Cytogenomic Profile of Uterine Leiomyoma: In Vivo vs. In Vitro Comparison
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Cell Cultures
2.3. Karyotyping of Cell Cultures
2.4. FISH Studies for Chromosomal Abnormalities
2.5. Q-FISH for Assessment of Telomere Length
2.6. DNA Isolation for Molecular Genetic Studies
2.7. Array Comparative Genomic Hybridisation
2.8. MED12 Mutation Analysis
3. Results
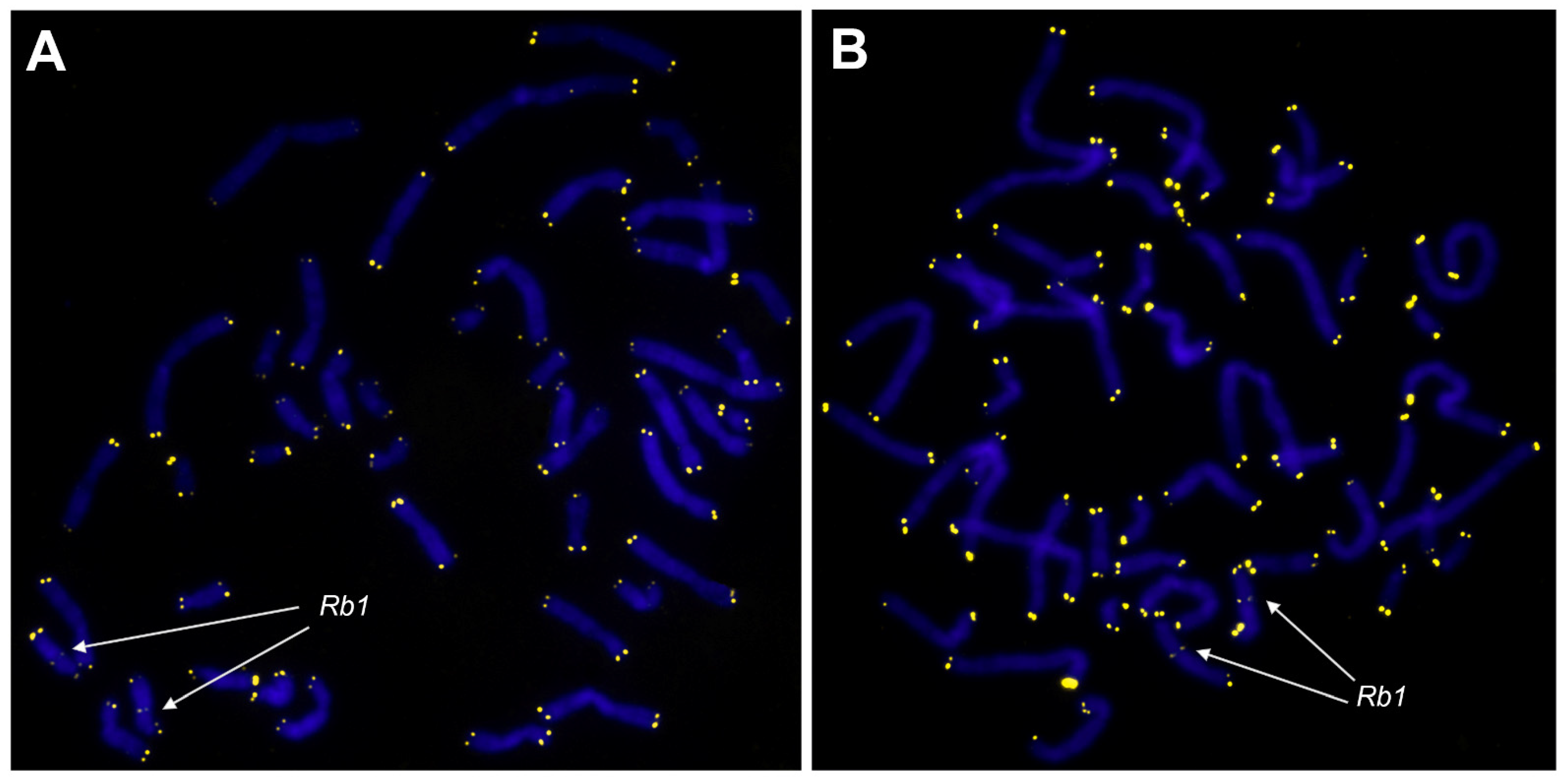
3.1. Karyotype Abnormalities in Cultured ULs Are Represented Primarily by Structural Chromosomal Rearrangements
3.2. Metaphase Chromosome Analysis of Cultured UL Cells Reveals More than One Clone in 50% of the ULs with an Abnormal Karyotype
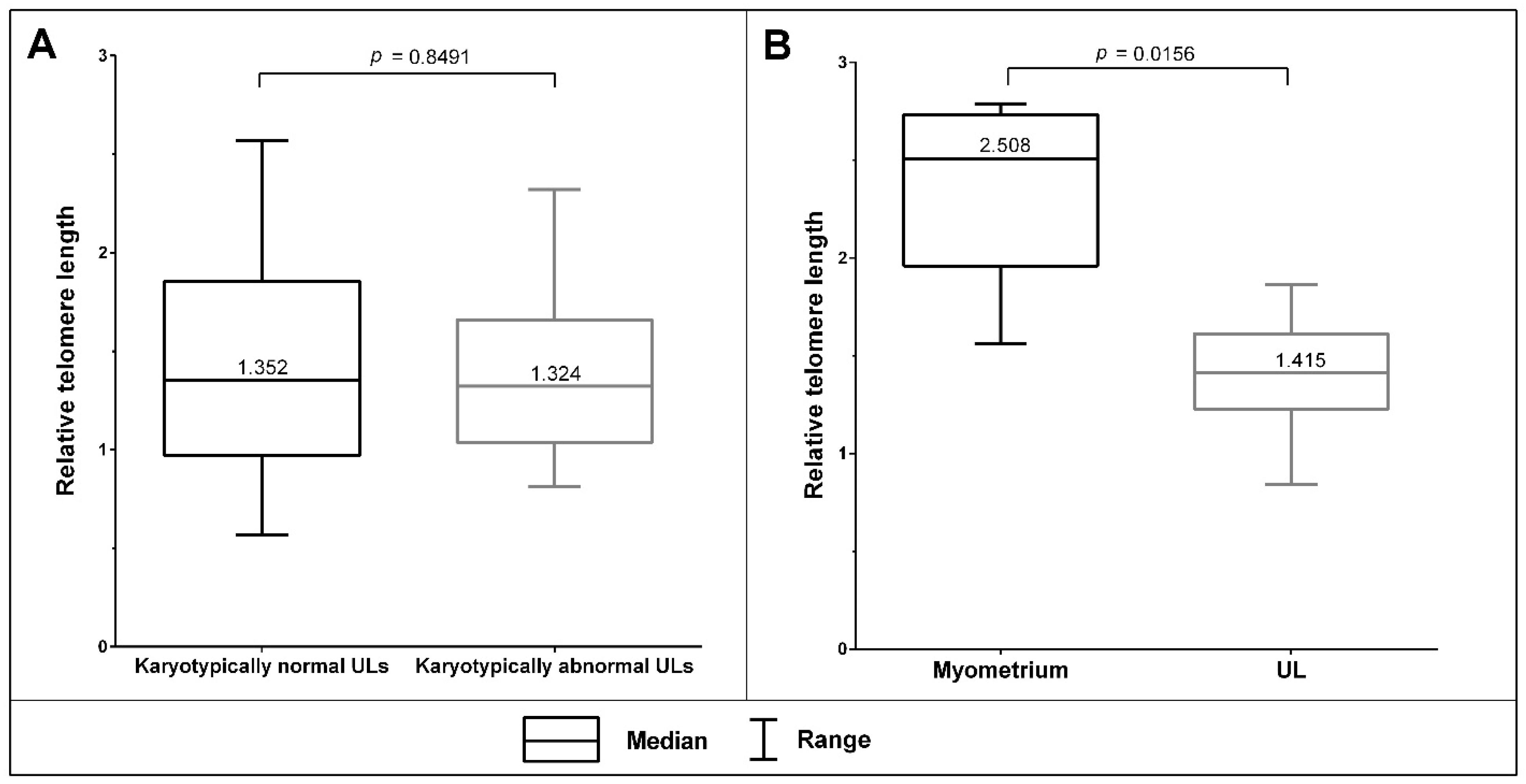
3.3. The Telomeric Regions of Metaphase Chromosomes Are Shorter in Both Karyotypically Normal and Abnormal UL Cells than in the Adjacent Myometrium Cells
3.4. Most Karyotypically Abnormal ULs Show Discordance between Results of Conventional Karyotyping of Cultured Cells and aCGH Results of Paired Uncultured Samples
3.5. Chromosomal Abnormalities Detected by Conventional Karyotyping and aCGH Are Present in the Interphase Cells of Uncultured ULs
3.6. The Initial Proportions of Normal and Abnormal Clones Established in Native ULs Alter after In Vitro Propagation
3.7. MED12 Gene Mutations Do Not Rule Out the Presence of Karyotype Abnormalities in ULs
3.8. A UL’s MED12 Status Is Not Associated with Differences in the Proportion of Cytogenetically Normal and Abnormal Clones between Cultured and Uncultured Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baird, D.D.; Dunson, D.B.; Hill, M.C.; Cousins, D.; Schectman, J.M. High cumulative incidence of uterine leiomyoma in black and white women: Ultrasound evidence. Am. J. Obstet. Gynecol. 2003, 188, 100–107. [Google Scholar] [CrossRef]
- Commandeur, A.E.; Styer, A.K.; Teixeira, J.M. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum. Reprod. Update 2015, 21, 593–615. [Google Scholar] [CrossRef] [PubMed]
- Wise, L.A.; Laughlin-Tommaso, S.K. Epidemiology of Uterine Fibroids: From Menarche to Menopause. Clin. Obstet. Gynecol. 2016, 59, 2–24. [Google Scholar] [CrossRef] [PubMed]
- Lippman, S.A.; Warner, M.; Samuels, S.; Olive, D.; Vercellini, P.; Eskenazi, B. Uterine fibroids and gynecologic pain symptoms in a population-based study. Fertil. Steril. 2003, 80, 1488–1494. [Google Scholar] [CrossRef]
- Stewart, E.A.; Cookson, C.L.; Gandolfo, R.A.; Schulze-Rath, R. Epidemiology of uterine fibroids: A systematic review. BJOG 2017, 124, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Parazzini, F.; Tozzi, L.; Bianchi, S. Pregnancy outcome and uterine fibroids. Best Pract. Res. Clin. Obstet. Gynaecol. 2016, 34, 74–84. [Google Scholar] [CrossRef]
- Whynott, R.M.; Vaught, K.C.C.; Segars, J.H. The Effect of Uterine Fibroids on Infertility: A Systematic Review. Semin. Reprod. Med. 2017, 35, 523–532. [Google Scholar] [CrossRef]
- Giuliani, E.; As-Sanie, S.; Marsh, E.E. Epidemiology and management of uterine fibroids. Int. J. Gynaecol. Obstet. 2020, 149, 3–9. [Google Scholar] [CrossRef]
- Linder, D.; Gartler, S.M. Glucose-6-phosphate dehydrogenase mosaicism: Utilization as a cell marker in the study of leiomyomas. Science 1965, 150, 67–69. [Google Scholar] [CrossRef]
- Mashal, R.D.; Fejzo, M.L.; Friedman, A.J.; Mitchner, N.; Nowak, R.A.; Rein, M.S.; Morton, C.C.; Sklar, J. Analysis of androgen receptor DNA reveals the independent clonal origins of uterine leiomyomata and the secondary nature of cytogenetic aberrations in the development of leiomyomata. Genes Chromosomes Cancer 1994, 11, 1–6. [Google Scholar] [CrossRef]
