | Ki | LE | LE_Scale | FQ | LELP |
|---|---|---|---|---|---|---|---|
| NANPDB86 | 24 | 2.79 | 5.87 × 10−7 | 0.354 | 0.404 | 0.876 | 7.88 |
| NANPDB95 | 24 | 2.94 | 1.15 × 10−6 | 0.338 | 0.404 | 0.837 | 8.70 |
| NANPDB95 | 24 | 2.94 | 1.15 × 10−6 | 0.338 | 0.404 | 0.837 | 8.70 |
| NANPDB142 | 25 | 2.93 | 1.37 × 10−6 | 0.320 | 0.391 | 0.818 | 9.16 |
| NANPDB205 | 20 | 1.81 | 8.23 × 10−7 | 0.415 | 0.467 | 0.889 | 4.36 |
| NANPDB397 | 24 | 2.66 | 1.15 × 10−6 | 0.338 | 0.404 | 0.837 | 7.87 |
| NANPDB2412 | 23 | 3.25 | 9.74 × 10−7 | 0.357 | 0.418 | 0.854 | 9.10 |
| NANPDB2476 | 22 | 3.55 | 1.37 × 10−6 | 0.364 | 0.433 | 0.841 | 9.75 |
| NANPDB3355 | 24 | 2.43 | 9.74 × 10−7 | 0.342 | 0.404 | 0.847 | 7.11 |
| NANPDB4048 | 23 | 3.61 | 9.74 × 10−7 | 0.357 | 0.418 | 0.854 | 10.11 |
| ZINC000014612849 | 25 | 2.22 | 1.15 × 10−6 | 0.324 | 0.391 | 0.829 | 6.85 |
| ZINC000033831303 | 23 | 3.37 | 1.37 × 10−6 | 0.348 | 0.418 | 0.833 | 9.68 |
| ZINC000095486250 | 21 | 3.68 | 1.15 × 10−6 | 0.386 | 0.449 | 0.860 | 9.53 |
| Compound ID | Binding Affinity from Docking [kcal/mol (kJ/mol)] | van der Waal Energy (kJ/mol) | Electrostatic Energy (kJ/mol) | Polar Solvation Energy (kJ/mol) | SASA Energy (kJ/mol) | Binding Energy (kJ/mol) |
|---|---|---|---|---|---|---|
| NANPDB2412 | −8.2 (−34.3088) | −112.794 ± 31.343 | −4.338 ± 7.888 | 63.305 ± 25.933 | −13.955 ± 3.243 | −67.782 ± 17.041 |
| NANPDB2476 | −8.0 (−33.472) | −72.353 ± 15.702 | −8.393 ± 9.299 | 46.887 ± 21.330 | −10.531 ± 2.288 | −44.390 ± 19.503 |
| NANPDB4048 | −8.2 (−34.3088) | −122.063 ± 24.789 | −3.170 ± 8.186 | 68.675 ± 23.656 | −15.854 ± 2.967 | −72.413 ± 15.915 |
| ZINC000095486250 | −8.1 (−33.8904) | −133.848 ± 15.162 | −6.489 ± 7.863 | 62.413 ± 10.653 | −16.289 ± 1.014 | −94.213 ± 12.755 |
| Amodiaquine | −7.0 (−29.288) | −150.934 ± 19.558 | −6.282 ± 8.679 | 83.311 ± 13.703 | −18.495 ± 1.647 | −92.400 ± 15.855 |
| EGCG | −8.1 (−33.8904) | −110.393 ± 27.459 | −46.227 ± 20.847 | 126.216 ± 35.236 | −14.160 ± 3.019 | −44.564 ± 23.104 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darko, L.K.S.; Broni, E.; Amuzu, D.S.Y.; Wilson, M.D.; Parry, C.S.; Kwofie, S.K. Computational Study on Potential Novel Anti-Ebola Virus Protein VP35 Natural Compounds. Biomedicines 2021, 9, 1796. https://doi.org/10.3390/biomedicines9121796
Darko LKS, Broni E, Amuzu DSY, Wilson MD, Parry CS, Kwofie SK. Computational Study on Potential Novel Anti-Ebola Virus Protein VP35 Natural Compounds. Biomedicines. 2021; 9(12):1796. https://doi.org/10.3390/biomedicines9121796
Chicago/Turabian StyleDarko, Louis K. S., Emmanuel Broni, Dominic S. Y. Amuzu, Michael D. Wilson, Christian S. Parry, and Samuel K. Kwofie. 2021. "Computational Study on Potential Novel Anti-Ebola Virus Protein VP35 Natural Compounds" Biomedicines 9, no. 12: 1796. https://doi.org/10.3390/biomedicines9121796
APA StyleDarko, L. K. S., Broni, E., Amuzu, D. S. Y., Wilson, M. D., Parry, C. S., & Kwofie, S. K. (2021). Computational Study on Potential Novel Anti-Ebola Virus Protein VP35 Natural Compounds. Biomedicines, 9(12), 1796. https://doi.org/10.3390/biomedicines9121796