- Hodges, L.C.; Hunter, D.S.; Bergerson, J.S.; Fuchs-Young, R.; Walker, C.L. An in vivo/in vitro model to assess endocrine disrupting activity of xenoestrogens in uterine leiomyoma. Ann. N. Y. Acad. Sci. 2001, 948, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.L. Role of hormonal and reproductive factors in the etiology and treatment of uterine leiomyoma. Recent Prog. Horm. Res. 2002, 57, 277–294. [Google Scholar] [CrossRef]
- Zhou, S.; Yi, T.; Shen, K.; Zhang, B.; Huang, F.; Zhao, X. Hypoxia: The driving force of uterine myometrial stem cells differentiation into leiomyoma cells. Med. Hypotheses 2011, 77, 985–986. [Google Scholar] [CrossRef]
- Greathouse, K.L.; Bredfeldt, T.; Everitt, J.I.; Lin, K.; Berry, T.; Kannan, K.; Mittelstadt, M.L.; Ho, S.M.; Walker, C.L. Environmental estrogens differentially engage the histone methyltransferase EZH2 to increase risk of uterine tumorigenesis. Mol. Cancer Res. 2012, 10, 546–557. [Google Scholar] [CrossRef]
- Shen, Y.; Xu, Q.; Ren, M.; Feng, X.; Cai, Y.; Gao, Y. Measurement of phenolic environmental estrogens in women with uterine leiomyoma. PLoS ONE 2013, 8, e79838. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Diamond, M.P.; Al-Hendy, A. Early Life Adverse Environmental Exposures Increase the Risk of Uterine Fibroid Development: Role of Epigenetic Regulation. Front. Pharmacol. 2016, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Efimova, O.A.; Koltsova, A.S.; Krapivin, M.I.; Tikhonov, A.V.; Pendina, A.A. Environmental Epigenetics and Genome Flexibility: Focus on 5-Hydroxymethylcytosine. Int. J. Mol. Sci. 2020, 21, 3223. [Google Scholar] [CrossRef] [PubMed]
- Baranov, V.S.; Ivaschenko, T.E.; Yarmolinskaya, M.I. Comparative systems genetics view of endometriosis and uterine leiomyoma: Two sides of the same coin? Syst. Biol. Reprod. Med. 2016, 62, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Osinovskaya, N.S.; Malysheva, O.V.; Shved, N.Y.; Ivashchenko, T.E.; Sultanov, I.Y.; Efimova, O.A.; Yarmolinskaya, M.I.; Bezhenar, V.F.; Baranov, V.S. Frequency and Spectrum of MED12 Exon 2 Mutations in Multiple Versus Solitary Uterine Leiomyomas from Russian Patients. Int. J. Gynecol. Pathol. 2016, 35, 509–515. [Google Scholar] [CrossRef]
- Baranov, V.S.; Osinovskaya, N.S.; Yarmolinskaya, M.I. Pathogenomics of Uterine Fibroids Development. Int. J. Mol. Sci. 2019, 20, 6151. [Google Scholar] [CrossRef]
- Machado-Lopez, A.; Simón, C.; Mas, A. Molecular and Cellular Insights into the Development of Uterine Fibroids. Int. J. Mol. Sci. 2021, 22, 8483. [Google Scholar] [CrossRef]
- Holzmann, C.; Markowski, D.N.; Koczan, D.; Küpker, W.; Helmke, B.M.; Bullerdiek, J. Cytogenetically normal uterine leiomyomas without MED12-mutations-a source to identify unknown mechanisms of the development of uterine smooth muscle tumors. Mol. Cytogenet. 2014, 7, 88. [Google Scholar] [CrossRef][Green Version]
- Holzmann, C.; Markowski, D.N.; Bartnitzke, S.; Koczan, D.; Helmke, B.M.; Bullerdiek, J. A rare coincidence of different types of driver mutations among uterine leiomyomas (UL). Mol. Cytogenet. 2015, 8, 76. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kämpjärvi, K.; Mäkinen, N.; Mehine, M.; Välipakka, S.; Uimari, O.; Pitkänen, E.; Heinonen, H.R.; Heikkinen, T.; Tolvanen, J.; Ahtikoski, A.; et al. MED12 mutations and FH inactivation are mutually exclusive in uterine leiomyomas. Br. J. Cancer 2016, 114, 1405–1411. [Google Scholar] [CrossRef]
- Markowski, D.N.; Bartnitzke, S.; Löning, T.; Drieschner, N.; Helmke, B.M.; Bullerdiek, J. MED12 mutations in uterine fibroids—Their relationship to cytogenetic subgroups. Int. J. Cancer 2012, 131, 1528–1536. [Google Scholar] [CrossRef]
- Galindo, L.J.; Hernández-Beeftink, T.; Salas, A.; Jung, Y.; Reyes, R.; de Oca, F.M.; Hernández, M.; Almeida, T.A. HMGA2 and MED12 alterations frequently co-occur in uterine leiomyomas. Gynecol. Oncol. 2018, 150, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Mehine, M.; Kaasinen, E.; Heinonen, H.R.; Mäkinen, N.; Kämpjärvi, K.; Sarvilinna, N.; Aavikko, M.; Vähärautio, A.; Pasanen, A.; Bützow, R.; et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc. Natl. Acad. Sci. USA 2016, 113, 1315–1320. [Google Scholar] [CrossRef] [PubMed]
- Pendina, A.A.; Efimova, O.A.; Tikhonov, A.V.; Chiryaeva, O.G.; Fedorova, I.D.; Koltsova, A.S.; Krapivin, M.I.; Parfenyev, S.E.; Kuznetzova, T.V.; Baranov, V.S. Immunofluorescence Staining for Cytosine Modifications Like 5-Methylcytosine and Its Oxidative Derivatives and FISH. In Fluorescence In Situ Hybridization (FISH), 2nd ed.; Liehr, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 337–346. [Google Scholar] [CrossRef]
- Kol’tsova, A.S.; Pendina, A.A.; Efimova, O.A.; Kaminskaya, A.N.; Tikhonov, A.V.; Osinovskaya, N.S.; Sultanov, I.Y.; Shved, N.Y.; Kakhiani, M.I.; Baranov, V.S. Differential DNA Hydroxymethylation in Human Uterine Leiomyoma Cells Depending on the Phase of Menstrual Cycle and Presence of MED12 Gene Mutations. Bull. Exp. Biol. Med. 2017, 163, 646–649. [Google Scholar] [CrossRef]
- Koltsova, A.S.; Efimova, O.A.; Pendina, A.A.; Chiryaeva, O.G.; Osinovskaya, N.S.; Shved, N.Y.; Yarmolinskaya, M.I.; Polenov, N.I.; Kunitsa, V.V.; Sagurova, Y.M.; et al. Uterine Leiomyomas with an Apparently Normal Karyotype Comprise Minor Heteroploid Subpopulations Differently Represented in vivo and in vitro. Cytogenet. Genome Res. 2021, 161, 43–51. [Google Scholar] [CrossRef]
- Liehr, T. Fluorescence In Situ Hybridization (FISH), 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar] [CrossRef]
- Pendina, A.A.; Krapivin, M.I.; Efimova, O.A.; Tikhonov, A.V.; Mekina, I.D.; Komarova, E.M.; Koltsova, A.S.; Gzgzyan, A.M.; Kogan, I.Y.; Chiryaeva, O.G.; et al. Telomere Length in Metaphase Chromosomes of Human Triploid Zygotes. Int. J. Mol. Sci. 2021, 22, 5579. [Google Scholar] [CrossRef]
- Krapivin, M.I.; Tikhonov, A.V.; Efimova, O.A.; Pendina, A.A.; Smirnova, A.A.; Chiryaeva, O.G.; Talantova, O.E.; Petrova, L.I.; Dudkina, V.S.; Baranov, V.S. Telomere Length in Chromosomally Normal and Abnormal Miscarriages and Ongoing Pregnancies and Its Association with 5-hydroxymethylcytosine Patterns. Int. J. Mol. Sci. 2021, 22, 6622. [Google Scholar] [CrossRef]
- Dzhemlikhanova, L.K.; Efimova, O.A.; Osinovskaya, N.S.; Parfenyev, S.E.; Niauri, D.A.; Sultanov, I.Y.; Malysheva, O.V.; Pendina, A.A.; Shved, N.Y.; Ivashchenko, T.E.; et al. Catechol-O-methyltransferase Val158Met polymorphism is associated with increased risk of multiple uterine leiomyomas either positive or negative for MED12 exon 2 mutations. J. Clin. Pathol. 2017, 70, 233–236. [Google Scholar] [CrossRef]
- Pendina, A.A.; Koltsova, A.S.; Efimova, O.A.; Malysheva, O.V.; Osinovskaya, N.S.; Sultanov, I.Y.; Tikhonov, A.V.; Shved, N.Y.; Chiryaeva, O.G.; Simareva, A.D.; et al. Case of chromothripsis in a large solitary non-recurrent uterine leiomyoma. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 219, 134–136. [Google Scholar] [CrossRef]
- Nibert, M.; Heim, S. Uterine leiomyoma cytogenetics. Genes Chromosomes Cancer 1990, 2, 3–13. [Google Scholar] [CrossRef]
- Pandis, N.; Heim, S.; Bardi, G.; Flodérus, U.M.; Willén, H.; Mandahl, N.; Mitelman, F. Chromosome analysis of 96 uterine leiomyomas. Cancer Genet. Cytogenet. 1991, 55, 11–18. [Google Scholar] [CrossRef]
- Rein, M.S.; Friedman, A.J.; Barbieri, R.L.; Pavelka, K.; Fletcher, J.A.; Morton, C.C. Cytogenetic abnormalities in uterine leiomyomata. Obstet. Gynecol. 1991, 77, 923–926. [Google Scholar] [PubMed]
- Sandberg, A.A. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: Leiomyoma. Cancer Genet. Cytogenet. 2005, 158, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Surti, U. Subgroups of uterine leiomyomas based on cytogenetic analysis. Hum. Pathol. 1991, 22, 1009–1016. [Google Scholar] [CrossRef]
- Cho, Y.L.; Bae, S.; Koo, M.S.; Kim, K.M.; Chun, H.J.; Kim, C.K.; Ro, D.Y.; Kim, J.H.; Lee, C.H.; Kim, Y.W.; et al. Array comparative genomic hybridization analysis of uterine leiomyosarcoma. Gynecol. Oncol. 2005, 99, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Meadows, K.L.; Andrews, D.M.; Xu, Z.; Carswell, G.K.; Laughlin, S.K.; Baird, D.D.; Taylor, J.A. Genome-wide analysis of loss of heterozygosity and copy number amplification in uterine leiomyomas using the 100K single nucleotide polymorphism array. Exp. Mol. Pathol. 2011, 91, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Brosens, I.; Johannisson, E.; Dal Cin, P.; Deprest, J.; Van den Berghe, H. Analysis of the karyotype and desoxyribonucleic acid content of uterine myomas in premenopausal, menopausal, and gonadotropin-releasing hormone agonist-treated females. Fertil. Steril. 1996, 66, 376–379. [Google Scholar] [CrossRef]
- Mehine, M.; Kaasinen, E.; Mäkinen, N.; Katainen, R.; Kämpjärvi, K.; Pitkänen, E.; Heinonen, H.R.; Bützow, R.; Kilpivaara, O.; Kuosmanen, A.; et al. Characterization of uterine leiomyomas by whole-genome sequencing. N. Engl. J. Med. 2013, 369, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Ozisik, Y.Y.; Meloni, A.M.; Powell, M.; Surti, U.; Sandberg, A.A. Chromosome 7 biclonality in uterine leiomyoma. Cancer Genet. Cytogenet. 1993, 67, 59–64. [Google Scholar] [CrossRef]
- Ishwad, C.S.; Ferrell, R.E.; Davare, J.; Meloni, A.M.; Sandberg, A.A.; Surti, U. Molecular and cytogenetic analysis of chromosome 7 in uterine leiomyomas. Genes Chromosomes Cancer 1995, 14, 51–55. [Google Scholar] [CrossRef]
- Vanni, R.; Marras, S.; Schoenmakers, E.F.; Dal Cin, P.; Kazmierczak, B.; Senger, G.; Bullerdiek, J.; Van de Ven, W.J.; Van den Berghe, H. Molecular cytogenetic characterization of del(7q) in two uterine leiomyoma-derived cell lines. Genes Chromosomes Cancer 1997, 18, 155–161. [Google Scholar] [CrossRef]
- Zeng, W.R.; Scherer, S.W.; Koutsilieris, M.; Huizenga, J.J.; Filteau, F.; Tsui, L.C.; Nepveu, A. Loss of heterozygosity and reduced expression of the CUTL1 gene in uterine leiomyomas. Oncogene 1997, 14, 2355–2365. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ligon, A.H.; Morton, C.C. Genetics of uterine leiomyomata. Genes Chromosomes Cancer 2000, 28, 235–245. [Google Scholar] [CrossRef]
- Da Silva-Santiago, S.C.; Pacheco, C.; Rocha, T.C.; Brasil, S.M.; Pacheco, A.C.; Silva, M.M.; Araújo, F.F.; De Vasconcelos, E.J.; De Oliveira, D.M. The linked human imprintome v1.0: Over 120 genes confirmed as imprinted impose a major review on previous censuses. Int. J. Data Min. Bioinform. 2014, 10, 329–356. [Google Scholar] [CrossRef]
- GeneImprint. Available online: https://www.geneimprint.com/site/genes-by-species.Homo+sapiens (accessed on 11 October 2011).
- Kim, J.; Bretz, C.L.; Lee, S. Epigenetic instability of imprinted genes in human cancers. Nucleic Acids Res. 2015, 43, 10689–10699. [Google Scholar] [CrossRef]
- Mackinnon, R.N.; Campbell, L.J. The role of dicentric chromosome formation and secondary centromere deletion in the evolution of myeloid malignancy. Genet. Res. Int. 2011, 2011, 643628. [Google Scholar] [CrossRef]
- Maciejowski, J.; de Lange, T. Telomeres in cancer: Tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017, 18, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Cleal, K.; Jones, R.E.; Grimstead, J.W.; Hendrickson, E.A.; Baird, D.M. Chromothripsis during telomere crisis is independent of NHEJ, and consistent with a replicative origin. Genome Res. 2019, 29, 737–749. [Google Scholar] [CrossRef]
- Maciejowski, J.; Chatzipli, A.; Dananberg, A.; Chu, K.; Toufektchan, E.; Klimczak, L.J.; Gordenin, D.A.; Campbell, P.J.; de Lange, T. APOBEC3-dependent kataegis and TREX1-driven chromothripsis during telomere crisis. Nat. Genet. 2020, 52, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Kyo, S.; Kanaya, T.; Ishikawa, H.; Ueno, H.; Inoue, M. Telomerase activity in gynecological tumors. Clin. Cancer Res. 1996, 2, 2023–2028. [Google Scholar] [PubMed]
- Bonatz, G.; Frahm, S.O.; Andreas, S.; Heidorn, K.; Jonat, W.; Parwaresch, R. Telomere shortening in uterine leiomyomas. Am. J. Obstet. Gynecol. 1998, 179 Pt 1, 591–596. [Google Scholar] [CrossRef]
- Ulaner, G.A.; Hu, J.F.; Vu, T.H.; Oruganti, H.; Giudice, L.C.; Hoffman, A.R. Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium. Int. J. Cancer 2000, 85, 330–335. [Google Scholar] [CrossRef]
- Carney, S.A.; Tahara, H.; Swartz, C.D.; Risinger, J.I.; He, H.; Moore, A.B.; Haseman, J.K.; Barrett, J.C.; Dixon, D. Immortalization of human uterine leiomyoma and myometrial cell lines after induction of telomerase activity: Molecular and phenotypic characteristics. Lab. Investig. 2002, 82, 719–728. [Google Scholar] [CrossRef]
- MacKenzie, D., Jr.; Watters, A.K.; To, J.T.; Young, M.W.; Muratori, J.; Wilkoff, M.H.; Abraham, R.G.; Plummer, M.M.; Zhang, D. ALT Positivity in Human Cancers: Prevalence and Clinical Insights. Cancers 2021, 13, 2384. [Google Scholar] [CrossRef]
- Nilbert, M.; Heim, S.; Mandahl, N.; Flodérus, U.M.; Willén, H.; Mitelman, F. Karyotypic rearrangements in 20 uterine leiomyomas. Cytogenet. Cell Genet. 1988, 49, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, K.; Fujii, S.; Konishi, I.; Nanbu, Y.; Nonogaki, H.; Mori, T. Mitotic activity in uterine leiomyomas during the menstrual cycle. Am. J. Obstet. Gynecol. 1989, 160, 637–641. [Google Scholar] [CrossRef]
- Hodge, J.C.; Park, P.J.; Dreyfuss, J.M.; Assil-Kishawi, I.; Somasundaram, P.; Semere, L.G.; Quade, B.J.; Lynch, A.M.; Stewart, E.A.; Morton, C.C. Identifying the molecular signature of the interstitial deletion 7q subgroup of uterine leiomyomata using a paired analysis. Genes Chromosomes Cancer 2009, 48, 865–885. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Miharu, N.; Okamoto, E.; Samura, O.; Hara, T.; Ohama, K. Detection of chromosomal abnormalities of chromosome 12 in uterine leiomyoma using fluorescence in situ hybridization. Jpn. J. Hum. Genet. 1996, 41, 193–202. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xing, Y.P.; Powell, W.L.; Morton, C.C. The del(7q) subgroup in uterine leiomyomata: Genetic and biologic characteristics. Further evidence for the secondary nature of cytogenetic abnormalities in the pathobiology of uterine leiomyomata. Cancer Genet. Cytogenet. 1997, 98, 69–74. [Google Scholar] [CrossRef]
- Pandis, N.; Heim, S.; Bardi, G.; Flodérus, U.M.; Willén, H.; Mandahl, N.; Mitelman, F. Parallel karyotypic evolution and tumor progression in uterine leiomyoma. Genes Chromosomes Cancer 1990, 2, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Ozisik, Y.Y.; Meloni, A.M.; Surti, U.; Sandberg, A.A. Deletion 7q22 in uterine leiomyoma. A cytogenetic review. Cancer Genet. Cytogenet. 1993, 71, 1–6. [Google Scholar] [CrossRef]
- Mehine, M.; Heinonen, H.R.; Sarvilinna, N.; Pitkänen, E.; Mäkinen, N.; Katainen, R.; Tuupanen, S.; Bützow, R.; Sjöberg, J.; Aaltonen, L.A. Clonally related uterine leiomyomas are common and display branched tumor evolution. Hum. Mol. Genet. 2015, 24, 4407–4416. [Google Scholar] [CrossRef]
- Fletcher, J.A.; Morton, C.C.; Pavelka, K.; Lage, J.M. Chromosome aberrations in uterine smooth muscle tumors: Potential diagnostic relevance of cytogenetic instability. Cancer Res. 1990, 50, 4092–4097. [Google Scholar]
- Packenham, J.P.; du Manoir, S.; Schrock, E.; Risinger, J.I.; Dixon, D.; Denz, D.N.; Evans, J.A.; Berchuck, A.; Barrett, J.C.; Devereux, T.R.; et al. Analysis of genetic alterations in uterine leiomyomas and leiomyosarcomas by comparative genomic hybridization. Mol. Carcinog. 1997, 19, 273–279. [Google Scholar] [CrossRef]
- Levy, B.; Mukherjee, T.; Hirschhorn, K. Molecular cytogenetic analysis of uterine leiomyoma and leiomyosarcoma by comparative genomic hybridization. Cancer Genet. Cytogenet. 2000, 121, 1–8. [Google Scholar] [CrossRef]
- Croce, S.; Ribeiro, A.; Brulard, C.; Noel, J.C.; Amant, F.; Stoeckle, E.; Devouassoux-Shisheborah, M.; Floquet, A.; Arnould, L.; Guyon, F.; et al. Uterine smooth muscle tumor analysis by comparative genomic hybridization: A useful diagnostic tool in challenging lesions. Mod. Pathol. 2015, 28, 1001–1010. [Google Scholar] [CrossRef]
- Liegl-Atzwanger, B.; Heitzer, E.; Flicker, K.; Müller, S.; Ulz, P.; Saglam, O.; Tavassoli, F.; Devouassoux-Shisheboran, M.; Geigl, J.; Moinfar, F. Exploring chromosomal abnormalities and genetic changes in uterine smooth muscle tumors. Mod. Pathol. 2016, 29, 1262–1277. [Google Scholar] [CrossRef] [PubMed]
- Croce, S.; Chibon, F. Molecular prognostication of uterine smooth muscle neoplasms: From CGH array to CINSARC signature and beyond. Genes Chromosomes Cancer 2021, 60, 129–137. [Google Scholar] [CrossRef]
- Mäkinen, N.; Kämpjärvi, K.; Frizzell, N.; Bützow, R.; Vahteristo, P. Characterization of MED12, HMGA2, and FH alterations reveals molecular variability in uterine smooth muscle tumors. Mol. Cancer 2017, 16, 101. [Google Scholar] [CrossRef] [PubMed]
- Vanni, R.; Van Roy, N.; Lecca, U.; Speleman, F. Uterine leiomyoma cytogenetics. III. Interphase cytogenetic analysis of karyotypically normal uterine leiomyoma excludes possibility of undetected trisomy 12. Cancer Genet. Cytogenet. 1992, 62, 40–42. [Google Scholar] [CrossRef]
- Han, K.; Lee, W.; Harris, C.P.; Simsiman, R.C.; Lee, K.; Kang, C.; Meisner, L.F. Comparison of chromosome aberrations in leiomyoma and leiomyosarcoma using FISH on archival tissues. Cancer Genet. Cytogenet. 1994, 74, 19–24. [Google Scholar] [CrossRef]
- Hayashi, S.; Miharu, N.; Okamoto, E.; Samura, O.; Hara, T.; Ohama, K. Detection of chromosomal abnormalities in uterine leiomyoma using conventional cytogenetic method and interphase fluorescence in situ hybridization. Cancer Genet. Cytogenet. 1996, 89, 98–104. [Google Scholar] [CrossRef]
- Hodge, J.C.; Pearce, K.E.; Clayton, A.C.; Taran, F.A.; Stewart, E.A. Uterine cellular leiomyomata with chromosome 1p deletions represent a distinct entity. Am. J. Obstet. Gynecol. 2014, 210, 572.e1–572.e7. [Google Scholar] [CrossRef] [PubMed]
- Chill, H.H.; Karavani, G.; Rachmani, T.; Dior, U.; Tadmor, O.; Shushan, A. Growth pattern of uterine leiomyoma along pregnancy. BMC Womens Health 2019, 19, 100. [Google Scholar] [CrossRef]
- Delli Carpini, G.; Verdecchia, V.; Papiccio, M.; Grelloni, C.; Ciavattini, A. Comparison of uterine fibroids’ growth pattern during pregnancy according to fetal sex: An observational study. Biol. Sex Differ. 2019, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.D.; Patchel, S.A.; Saldana, T.M.; Umbach, D.M.; Cooper, T.; Wegienka, G.; Harmon, Q.E. Uterine fibroid incidence and growth in an ultrasound-based, prospective study of young African Americans. Am. J. Obstet. Gynecol. 2020, 223, 402.e1–402.e18. [Google Scholar] [CrossRef]
- Davis, B.J.; Risinger, J.I.; Chandramouli, G.V.; Bushel, P.R.; Baird, D.D.; Peddada, S.D. Gene expression in uterine leiomyoma from tumors likely to be growing (from black women over 35) and tumors likely to be non-growing (from white women over 35). PLoS ONE 2013, 8, e63909. [Google Scholar] [CrossRef]
- Bulun, S.E.; Moravek, M.B.; Yin, P.; Ono, M.; Coon, J.S., 5th; Dyson, M.T.; Navarro, A.; Marsh, E.E.; Zhao, H.; Maruyama, T.; et al. Uterine Leiomyoma Stem Cells: Linking Progesterone to Growth. Semin. Reprod. Med. 2015, 33, 357–365. [Google Scholar] [CrossRef]
- Roix, J.J.; McQueen, P.G.; Munson, P.J.; Parada, L.A.; Misteli, T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat. Genet. 2003, 34, 287–291. [Google Scholar] [CrossRef]
- Martin, L.D.; Harizanova, J.; Zhu, G.; Righolt, C.H.; Belch, A.R.; Mai, S.; Pilarski, L.M. Differential positioning and close spatial proximity of translocation-prone genes in nonmalignant B-cells from multiple myeloma patients. Genes Chromosomes Cancer 2012, 51, 727–742. [Google Scholar] [CrossRef]
- Osborne, C.S. Molecular pathways: Transcription factories and chromosomal translocations. Clin. Cancer Res. 2014, 20, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Ugarte, G.D.; Vargas, M.F.; Medina, M.A.; León, P.; Necuñir, D.; Elorza, A.A.; Gutiérrez, S.E.; Moon, R.T.; Loyola, A.; De Ferrari, G.V. Wnt signaling induces transcription, spatial proximity, and translocation of fusion gene partners in human hematopoietic cells. Blood 2015, 126, 1785–1789. [Google Scholar] [CrossRef] [PubMed]
- Fritz, A.J.; Sehgal, N.; Pliss, A.; Xu, J.; Berezney, R. Chromosome territories and the global regulation of the genome. Genes Chromosomes Cancer 2019, 58, 407–426. [Google Scholar] [CrossRef]
- Gross, K.L.; Neskey, D.M.; Manchanda, N.; Weremowicz, S.; Kleinman, M.S.; Nowak, R.A.; Ligon, A.H.; Rogalla, P.; Drechsler, K.; Bullerdiek, J.; et al. HMGA2 expression in uterine leiomyomata and myometrium: Quantitative analysis and tissue culture studies. Genes Chromosomes Cancer 2003, 38, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Hodge, J.C.; Kim, T.M.; Dreyfuss, J.M.; Somasundaram, P.; Christacos, N.C.; Rousselle, M.; Quade, B.J.; Park, P.J.; Stewart, E.A.; Morton, C.C. Expression profiling of uterine leiomyomata cytogenetic subgroups reveals distinct signatures in matched myometrium: Transcriptional profilingof the t(12;14) and evidence in support of predisposing genetic heterogeneity. Hum. Mol. Genet. 2012, 21, 2312–2329. [Google Scholar] [CrossRef] [PubMed]
- Klemke, M.; Meyer, A.; Nezhad, M.H.; Bartnitzke, S.; Drieschner, N.; Frantzen, C.; Schmidt, E.H.; Belge, G.; Bullerdiek, J. Overexpression of HMGA2 in uterine leiomyomas points to its general role for the pathogenesis of the disease. Genes Chromosomes Cancer 2009, 48, 171–178. [Google Scholar] [CrossRef] [PubMed]




| Case | Probe Mix | Cytogenetic Localisation | Manufacturer | Analysis (Detection of Chromosomal Abnormality) |
|---|---|---|---|---|
| 1 | CEP 12 (D12Z3) | 12p11.1–q11 Alpha Satellite DNA | Abbott Laboratories | There may be three signals of the CEP 12 (D12Z3) probe. |
| 2 | MCB6-2 MCB10-5a MCB16-1a | 6p24.1–p21.33 10q22.1–q25.2 16p13.3–p11.1 | Homemade | The signals of MCB10–5a and MCB16–1a probes may become juxtaposed on one chromosome because of the t(6;10;16)(p21;q22;q13). |
| 3 | MCB10-4a MCB12-5 | 10q11.23–10q23.1 12q15–12q23.3 | Homemade | The signals may become juxtaposed on one chromosome because of the t(4;?;12;10). |
| 4 | LSI ELN/LSI D7S486, D7S522 | 7q11.23, 7q31 | Abbott Laboratories | The signals of LSI ELN and LSI D7S486, D7S522 probes may become juxtaposed on one chromosome because of the del(7)(q21.11q23). |
| 5 | MCB7-5 MCB12-5 MCB14-1a | 7q21.11–7q31.2 12q15–12q23.3 14q13.3–14q23.3 | Homemade | The signals of MCB12–5 and MCB14–1a probes may become juxtaposed on one chromosome because of the t(12;14)(q15;q24); one signal of MCB7–5 probe may be absent because of the del(7)(q21.11q33). |
| 6 | MCB1-1 MCB1-4 | 1p35.2–1pter 1p12–1p31.1 | Homemade | (1) One additional signal of both MCB1–1 and MCB1–4 probes may be present, and they may become juxtaposed because of the inv(1)(p22p36) and the t(1;10)(p36;q26). (2) One signal of the MCB1–1 probe may be absent because of the del(1)(p33). |
| Case | Conventional Karyotyping of CTS (with Subsequent FISH on Metaphase Chromosomes) | aCGH of UTS | MED12 Status of UTS | Interphase FISH on CTS and UTS |
|---|---|---|---|---|
| 1 | 47,XX, + 12[18] | arr(X,1-22) × 2 | wt/c.131G > A, pG44D | UTS: nuc ish(D12Z3×3)[82/1000] CTS: nuc ish(D12Z3×3)[985/1001] |
| 2 | 46,XX,t(6;10;16)(p21;q22;p13)[13] | arr(X,1-22) × 2 | wt/c.131G > A, pG44D | UTS: nuc ish(MCB6-2,MCB10-5a,MCB16-1a)×2(MCB10-5a con MCB16-1a×1)[454/555] CTS: nuc ish(MCB6-2,MCB10-5a,MCB16-1a)×2(MCB10-5a con MCB16-1a×1)[448/510] |
| 3 | 46,XX,del(7)(q22.1q31.2),t(4;?;12;10)(p11;?;q15;q22)[15] | arr[GRCh37] 7q22.1q31.2(98726412_115199215)×1[0,7] | wt/wt | UTS: nuc ish(MCB10-4a,MCB12-5)×2(MCB10-4a con MCB12-5×1)[391/1000] CTS: nuc ish(MCB10-4a,MCB12-5)×2(MCB10-4a con MCB12-5×1)[286/1000] |
| 4 | 46,XX,del(7)(q21.11q22.3)[7]/46,XX[2] | arr[GRCh37] 7q21.11q22.3(83605684_105796277)×1 | wt/c.131G > A, pG44D | UTS: nuc ish(ELN,D7S486)×2(ELN con D7S486×1)[350/1016] CTS: nuc ish(ELN,D7S486)×2(ELN con D7S486×1)[297/1028] |
| 5 | 46,XX,del(7)(q21.1q35),t(12;14)(q15;q23)[15] | arr[GRCh37] 7q21.11q35(78201649_146170074)×1[0,6] | wt/wt | UTS: nuc ish(MCB7-5×1,MCB12-5×2,MCB14-1a×2)(MCB12-5 con MCB14-1a×1)[445/1000],(MCB7-5,MCB12-5,MCB14-1a)×2(MCB12-5 con MCB14-1a×1)[186/1000] CTS: nuc ish(MCB7-5×1,MCB12-5×2,MCB14-1a×2)(MCB12-5 con MCB14-1a×1)[766/1000],(MCB7-5,MCB12-5,MCB14-1a)×2(MCB12-5 con MCB14-1a×1)[72/1000] |
| 6 | 46,XX,inv(1)(p36p21),t(1;10)(p36;q26)[9]/46,XX[21] | arr[GRCh37] (1,8,14)cx[0,6] | wt/wt | UTS: nuc ish(MCB1-1×1,MCB1-4×2)[126/1000],(MCB1-1,MCB1-4)×3(MCB1-1 con MCB1-4×1)[40/1000] CTS: nuc ish(MCB1-1,MCB1-4)×3(MCB1-1 con MCB1-4×1)[35/1000],(MCB1-1×1,MCB1-4×2)[23/1000] |
| 7 | 46,XX,del(1)(p34p32),del(3)(q26),del(16)(q12q24),t(1;17)(p35;q25),t(2;9)(p16;q21)[25]/46,XX[14] | arr[GRCh37] 1p34.3p32.3(36643269_52009701)×1[0,8],3q13.31q21.1(116742856_122583187)×1[0,8],3q24q26.33(147591180_180696172)×1[0,8],16q12.1q22.1(48779768_69195217)×1[0,8],16q23.2q24.1(79678725_85191053)×1[0,8],(19)×1[0,4] | wt/wt | no |
| 8 | 45,XX,der(1)t(1;1),der(3)t(1;3),der(13)t(1;3;13),-1[12] | arr[GRCh37] 1p35.1p34.3(34079411_36223052)×1[0,7],1p13.2p12(113262062_120527194)×1[0,7],1q24.3q25.1(172004633_174625860)×1[0,7],1q32.1q44(205440703_249208145)×1[0,7],3q25.32q25.33(158746175_160073609)×1[0,7],3q26.31q29(173336557_197771082)×1[0,7],13q13.1q33.1(32365197_101748020)×1[0,7] | wt/wt | no |
| 9 | 45,X,-X,der(2)t(2;11)(2p16→2q24::11p15.1→11pter),der(6)(6pter→6p24::6p21→6q14.1::6q22.1→6qter),der(9)t(X;9)(Xpter→Xp11.1::Xq11.1→Xq26::9p22→9qter),der(11)t(2;11)(2qter→2q35::2p24→2p16::2q35→2q24::11p15.1→11qter),der(14)t(6;14)(14pter→14q22::6p24→6p?23::6p?22.1→6p?21),der(16)t(16;6;14)(16p→16qter::6p?22.3→6p?22.1::14q22→14q31.3::6p?23→6p?22::14q31.3→14qter)[cp25] | arr[GRCh37] 2p25.1(8446980_8682420)×1[0,7],2p24.3(12646763_13128408)×1[0,7],2p24.3(14202109_14930579)×1[0,6],2p21(42190927_43424758)×1[0,7],2p21(45127208_45383273)×1[0,7],2q35(218424429_219280813)×1[0,7],2q36.1(224312511_224706849)×1[0,7],6q14.1(81386902_81640806)×1 [0,7],6q14.1(82451363_82888958)×1[0,7],6q14.1q22.1(83806540-114940922)×1[0,7],6q24.1(139618109_140460727)×1[0,7],14q24.1(69062128_69249764)×1[0,7,16p11.2p11.1(32637849_34721199)×3,16q24.2(87466743_87533166)×1 | wt/wt | no |
| 10 | 46,XX,t(6;6)(p21;p25)[2]/46,XX,ins(13;6)(q14;q23q13)[8]/46,XX[90] | arr(X,1-22)×2 | wt/c.107T > G, pL36R | no |
| 11 | 46,XX,del(1)(p32p13)[2]/46,XX[24] | arr[GRCh37] 1q41q43(220523143_241093232)×1[0,5],12q22(93893845_94451632)×1[0,8],Xp22.31(6456036_8152935)×3 | wt/wt | no |
| 12 | 46,XX,del(1)(p36)[2]/46,XX[28] | arr(X,1-22)×2 | wt/c.131G > A, pG44D | no |
| Case | Patient’s Age, Years | Hormonal Treatment before Myomectomy | Menstrual Cycle Phase at the Time of Myomectomy | Solitary (S) or Multiple (M) ULs | Diameter of Analyzed UL Nodule, cm | Localisation of Analyzed UL Nodule (FIGO) | Time Elapsed between UL Diagnosis and Myomectomy | Rapid UL Growth within One Year before Myomectomy | Histological Examination |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 32 | No | Proliferative | M | 7 | 5 | 6 years | yes | Leiomyoma with necrosis |
| 2 | 45 | Ulipristal acetate | Hormonal treatment | S | 8 | 4 | 5 years | yes | Leiomyoma with oedema and hyalinosis |
| 3 | 40 | No | Proliferative | S | 9 | 6 | 5 years | no | Leiomyoma with edema |
| 4 | 43 | Buserelin acetate | Hormonal treatment | M | 8 | 4 | 13 years | yes | Leiomyoma with fibrohyalinosis |
| 5 | 29 | No | Proliferative | S | 8 | 5 | 1 year | yes | Leiomyoma |
| 6 | 42 | No | Secretory | S | 5 | 4 | 7 months | no | Leiomyoma with hyalinosis |
| 7 | 31 | No | Secretory | M | 9 | 6 | 2 months | no | Leiomyoma |
| 8 | 44 | Triptorelin acetate | Hormonal treatment | S | 7 | 4 | 5 years | no | Leiomyoma with sclerosis and hyalinosis |
| 9 | 41 | No | Proliferative | M | 10 | 6 | 6 years | yes | Leiomyoma |
| 10 | 32 | No | Proliferative | S | 5 | 4 | 1 year | no | Leiomyoma |
| 11 | 36 | Cyproterone + Ethinylestradiol | Hormonal treatment | S | 5 | 4 | 5 years | no | Leiomyoma |
| 12 | 37 | No | Secretory | M | 5 | 2 | 2 years | no | Leiomyoma |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koltsova, A.S.; Efimova, O.A.; Malysheva, O.V.; Osinovskaya, N.S.; Liehr, T.; Al-Rikabi, A.; Shved, N.Y.; Sultanov, I.Y.; Chiryaeva, O.G.; Yarmolinskaya, M.I.; et al. Cytogenomic Profile of Uterine Leiomyoma: In Vivo vs. In Vitro Comparison. Biomedicines 2021, 9, 1777. https://doi.org/10.3390/biomedicines9121777
Koltsova AS, Efimova OA, Malysheva OV, Osinovskaya NS, Liehr T, Al-Rikabi A, Shved NY, Sultanov IY, Chiryaeva OG, Yarmolinskaya MI, et al. Cytogenomic Profile of Uterine Leiomyoma: In Vivo vs. In Vitro Comparison. Biomedicines. 2021; 9(12):1777. https://doi.org/10.3390/biomedicines9121777
Chicago/Turabian StyleKoltsova, Alla S., Olga A. Efimova, Olga V. Malysheva, Natalia S. Osinovskaya, Thomas Liehr, Ahmed Al-Rikabi, Natalia Yu. Shved, Iskender Yu. Sultanov, Olga G. Chiryaeva, Maria I. Yarmolinskaya, and et al. 2021. "Cytogenomic Profile of Uterine Leiomyoma: In Vivo vs. In Vitro Comparison" Biomedicines 9, no. 12: 1777. https://doi.org/10.3390/biomedicines9121777
APA StyleKoltsova, A. S., Efimova, O. A., Malysheva, O. V., Osinovskaya, N. S., Liehr, T., Al-Rikabi, A., Shved, N. Y., Sultanov, I. Y., Chiryaeva, O. G., Yarmolinskaya, M. I., Polenov, N. I., Kunitsa, V. V., Kakhiani, M. I., Tral, T. G., Tolibova, G. K., Bespalova, O. N., Kogan, I. Y., Glotov, A. S., Baranov, V. S., & Pendina, A. A. (2021). Cytogenomic Profile of Uterine Leiomyoma: In Vivo vs. In Vitro Comparison. Biomedicines, 9(12), 1777. https://doi.org/10.3390/biomedicines9121777








